Supplemental Digital Content is available in the text
Keywords: differentially expressed genes, environmental risk factors, Kashin–Beck disease
Abstract
As environmental risk factors (ERFs) play an important role in the pathogenesis of Kashin–Beck disease (KBD), it is important to identify the interaction between ERFs and differentially expression genes (DEGs) in KBD. The environmental response genes (ERGs) were analyzed in cartilage of KBD in comparison to normal controls.
We searched 5 English and 3 Chinese databases from inception to September 2015, to identify case–control studies that examined ERFs for KBD using integrative meta-analysis and systematic review. Total RNA was isolated from articular cartilage of KBD patients and healthy controls. Human whole genome microarray chip (Agilent) was used to analyze the amplified, labeled, and hybridized total RNA, and the validated microarray data were partially verified using real-time quantitative polymerase chain reaction (qRT-PCR). The ERGs were derived from the Comparative Toxicogenomics Database. The identified ERGs were subjected to KEGG pathway enrichment, biological process (BP), and interaction network analyses using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7, and STRING.
The trace elements (selenium and iodine), vitamin E, and polluted grains (T-2 toxin/HT-2 toxin, deoxynivalenol, and nivalenol) were identified as the ERFs for KBD using meta-analysis and review. We identified 21 upregulated ERGs and 7 downregulated ERGs in cartilage with KBD compared with healthy controls, which involved in apoptosis, metabolism, and growth and development. KEGG pathway enrichment analysis found that 2 significant pathways were involved with PI3K-Akt signaling pathway and P53 signaling pathway, and gene ontology function analysis found 3 BPs involved with apoptosis, death, and cell death in KBD cartilage.
According to previous results and our own research, we suggest that the trace element selenium and vitamin E induce PI3K-Akt signaling pathway and the mycotoxins (T-2 toxin/HT-2 toxin and DON) induce P53 signaling pathway, contributing to the development of KBD, and chondrocyte apoptosis and cell death.
1. Introduction
Kashin–Beck disease (KBD) is an endemic, chronic osteochondropathy, which develops mainly in children ranged 5 to 15 years old. The main clinical symptoms of KBD are joint pain, enlargened limb joints, short fingers, short limbs, and even deformation.[1–4] However, all these clinical signs are secondary lesions in the late stage. The pathological changes in the earlier stage of KBD involve epiphyseal cartilage and articular cartilage cell necrosis.[3,4] A mass of epidemiological investigations during a long time has confirmed that environmental risk factors (ERFs) contribute to the deep cartilage cells necrosis of KBD, and has put forward more than 50 kinds of hypotheses. Some hypotheses have been obsolete with the in-depth study of the etiology of KBD. Now, most studies concentrate on 3 hypotheses: biogeochemistry theory (represented by selenium deficiency), food poisoning by mycotoxin (mainly for T-2 toxin), and water poisoning by organic compounds (mainly for humic acid).[5–9] Still, none of the hypotheses can alone explain the etiology of KBD successfully. Thus, some researchers have proposed that a variety of ERFs, such as the T-2 toxin infection under the condition of selenium deficiency, may lead to KBD.
Previously, we have performed integrative multivariate logistic regression analysis of ERFs for KBD, and found that the ERFs of low protein intake, polluted grain, and selenium deficiency may contribute to the onset of KBD together.[10,11] Meanwhile, the comprehensive gene expression profiles of human healthy and KBD cartilage samples have been compared by our research group, and we identified 55 upregulated and 24 downregulated genes between KBD and healthy controls.[12] However, there is little knowledge about the molecular mechanism between the ERFs and differential expression genes (DEGs).
Thus, in order to clarify the interaction networks between ERFs and DEGs, we identified 2 main ERFs groups, including the low level of trace elements (selenium and iodine), vitamin E, and polluted grains [T-2 toxin, deoxynivalenol (DON), and nivalenol (NIV)] through meta-analysis and systematic review based on the previous studies. Second, we selected the 55 upregulated and 24 downregulated genes recognized in KBD articular cartilage using Agilent Human 1A 22 k (Agilent Technologies, USA) high-density oligonucleotide array analysis. Third, the environmental response genes (ERGs) in KBD were identified according to the Comparative Toxicogenomics Database (http://ctdbase.org/), and combined the identified ERFs and DEGs. Thus, our results may help to reveal the molecular mechanism of interaction between ERFs and DEGs in KBD.
2. Methods
2.1. Environmental risk factors
The integrative meta-analysis was used to determine the current evidence on ERFs for KBD.[10] We searched 5 English and 3 Chinese databases from inception to September 2015 to identify case–control studies that examined ERFs for KBD using multivariate logistic analysis. DerSimonian and Laird effective models were applied in processing the data using pooled odds ratios (ORs) and 95% confidence interval (CI). Seven studies were identified with 3087 cases and 6402 controls. The main risk factors found to be significantly associated with the onset of KBD were food source, wheat, wheat germ necrosis rate, total volatile basic nitrogen, and low selenium in hair. We found that the combination of polluted grain, vitamin E, and trace element deficiency may be ERFs of KBD. We also refer to previously published review on the ERFs of KBD.[12] Therefore, we identified ERFs including the trace elements (selenium and iodine), vitamin E, and polluted grain (T-2 toxin/HT-2 toxin, DON, and NIV).
2.2. Differentially expressed genes
Cartilage samples were obtained from 9 KBD patients and 9 healthy donors (Table 1). All the KBD patients were graded as second or third degree according to the diagnosis criterion of KBD (GB16395-1996). KBD cartilage were collected from the discarded tissue during total knee replacement, and the healthy cartilage were obtained at necropsy, within 8 hours of death, from non-KBD afflicted donors, who had no history of arthritic diseases and who died due to traffic accidents. The Institutional Review Board of Xi’an Jiaotong University has approved the present investigation, and after investigation, KBD patients and/or their family members signed informed consent documents.
Table 1.
Comparison basic characteristics of KBD and healthy controls.

Total RNA was extracted from the cartilage specimens using the Agilent total RNA isolation mini kit (Agilent Technologies, Santa Clara, CA) in accordance with the manufacturer's protocol. The quality of total RNA was evaluated using a NanoDrop ND-1000 UV–Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE), then transcribed into cRNA, and control samples were labeled with fluorescent Cy3 dye, while KBD samples were labeled with Cy5, using an Amino Allyl MessageAmp aRNA Kit (Ambion, Austin, TX). Labeled complementary RNA was hybridized with the Agilent Human 1A 22 k oligonucleotide microarray (Agilent Technologies, G4110B) and the data were produced according to the methods previously reported.[12]
2.3. Identified environmental response genes
The ERGs list was accessed from the environmentally related genomic database named Comparative Toxicogenomics Database (http://ctdbase.org/). This database included 3096 vitamin E related response genes, 2249 selenium-related response genes, 2037 selenium compounds related response genes, 59 iodine-related response genes, 501 T-2 toxin/HT-2 toxin-related response genes, 1046 DON-related response genes, and 78 NIV-related response genes. To identify the expression levels of ERGs in KBD cartilage, the expression ratios of the above ERGs were calculated from the microarray data. Significantly differentially expressed ERGs were defined by expression ratios <0.5 or >2.0.
2.4. Data analysis
To further investigate the molecular functions and biological processes (BPs) of the ERGs expressions in articular cartilage of KBD, gene set enrichment analysis and protein–protein interactions analysis software were used to identify Kyoto Encyclopedia of Genes and Genomes (KEGG) database pathways, BPs, and interaction network diagrams in cartilage between KBD and healthy controls. The Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.7 was used to analyze the findings because it has an integrated and expanded back-end annotation database, advanced modular enrichment algorithms, and powerful exploratory ability in an integrated data-mining environment, and was used for interpretation of the findings.[13] For any given gene list, DAVID tools can identify enriched BPs, and visualize genes on KEGG pathway maps. STRING, which is a database of known and predicted protein interactions and covers more than 9.6 million proteins from more than 2000 organisms, was also used in the analyses. The STRING interactions include direct (physical) and indirect (functional) associations, which are derived from 4 sources: genomic context, high-throughput experiments, coexpression, and previous knowledge; and it integrates interaction data from these sources for a large number of organisms.[14]
In order to verify the reliability of gene expression by microarray data, 4 upregulated genes (expression rate >2.0) and 4 downregulated genes (expression rate <0.5) identified in the microarray data were randomly selected to be measured by real-time quantitative PCR (qRT-PCR). The selected genes were the following, the differential expression ratio KBD versus normal (DER) given in parentheses: TMSL8 (DER = 9.65), CASP8AP2 (DER = 5.43), VEGF (DER = 2.99), PAPSS2 (DER = 3.79), POSTN (DER = 0.27), CBR3 (DER = 0.36), BMF (DER = 0.37), and TACC1 (DER = 0.43). The total RNA extraction method was the same as for gene expression microarray. Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) was used to convert the isolated total RNA into cDNA. ABI 7500 Real-Time PCR Detection System (Applied Biosystems, Foster City, CA) was applied for amplification and detection of cDNA according to the manufacturer's protocol.
3. Results
3.1. Single gene expression analysis
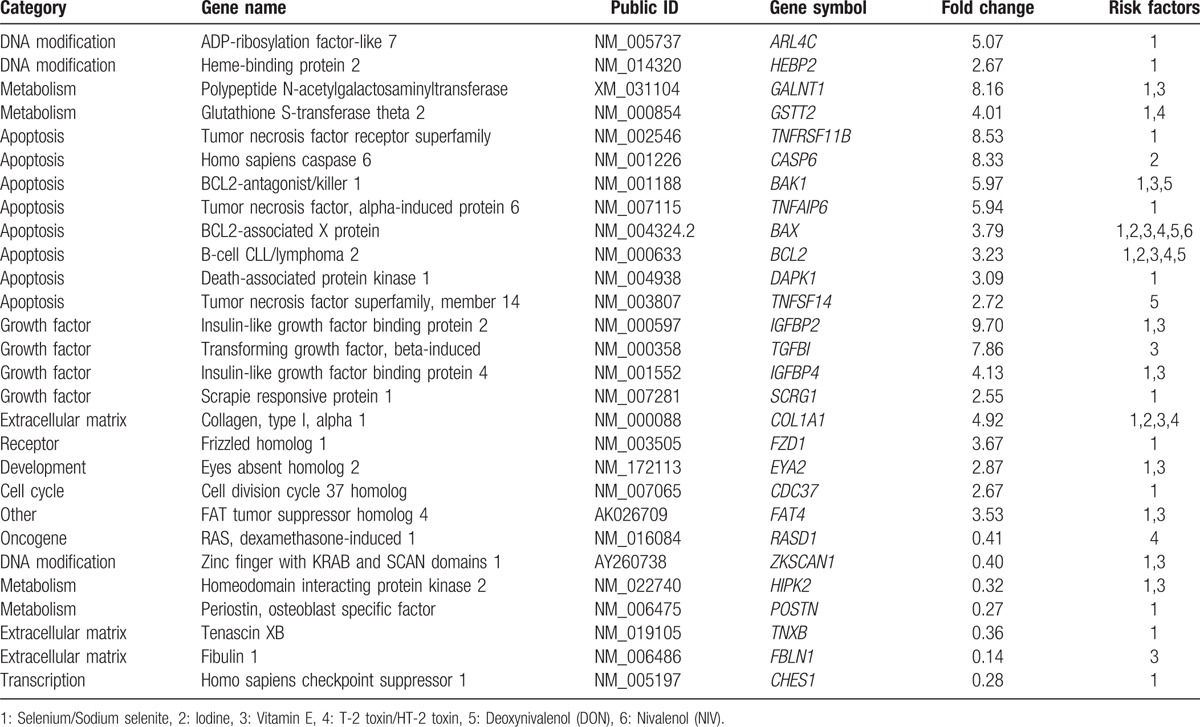
We identified 6 ERFs of trace elements (selenium and iodine), vitamin E, and polluted grain (T-2 toxin/HT-2 toxin, DON, and NIV). The selenium/sodium selenite was related with 18 upregulated and 6 downregulated, iodine with 3 upregulated, and vitamin E with 11 upregulated and 3 downregulated genes, while the T-2 toxin/HT-2 toxin was involved with 5 downregulated and 1 downregulated genes, DON with 5 upregulated, and NIV with 1 upregulated ones (Table 2). Thus, in total, the ERFs were involved with 21 upregulated and 7 downregulated genes listed in Table 3; the identified 28 DEGs participated in DNA modification, metabolism, apoptosis, growth factor, extracellular matrix, and so on. The interaction between ERFs and DEGs is shown in Appendix S1.
Table 2.
Identified differently expressed environmental response genes in the articular cartilage of KBD and healthy controls.
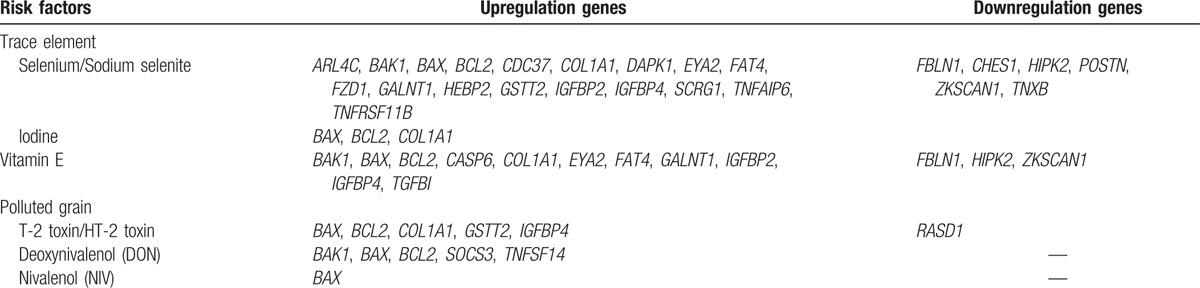
Table 3.
List the identified differently expressed environmental response genes in the articular cartilage of KBD and healthy controls.
3.2. KEGG pathway and biological processes
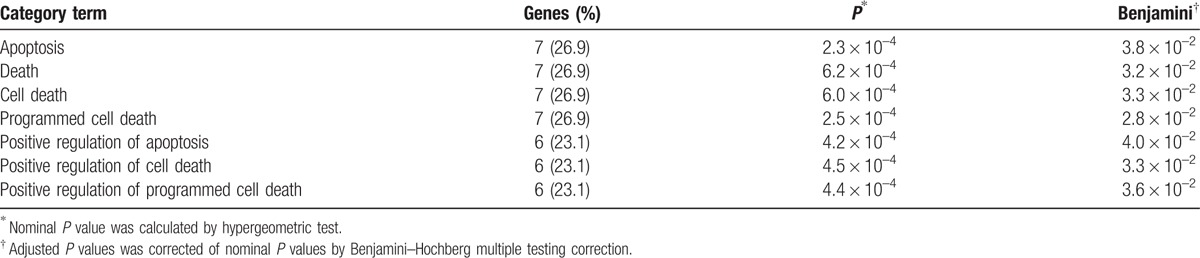
The 28 ERGs were taken for enrichment analysis using the DAVID and STRING software. The KEGG pathway enrichment analysis revealed apoptosis, and PI3K-Akt, p53, and Ras signaling pathways differentially expressed in KBD with a false discovery rate less than 0.05 (Table 3). According to the principle of gene ontology function analysis, we concluded the functional gene expression spectrum of KBD (Table 4). For KBD, we identified 7 significantly overrepresented BPs, including apoptosis, death, cell death, programmed cell death, positive regulation of apoptosis, positive regulation of cell death, and positive regulation of programmed cell death (Table 5) on the basis of Bonferroni-adjusted P values (P < 0.05).
Table 4.
The significantly enriched KEGG pathways in the articular cartilage of KBD and healthy controls.

Table 5.
The different expression of biological process in the articular cartilage of KBD and healthy controls.
3.3. Protein–protein interaction
STRING interaction network analysis was performed for the identified 28 ERGs in KBD, and 17 coded proteins were identified. Then, the protein–protein interaction (PPI) network was widened by adding 9 partners (BCL2L1, BCL2L11, HSP90AA1, TP53, MCL1, BAD, BIK, BECN1, and IGF1) using the STRING, which had similar physiological function with included REGs. The 24 nodes were imported into the cytoscape software to screen for the combined score of interactions more than 0.5, excluding the isolated nodes and distractions connection nodes. The degree of nodes more than 10 was considered as the key center node in KBD network in Fig. 1. The identified 6 genes (BAX, BCL2, BAD, BCL2L1, TP53, and CASP6) were relative with the chondrocyte apoptosis and cell death.
Figure 1.
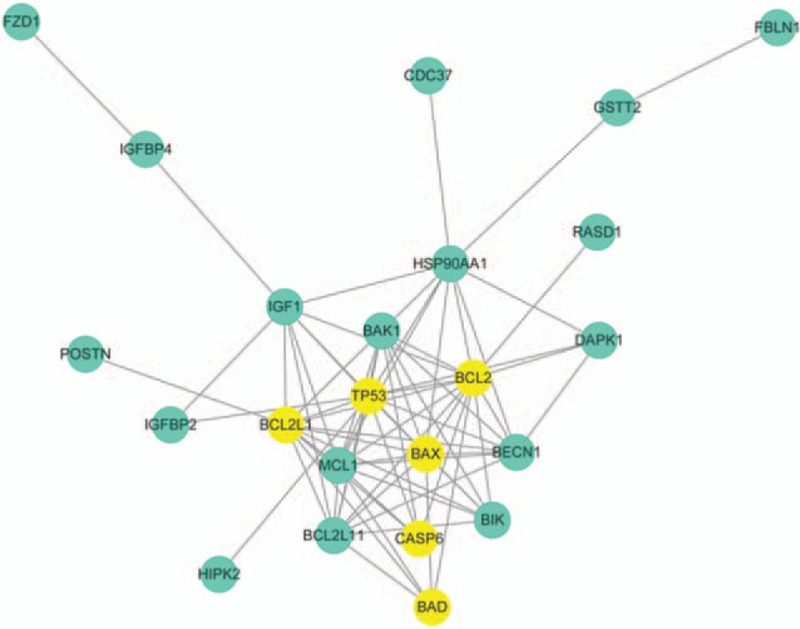
The protein–protein interaction (PPI) of identified 28 ERGs by STRING and Cytoscape software, and was also widened by adding 9 partners (BCL2L1, BCL2L11, HSP90AA1, TP53, MCL1, BAD, BIK, BECN1, and IGF1), which had similar physiological function with included DEGs. The intensity of the yellow genes relative with the chondrocyte apoptosis, and the PPI reveals that the 28 ERGs coded protein contributed to the chondrocyte apoptosis and cell death induced by the Ras, PI3K-Akt, and P53 signaling pathway.
3.4. qRT-PCR validation
The validity of microarray data is shown in Fig. 2. The identified TMSL8, CASP8AP2, PAPSS2, and VEGF were with higher expression, and identified POSTN, TACC1, CBR3, and BMF were with lower expression in the articular cartilage of KBD than healthy controls. The expression patterns of the 8 identified genes in microarray data were consistent with qRT-PCR, although high variations were evident in some of the genes.
Figure 2.
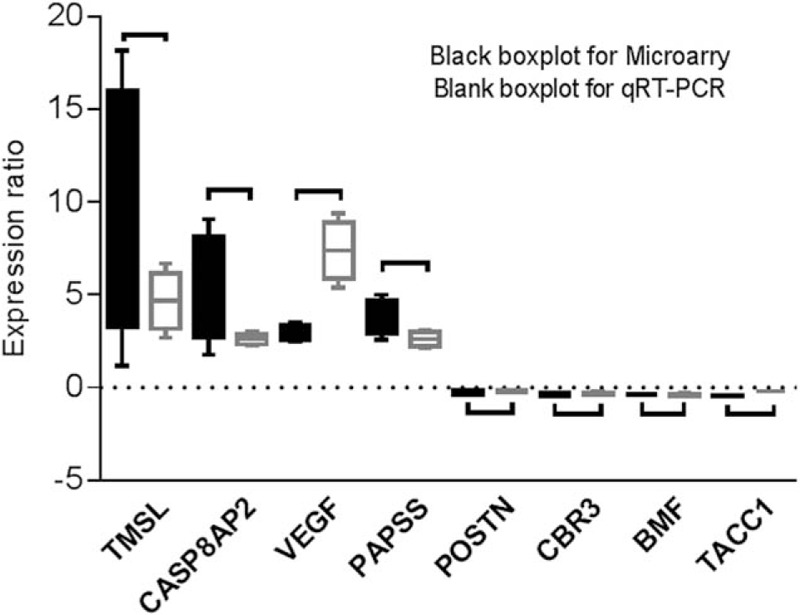
The expression values of identified 8 genes measured by microarray and qRT-PCR.
4. Discussion
A large number of previous integrative meta-analyses of epidemiological investigations have revealed that lack of trace element and food-polluted mycotoxin are the risk factors for KBD. So, it can be questioned whether the combination of both leads to the deep cartilage cell death of KBD. Previously, it has been shown that when miniature pigs were fed with food containing low selenium content (35 ng/kg) for 30 days, and then continued with the one with added T-2 toxin (1.5 mg/kg) for 105 days, numerous deep zone articular chondrocytes went to necrosis.[15] This result reveals that the food containing low amount of selenium and supplemented with T-2 toxin can lead to the similar articular chondrocyte cell deaths in animals as can be seen in patients with KBD. Our present study used comparative analysis of the interaction network between ERFs and differentially expressed genes from KBD articular cartilage compared with normal controls, using gene expression spectrum analysis technology and international ERG database. Four enrichment KEGG pathways and 7 related BPs were identified.
4.1. Ras and PI3K-Akt signaling pathways
The Ras signaling pathways can regulate cell proliferation, survival, growth, migration, differentiation, or cytoskeletal dynamics. Activated Ras (RAS-GTP) regulates multiple cellular functions through effectors, including RAF and phosphatidylinositol 3-kinase (PI3K).[16] The RAF pathway generally mediates the growth factor signaling[16] increasing the proliferation. The PI3K catalyzes the production of phosphatidylinositol-3,4,5-triphosphate (PIP3), which is considered as a second messenger, which helps to activate Akt. Akt can control key cellular processes, including apoptosis, protein synthesis, metabolism, and cell cycle by phosphorylating relevant substrates.[16]
The PI3K-Akt signaling pathway is activated by selenium and vitamin E and can regulate cellular transcription, translation, proliferation, growth, and survival. It has also been reported that low Se intake is associated with an increased risk of bone disease, which can lead to growth retardation and changes in bone metabolism.[17] In a study, which consisted of 232 KBD patients and 331 healthy controls indicating selenoprotein S G105A polymorphisms as an independent risk factor for KBD, it was shown that PI3K/Akt signaling and apoptosis was increased in whole blood and chondrocytes of AA and GA genotypes.[18] Thus, the finding suggested that activation of PI3K/Akt signaling pathway could lead to apoptosis and death of chondrocytes in KBD. The study also showed that tert-butylhydroperoxide-related oxidative damage of cultured C28/I2 human chondrocytes led to upregulation of PI3K/Akt signaling pathway.[18]
Vitamin E is an antioxidant, which together with selenium can protect from oxidative stress.[19] It modulates the enzymatic activity of specific enzymes involved in signal transduction, which includes protein kinase B (PKB/Akt), phosphatidylinositol-3-kinase alpha (PI3Kα), and phosphatidylinositol-3-kinase gamma (PI3Kγ).[20] During recent years, the research of PI3K signaling pathway has revealed that Akt and its downstream target protein is apparently a key regulator of bone formation and bone reconstruction, as androgens could promote MC3T3-E1 osteoblast proliferation when PI3K/Akt signal pathway was activated.[21]
Prevention of PI3K/Akt activation can be considered as a potential therapeutic target. Indeed, supplementary Na2SeO3 could downregulate the expression levels of PI3K/Akt and minimize the oxidative damage.[18] Further, supplementary selenium improved the function of antioxidant defenses, and supported immune surveillance and cell proliferation and differentiation.[17] There is also evidence that Na2SeO3 has a role in protecting chondrocytes from the anti-oxidative damage and inhibiting the PI3K/Akt signaling pathway.[18] Sodium selenite inhibited PI3K/AKT and extracellular signal regulated kinase signaling pathways, and reduced osteoblastic differentiation and decreased apoptosis was observed in vascular smooth muscle cells.[22]
Heat shock protein 90 (Hsp90) is an essential player in the signaling pathways of Ras/Raf and PI3K/AKT pathways. Inhibitors for Hsp90 are now under a major interest due to their potential antiproliferative properties in tumor cells, and many new molecules are under development. Their mode of action is degradation of Hsp90 client proteins, such as AKT and Raf, in the proteasomes.[23] Whether their effective inhibition of the activated Ras and PI3K/AKT would be useful for the KBD patients is an interesting question.
4.2. P53 signaling pathway
A number of stress signals of DNA damage, oxidative stress, and activated oncogenes can activate P53 signaling pathway. The P53 protein is employed as a transcriptional activator of p53-regulated genes, which regulate a variety of BPs, such as cell cycle arrest, cellular senescence, and apoptosis. P53 signaling pathway plays an important role in the mitochondrial membrane stability and DNA damage induced apoptosis. It has been reported that hyaluronan suppresses lidocaine-induced apoptosis of human chondrocytes in vitro through inhibiting the p53-dependent mitochondrial apoptotic pathway.[24] Moreover, current evidence indicates that there is an increased expression of P53 protein and mRNA in T-2 toxin-induced human chondrocytes.[25] In T-2 toxin-induced human chondrocytes, activated P53 by would then affect the expression of its downstream effectors, such as the Bcl-2 family proteins.[25] DON under certain concentration inhibited mRNA translation and the regulation of p38 and extracellularly regulated protein kinase activity, thus inducing phosphorylation of p53 and eventually lead to accelerate cell death.[26] P53 inhibitors (PFT-α) could significantly reduce the P53 phosphorylation induced by DON, and siRNA of PFT-α and P53 can inhibit the activation of caspase-3 induced by DON.[26] The effects of DON on the growth and metabolism of cultured chondrocytes have been reported, specifically that 1.0 μg/mL DON could lead to fatal chondrocyte damage after 24 hours. More chondrocyte damage was observed with higher concentration of DON (2.0 and 2.5 μg/mL) and longer exposure time (120 hours, specifically).[27]
Current evidence suggested that trace element and vitamin E induced PI3K-Akt signaling pathway and mycotoxin-induced P53 signaling pathway contributed to the chondrocytes apoptosis and necrosis in KBD together.
Acknowledgment
We thank National Natural Scientific Foundation of China (81472924, 81620108026) and the Fundamental Research Funds for the Central Universities in 2015 who supported our work.
Supplementary Material
Footnotes
Abbreviations: CI = confidence interval, DAVID = Database for Annotation, Visualization and Integrated Discovery, DEGs = differential expression genes, DON = deoxynivalenol, ERFs = environmental risk factors, ERGs = environmental response genes, KBD = Kashin–Beck disease, KEGG = Kyoto Encyclopedia of Genes and Genomes database, NIV = nivalenol, OR = odds ratios.
Funding/support: National Natural Scientific Foundation of China (81472924, 81620108026) and the Fundamental Research Funds for the Central Universities in 2015.
The authors declare no competing financial interests.
Supplemental Digital Content is available for this article.
References
- [1].Guo X, Ma WJ, Zhang F, et al. Recent advances in the research of an endemic osteochondropathy in China: Kashin-Beck disease. Osteoarthritis Cartilage 2014;22:1774–83. [DOI] [PubMed] [Google Scholar]
- [2].Mathieu F, Begaux F, Lan ZY, et al. Clinical manifestations of Kashin-Beck disease in Nyemo Valley, Tibet. Int Orthop 1997;21:151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Xiong G. Diagnostic, clinical and radiological characteristics of Kashin-Beck disease in Shaanxi Province, PR China. Int Orthop 2001;25:147–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fu Q, Cao J, Renner JB, et al. Radiographic features of hand osteoarthritis in adult Kashin-Beck Disease (KBD): the Yongshou KBD study. Osteoarthritis Cartilage 2015;23:868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ning YJ, Wang X, Ren L, et al. Effects of dietary factors on selenium levels of children to prevent Kashin–Beck disease during a high-prevalence period in an endemic area: a cohort study. Biol Trace Elem Res 2013;153:58–68. [DOI] [PubMed] [Google Scholar]
- [6].Yang L, Zhao GH, Yu FF, et al. Selenium and iodine levels in subjects with Kashin-Beck disease: a meta-analysis. Biol Trace Elem Res 2016;170:43–54. [DOI] [PubMed] [Google Scholar]
- [7].Guan F, Li S, Wang ZL, et al. Histopathology of chondronecrosis development in knee articular cartilage in a rat model of Kashin–Beck disease using T-2 toxin and selenium deficiency conditions. Rheumatol Int 2013;33:157–66. [DOI] [PubMed] [Google Scholar]
- [8].Li D, Han J, Guo X, et al. The effects of T-2 toxin on the prevalence and development of Kashin–Beck disease in China: a meta-analysis and systematic review. Toxicol Res 2016;5:731–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Peng A, Wang WH, Wang CX, et al. The role of humic substances in drinking water in Kashin-Beck disease in China. Environ Health Perspect 1999;107:293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yu F, Liu H, Guo X. Integrative multivariate logistic regression analysis of risk factors for Kashin-Beck disease. Biol Trace Elem Res 2016;174:274–9. [DOI] [PubMed] [Google Scholar]
- [11].Guo X. Progression and prospect of etiology and pathogenesis of Kashin-Beck disease. J Xi’an Jiaotong Univ (Med Sci) 2008;29:481–8. [Google Scholar]
- [12].Wang WZ, Guo X, Duan C, et al. Comparative analysis of gene expression profiles between the normal human cartilage and the one with endemic osteoarthritis. Osteoarthritis Cartilage 2009;17:83–90. [DOI] [PubMed] [Google Scholar]
- [13].Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [14].Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2014;43(Database issue):D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mo XD. Effects of T-2 toxin moniliformin and selenium on osteo-chondrosis of Chinese minipig. Chin J Control Endem Dis 1996;11:130–3. [Google Scholar]
- [16].De Luca A, Maiello MR, D’Alessio A, et al. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets 2012;16(Suppl 2):S17–27. [DOI] [PubMed] [Google Scholar]
- [17].Zeng H, Cao JJ, Combs GF. Selenium in bone health: roles in antioxidant protection and cell proliferation. Nutrients 2013;5:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Du XA, Wang HM, Dai XX, et al. Role of selenoprotein S (SEPS1)-105G> A polymorphisms and PI3K/Akt signaling pathway in Kashin-Beck disease. Osteoarthritis Cartilage 2015;23:210–6. [DOI] [PubMed] [Google Scholar]
- [19].Amara B, Karray A, Hakim A, et al. Dimethoate induces kidney dysfunction, disrupts membrane-bound ATPases and confers cytotoxicity through DNA damage. Protective effects of vitamin E and selenium. Biol Trace Elem Res 2013;156:230–42. [DOI] [PubMed] [Google Scholar]
- [20].Zingg JM. Vitamin E: a role in signal transduction. Annu Rev Nutr 2015;35:135–73. [DOI] [PubMed] [Google Scholar]
- [21].Kang HY, Cho CL, Huang KL, et al. Nongenomic androgen activation of phosphatidylinositol 3-kinase/Akt signaling pathway in MC3T3-E1 osteoblasts. J Bone Miner Res 2004;19:1181–90. [DOI] [PubMed] [Google Scholar]
- [22].Liu H, Li X, Qin F, et al. Selenium suppresses oxidative-stress-enhanced vascular smooth muscle cell calcification by inhibiting the activation of the PI3K/AKT and ERK signaling pathways and endoplasmic reticulum stress. J Biol Inorg Chem 2014;19:375–88. [DOI] [PubMed] [Google Scholar]
- [23].Xue N, Jin J, Liu D, et al. Antiproliferative effect of HSP90 inhibitor Y306zh against pancreatic cancer is mediated by interruption of Akt and MAPK signaling pathways. Curr Cancer Drug Targets 2014;14:671–83. [DOI] [PubMed] [Google Scholar]
- [24].Lee YJ, Kim SA, Lee SH. Hyaluronan suppresses lidocaine-induced apoptosis of human chondrocytes in vitro by inhibiting the p53-dependent mitochondrial apoptotic pathway. Acta Pharmacol Sin 2016;37:664–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen J, Cao JL, Chu YL, et al. T-2 toxin-induced apoptosis involving Fas, p53, Bcl-xL, Bcl-2, Bax and caspase-3 signaling pathways in human chondrocytes. J Zhejiang Univ Sci B 2008;9:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bensassi F, El Golli-Bennour E, Abid-Essefi S, et al. Pathway of deoxynivalenol-induced apoptosis in human colon carcinoma cells. Toxicology 2009;264:104–9. [DOI] [PubMed] [Google Scholar]
- [27].Sichun CJMXL, Shiyuan Z. Effect of deoxynivalenol (DON) on the growth metabolism of cultural chondrocytes. Chin J Endemiol 1993;12:275–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


