Abstract
Breast carcinoma is a heterogeneous disease that has exhibited rapid resistance to treatment in the last decade. Depending genotype and phenotype of breast cancer, there are discernible differences in DNA repair protein responses including DNA double strand break repair. It is a fact that different molecular sub-types of breast carcinoma activate these dedicated protein pathways in a distinct manner. The DNA double-strand damage repair machinery is manipulated by breast carcinoma to selectively repair the damage or insults inflicted by the genotoxic effects of chemotherapy or radiation therapy. The two DNA double-strand break repair pathways employed by breast carcinoma are homologous recombination and non-homologous end joining. In recent decades, therapeutic interventions targeting one or more factors involved in repairing DNA double-strand breaks inflicted by chemo/radiation therapy have been widely studied. Herein, this review paper summarizes the recent evidence and ongoing clinical trials citing potential therapeutic combinatorial interventions targeting DNA double-strand break repair pathways in breast carcinoma.
Keywords: Genotoxic drug, DNA repair, Breast carcinoma, Drug therapy, Radiotherapy
INTRODUCTION
Breast carcinoma is a heterogeneous cancer caused by various factors, including genetic, reproductive, environmental and lifestyle factors.1–3 These clinical behavioral changes observed in breast carcinoma may arise through genetic aberrations, epigenetic modifications and precise transcriptional regulation.2,3
Currently, the genotoxic effects of chemotherapy and radiation therapy lead to problems with responsiveness and resistance in breast carcinoma. One plausible mechanism for these effects is the role of abnormal and compensatory DNA repair pathways among genotypically different breast carcinoma cells.2,4–7 There are several genes involved in maintaining genomic integrity through DNA repair, cell cycle checkpoint control and the regulation of key mitotic steps. Experimental data from genetic and epigenetic studies of DNA repair genes has revealed that slight defects in DNA repair capacities are linked to breast cancer risk.8–10 The overexpression of double-strand break (DSB) repair enzymes, the absence of surveillance factors and the mutation or loss of heterozygosity (LOH) in any of these genes contributes to the pathogenesis of sporadic breast cancers.10 Non-homologous end joining (NHEJ) and homologous recombination (HR) are the two major repair mechanisms for DNA DSBs, but the choice between HR and NHEJ is controlled during the cell cycle.8–11
Several inhibitors/drugs/silencing approaches are currently being used to target DNA DSB repair methods including HR and NHEJ in breast carcinoma and other cancer types.8,9,11–16 In this review, the authors attempted to summarize the efficacy of promising combinatorial therapies in breast carcinoma including chemo/radiotherapy combined with specific DNA DSB repair protein inhibitors.
DNA DAMAGE RESPONSE AND BREAST CANCER PHENOTYPE
Recently, the molecular characterization of breast tumors has shown that breast tumors exhibit significant heterogeneity and are endowed with a powerful feature to change their phenotypic behavior. The heterogeneous nature of breast carcinoma is mostly attributed to genetic and epigenetic contributions.17–19 In addition to outside genomic insults, replication and inherent processes lead to many unavoidable changes including incorrect nucleotide base modifications, DNA strand breaks and chemical modifications of nucleotide bases.4–7 There is a growing consensus that both normal and carcinoma cells activate strategies to thwart conditions of unfavorable genomic instability. These changes to the genetic material play a role in stimulating surveillance efforts in all cells including normal and breast carcinoma cells.20 The DNA repair machinery is described as a built in cellular weapon to maintain genomic integrity by repairing DNA damage and inducing cell cycle arrest in carcinoma.4–6,15,21–24
Some individuals are genetically predisposed to breast cancer due to certain dedicated cellular proliferation and DNA repair genes such as BRCA1, BRCA2, and p53. Triple negative breast cancer (TNBC), also known as basal-like breast cancer, is reported to be caused by a germline mutation in the BRCA1 gene. It is understood that mutations in BRCA1 and BRCA2 cause impaired HR, inactivation of the base excision repair (BER) pathway and epigenetic alterations that may lead to genomic instability.2,25 It is commonly known that breast cancers are recognized for their abnormalities in DNA damage repair and BRCA1 inactivation through mutations or epigenetic modifications. Breast cancer phenotypes are also broadly classified as inherited versus sporadic. The genetic contributions to breast cancer are inherited through autosomal dominant transmission of germline mutations in BRCA1 and BRCA2. Based on germline mutations, 5% to 10% of breast cancers are classified as hereditary breast carcinoma and genes such as BRCA1, BRCA2, CHEK2, BRIP1, RAD51, and ataxia telangiectasia mutated (ATM) are associated with breast cancer.26 Among the DNA repair response players, BRCA1 and BRCA2 are key players in the error-free HR DNA repair system and others are involved in most DSB repair pathways. BRCA1 and BRCA2 are recognized for their clear role in transcription, DSB repair, recombination, tumor suppression and the maintenance of genomic stability.27 There is evidence of single nucleotide polymorphisms (in DNA repair genes including BRCA1, BRCA2, RAD50, TP53, ATM, CHEK2, PALB2, and BRIP1, which may be linked with the development of breast cancer.28,29
DOUBLE-STRAND DNA BREAK REPAIR PATHWAY
In cellular replication, the correct segregation of genomic material is intended, which requires coordinated genomic stability coupled with precision and a thorough check for DNA damage.20 Thus, the DNA damage repair machinery in both normal and breast carcinoma plays a role in the maintenance of genomic integrity and stability to thwart genomic insults.6,7,24 The well-crafted DNA damage repair machinery regulates a specific family of repair genes that are recruited when particular lesions are sensed. The onset of carcinoma changes the DNA damage response and enhances genotoxic stress, which is a target for therapeutic agents in carcinoma.6,24
In breast carcinoma, a dedicated set of DNA repair pathways such as BER, nucleotide excision repair (NER) and mismatch repair (MMR) are directed against single-strand DNA damage caused by genotoxic drugs and oxidative stress.5,8,30 BER focuses on the recognition and removal of misfit or damaged nucleotide bases by DNA glycosylase. In the case of NER, DNA repair is achieved using a cut and paste model by scanning the whole genome for distortions in the DNA double helix.5,30 In MMR, mismatches induce a single-stranded incision site where nuclease, ligase and protease act to remove the incorrectly placed DNA nucleotide base.5,8,30
Improper DNA replication, radiation and radiomimetic chemicals cause DSBs, which are repaired through the HR and NHEJ pathways.30–33 According to study findings, DSBs are repaired through classical non-homologous end joining (C-NHEJ), microhomology-mediated end joining (MMEJ or Alt-MMEJ), and HR methods. This joining of DNA strands utilizes various protein families to repair the damage.8,13,15,34
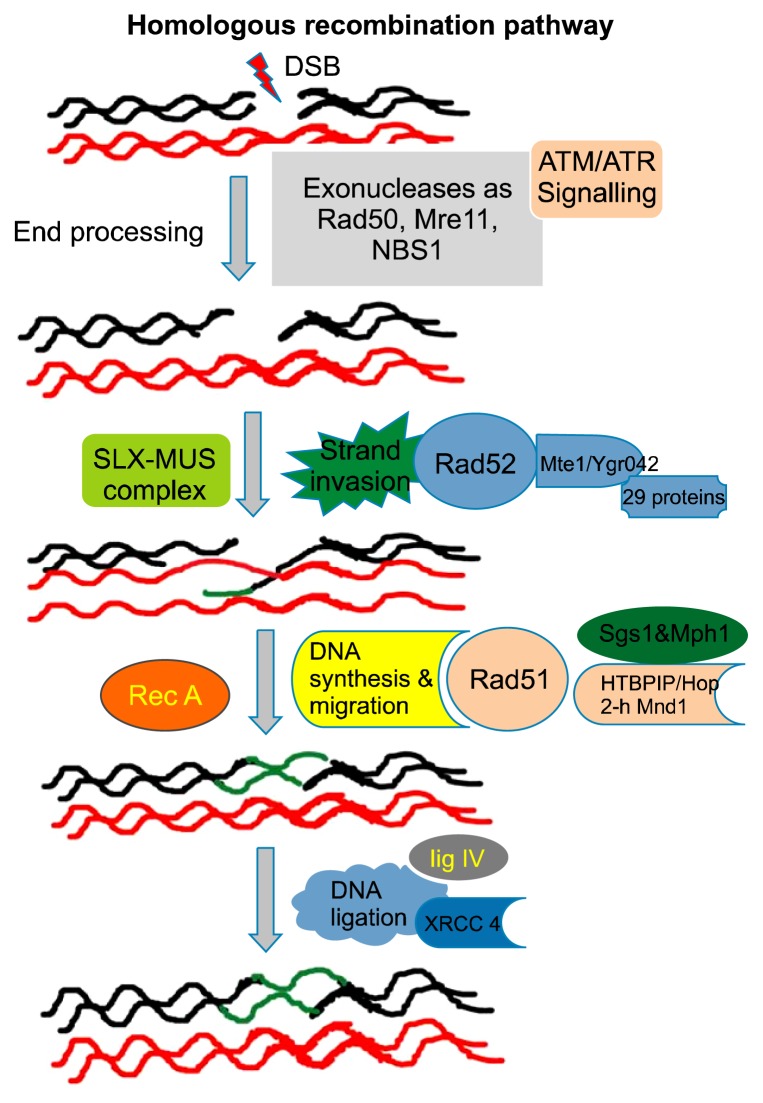
The HR-mediated repair pathways are facilitated by identical or nearly identical DNA sequences that are used as a template to fill in the gap. The set of players dedicated to the HR repair process is closely related to the machinery responsible for crossing-over during meiosis. DSBs due to aberrant replication may cause single-strand breaks or unrepaired lesions, which leads to collapse of the replication fork. These breaks are repaired through the process of recombination, which is shown in a schematic diagram in Figure 1.13,34 DSB repair through HR is initiated by a 5′ to 3′ strand resection (DSB resection), with nucleases generating the 3′ single-strand DNA (3′ssDNA) at DSB sites. In the case of homologous-mediated correction of DNA DSBs, the first step is DNA resection. During DNA resection, C-terminal binding protein (CTBP)-interacting protein (CTIP) (also known as RB binding protein 8 [RBBP8]) interacts with the MRN (comprised of meiotic recombination 11 homologue A [Mre11A], RAD50, and Nijmegen breakage syndrome 1 [NBS1]) complex to promote DNA end resection and the generation of single-stranded DNA.35,36 Several proteins involved in HR re-localize into discrete nuclear foci when DNA damage occurs. In total, 29 proteins have been identified to co-localize with the recombination repair protein Rad52 in response to DNA damage. A protein with an unknown function Mte1/Ygr042 has been shown to co-localize with Rad52.35–37
Figure 1.
This diagram presents a schematic illustration of the homologous recombination process in DNA double-strand break (DSB) repair. This repair pathway consists of a series of several steps. After the formation of the DSB, the meiotic recombination 11 homologue A (Mre11)–RAD50–Nijmegen breakage syndrome 1 (NBS1) (MRN) complex detects and binds the broken DNA ends, which leads to the recruitment of ataxia telangiectasia mutated (ATM) and the initiation of DSBR. In the next step, resection (cutting back) of 5′ DNA end of either side of the DSB occurs, resulting in the exposure of the two regions of the single-stranded DNA (ssDNA). Next, RAD51 binds to the DNA and forms a nucleoprotein filament that has the ability to invade the DNA double helix at an intact, homologous stretch of DNA. DNA polymerases use the homologous DNA sequence as a template and the invaded ssDNA as a primer to synthesize new DNA. DNA ligases and endonucleases resolve the complex DNA structures that form, which consequently results in the repair of the DSB. ATR, ataxia telangiectasia and Rad3-related kinase.
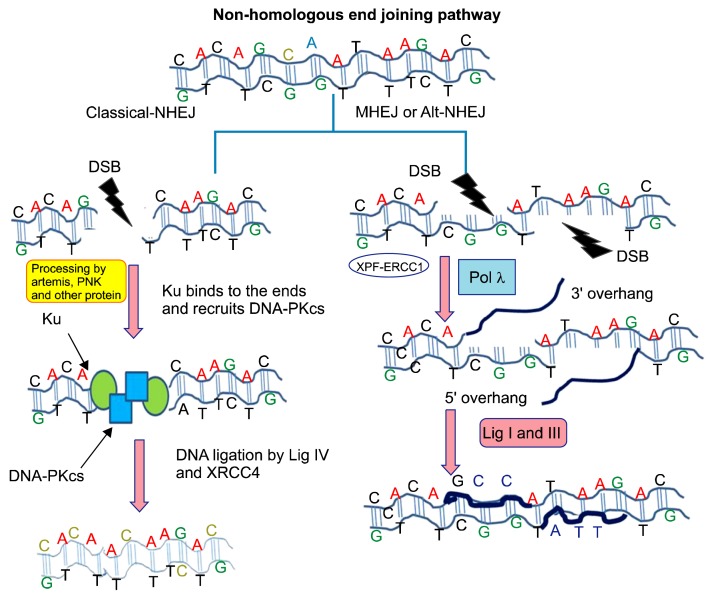
In terms of the NHEJ repair pathway, the severed DNA ends are directly joined through the help of ligase IV, which is a dedicated DNA ligase that forms a complex with the cofactor X-ray repair cross complementing 4 (XRCC4). To facilitate NHEJ, short homologous sequences, called microhomologies, present on the single-stranded tails of the DNA ends serve as a guide sequence for repair.13,35–37 The NHEJ pathway can be accurate or error prone depending on the availability of the guiding sequence when the HR pathways are unable to resolve the DNA break.13,34,35 In addition to C-NHEJ, there are reports of alternative non-homologous end-joining (Alt-NHEJ), which is also referred to as MMEJ. This repair pathway represents one of the dedicated mechanisms by which DNA DSBs are repaired. The difference between MMEJ and CNEHJ is that MMEJ selects 2–5 base pair (bp) micro-homologous sequences during the repair process. Simultaneously, MMEJ is also described as a Ku protein and DNA-PK-independent repair mechanism.13,38–40 A schematic illustration of the NHEJ pathway is depicted in Figure 2.13,34–40 The repair of DNA DSBs is believed to be a cell cycle-dependent repair mechanism. In the G1 phase, the repair of DSBs occurs through NHEJ or MMEJ due to the absence of sister chromatids. The NHEJ pathway is believed to be an error-prone pathway and often involves the loss of the DNA sequence at the break. In the late S and G2 phases, although DNA end-joining pathways remain functional, there is an increase in the repair of DSBs through HR, which is mostly error-free.13,34–40
Figure 2.
This figure illustrates the non-homologous end joining (NHEJ) DNA double-strand break (DSB) repair pathway. In the classical NHEJ pathway, after the formation of the DSB, the Ku complex is recruited. Subsequently, proteins such as DNA dependent protein kinase catalytic subunit (DNA-PKcs) ultimately lead to blunt double-strand ligation through the help of DNA ligase IV and X-ray repair cross complementing 4 (XRCC4), which seals the DSB. In the alternative NHEJ pathway, the DSB is processed by ERCC excision repair 1 (ERCC1) and DNA polymerase gamma. In the next step, ligation is completed by DNA ligase I and DNA ligase III to fill the DSB gap.
SMALL MOLECULE INHIBITION AND HOMOLOGOUS RECOMBINATION
HR repairs DSBs to maintain genome integrity. Tumors carrying BRCA1- and BRCA2-inactivating mutations have been reported to become lethal through RAD52 inhibition either by a specific shRNA or a small peptide aptamer.41 To achieve better outcomes in breast carcinoma, DNA repair protein inhibition strategies are highly necessary. In the same direction, imipramine blue, which is an inhibitor of forkhead box protein M1 (FOXM1)-mediated HR DNA repair pathways, inhibits breast cancer growth.42 Ataxia telangiectasia and Rad3-related kinase, a member of the phosphatidylinositol-3-related protein kinases, is the basis for strategies to silence several types of cancer because these kinases are responsible for the DNA damage response during chemotherapy and radiation therapy in cancer patients.43 Few drug inhibitors targeting ATM kinases have been reported in preclinical studies, which limits their scope of use in cancer therapeutic interventions.44 Recently, modulation of the chromatin landscape has been highlighted by histone DNA methyltransferase inhibitors such as Vorinostat and Chaetocin in radio-sensitization of resistant breast carcinoma.45 To breach the barriers in radiosensitivity in breast carcinoma, miR-15 has been reported to augment cell death in response to radiation treatment. 46 There is a growing idea that one of the clinical implications of inhibiting the DNA cytosine deaminase APOBEC3B may be easing the problem of genotoxic drug resistance in breast cancers.47
There are reports that the protein Survivin has emerged as a potential target of inhibition to increase anthracycline responsiveness against breast cancers.48 Consistent with several key protein players that protect genomic integrity, RAD51 is engaged in DSB repair and has been proposed as a target for improved chemotherapy response in breast carcinoma. In recent years, authors have reported on the specific role of microRNA (miRNA)-155 to decrease the expression of RAD51, which enhances radiation sensitivity in breast carcinoma.49 The DNA repair protein RAD51, which is overexpressed in many cancer types, has been reported to be downregulated by the small molecule inhibitor BO2.50 During the G phase of the cell cycle, HR leads to the LOH. MiRNAs such as miR-1255b, miR-148b, and miR-193b suppress the HR pathway in the G1 phase. These miRNAs target the transcripts of HR repair factors such as BRCA1, BRCA2, and RAD51. Hence, inhibiting miR-1255b, miR-148b, and miR-193b may lead to impaired DSB repair.51 Recently, a targeted liposomal-based ATM gene-specific siRNA-based drug therapy approach was tested in a breast carcinoma animal model. The application of siRNAs targeting ATM has been reported to be effective for breast cancer inhibition.52
Targeted approaches to inhibit target proteins, such as ribonucleotide reductase 1 and 2 (RRM1 and 2) and checkpoint kinase 1 (CHK1), are engaged in DNA repair, proliferation and cell cycle process within TNBC. Using siRNA-based approaches, these breast carcinoma types demonstrated increased potentiation against the genotoxic effects of gemcitabine-based therapies.53 To sensitize breast carcinoma against genotoxic drugs such as cisplatin and Doxorubicin, approaches such as silencing RAD50 or exploring potential small molecule inhibitors have been discussed.54 In the triple negative MDA-MB-468 breast carcinoma study, a new class of inhibitor drugs (ZRBA1) targeting the epidermal growth factor receptor TK domain has been reported to increase radiation sensitization.55 The repair response associated with ionizing radiation (IR)-induced DNA DSBs is linked to sensitization of tumor cells to IR. To induce tumor sensitization, an artificial miRNA (amiR) is engineered to target the 3′-un-translated regions of XRCC2 (an homologous recombinational repair factor) or XRCC4 (an NHEJ factor). Combining amiR and siRNA techniques is suggested to target the gene-coding region, which can improve the efficiency of gene silencing to achieve more robust radiosensitization.56
SMALL MOLECULE INHIBITION AND NON-HOMOLOGOUS END JOINING
The NHEJ is distinctly involved in the removal of DSBs from the genome. Recently, researchers have attempted to block DSB repair proteins to retard carcinoma growth and proliferation. In line with inhibitor-based therapies, there are several reports on the development of the inhibitors SCR-7 and SCR-17, which are directed against DNA ligase IV and DNA ligase I in the NHEJ repair system.12,16 Reports on the phenotypic screening of drug inhibitors such as ARTIK-52, which is an androgen receptor inhibitor, show an enhanced replication-dependent DNA damage repair response and p53 activation in breast carcinoma cells.57 Recently, two Forkhead transcription factors forkhead box O3 (FOXO3a) and FOXM1 have been described for their explicit contribution in the DNA damage response. Therefore, ways to explore inhibitors against these two transcription factors are being attempted to augment senescence and cell death in carcinoma.58 Cell cycle checkpoints are potential cellular targets in several carcinomas including the DNA damage response and survival strategies in breast cancer. There are reports that Palbociclib (PD0332991), a specific inhibitor of CDK4 (IC50 = 11 nM) and CDK6 (IC50 = 16 nM), suppresses growth in estrogen receptor-positive breast cancer.59,60
Recently, Phase II clinical studies in estrogen receptor-positive breast cancer have been conducted for the CDK4/6-selective inhibitors LEE011 (ribociclib) and LY2835219 (abemaciclib), which have the potential to retard tumor growth. These inhibitors show promise for combinatorial drug therapy with both DNA repair protein and cell cycle check point inhibitors to combat the problem of chemotherapy and radiation therapy resistance.15,61
In BRCA1-BER-deficient breast carcinoma, ATM and DNA-PKcs may be inhibited as a better treatment strategy. It has been suggested that BRCA1-BER-deficient cells may show improved sensitivity to treatment when cisplatin is concomitantly administered with ATM and DNA-PKcs inhibitors. These types of drugs/inhibitors combination treatment options will provide better options for personalized therapy.15,62 In preliminary findings, a new class of drugs, an imidazopyridine derivative, targeting the DNA damage response pathway was administered in combination with Doxorubicin to assess cytotoxicity in breast carcinoma. The results suggested that there is marked improvement in apoptosis in the breast carcinoma MCF-7 cell line.63
In view of the close association between DNA repair and genetic instability, there is evidence showing that components of the DNA DSBs repair machinery, including BRCA1, BRCA2, CHK1, DNA-PKcs, FANCA, and the MRE11/RAD50/NBN complex, are targets of Hsp90. Studies are in progress to explore inhibition of the functions of Hsp90 using small molecule inhibitors to disrupt the stabilization of DDR proteins and to act as a cell-specific and tumor-selective radiosensibilizer.64 It is believed that homology-directed repair and NHEJ of DNA DSB are antagonistically regulated by BRAC1 and 53BP1. Findings indicate that retention of 53BP1 at DSB sites is inhibited by the histone deacetylase (HDAC) inhibitor trichostatin A. In addition, another methylated histone residue H3K9me2 was identified to be suppressed by the histone lysine methyltransferase (HKMT) inhibitor UNC0638, which affects retention of the BRCA1/BARD1 complex.65 Recently, it is agreed that several cell survival and death pathways combine to act as a protective cover against genotoxic and radiation insults in cancer. To exploit this idea, an inhibitor (Nutlin-3a) targeting the protein-protein interactions of murine double minute 2 (Mdm2) with several key regulatory proteins in cancer has been studied for its potential accentuating effects of the DNA damaging drug Carboplatin in a breast carcinoma model.66 In view of the implications of signaling pathways, several findings indicate the potential of developing inhibitors targeting the Hedgehog (Hh) signaling pathway, which is linked to DNA repair pathways in human cancer types. The Hh signaling pathway may provide carcinoma cells better capabilities in terms of thwarting the effects of genotoxic- and radiation-mediated DNA damage.67 DNA repair strategies in breast carcinoma are strengthened by the combination or co-operation of multiple signaling cascade pathways. Recent information regarding the mechanism by which HDAC inhibitors induce miRNA-182 to target RAD51 suggests this pathway may be a novel pharmacological strategy to prevent acute myeloid leukemia (AML) cells from conducting HR. Further, these findings indicate that AML cells are sensitized by DNA-damaging agents that activate the HR repair machinery.68 There are promising reports from some ongoing clinical trials evaluating the inhibitors that are directly or indirectly associated with aberrant DNA repair. Among the many DNA repair proteins, there are reports on clinical trials using the small molecule inhibitor BI2536, which targets polo-like kinase 1. According to the clinical trial outcomes, these inhibitors demonstrated limited antitumor activity; however, studies are investigating other derivatives from the same class of inhibitor molecules.69–72 The list of reported clinical trials on various drugs/inhibitors are presented in Table 1.73–76
Table 1.
List of clinical trials investigating drugs/inhibitors dedicated to DNA repair abnormalities in breast cancer
| Serial No. | Title of clinical trial | Drugs/inhibitors | Intervention | Reference |
|---|---|---|---|---|
| 1 | PARP-inhibition and CTLA-4 Blockade in BRCA-deficient Ovarian Cancer | Olaparib and Tremelimumab | To cripple DNA double-strand break repair ability via PARP inhibitors in solid tumors such as ovarian cancer; the clinical trial of the PARP enzyme. | 73 |
| 2. | Phase II Single Arm Pilot Study of the Chk1/2 Inhibitor (LY2606368) in BRCA1/2 Mutation Associated Breast or Ovarian Cancer, Triple Negative Breast Cancer, High Grade Serous Ovarian Cancer, and Metastatic Castrate-Resistant Prostate Cancer. | LY2606368 | A Chk1/2 inhibitor in several solid tumors, including breast tumors having BRCA1/2 defects. | 74 |
| 3. | ABT-888 and Temozolomide for Metastatic Breast Cancer and BRCA1/2 Breast Cancer | ABT-888 and temozolomide | To test PARP inhibitor ABT-188 to enhance the potential of chemotherapeutic agents such as temozolomide to induce cell death. | 75 |
| 4. | Veliparib and Atezolizumab Either Alone or in Combination in Treating Patients With Stage III–IV Triple Negative Breast Cancer | Atezolizumab Other: Laboratory Biomarker Analysis Drug: Veliparib |
A phase II multiple-arm, open-label, randomized study of PARP inhibition (Veliparib; ABT-888) and anti-PD-L1 therapy (Atezolizumab; MPDL3280A) either alone or in combination in homologous DNA repair deficient triple negative breast cancer | 76 |
CONCLUSION
In conclusion, current regimens of genotoxic drugs and radiation-based therapies are only marginally successful. At the same time, failures are widely witnessed in breast cancer treatment. In recent decades, several reports citing laboratory-based outcomes emphasize the importance of DNA DSB repair inhibitors as agents to increase the effectiveness of chemotherapy and radiation therapy treatment options in breast carcinoma. Further understanding the efficacy of DNA repair inhibitors combined with chemotherapy and radiation therapy will open the possibility to personalized therapy.
ACKNOWLEDGMENTS
We would like to thank the Cancer and Translational Research Lab for their constant support and motivation and our host institute Dr. D. Y Patil Biotechnology & Bioinformatics Institute, Dr. D. Y Patil Vidyapeeth, Pune for providing resources. The authors acknowledge the financial support from DST, SERB, New Delhi and DPU Pune.
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010;32:35–48. doi: 10.3233/BD-2010-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, et al. Homologous Recombination Deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22:3764–73. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benevolenskaya EV, Islam AB, Ahsan H, Kibriya MG, Jasmine F, Wolff B, et al. DNA methylation and hormone receptor status in breast cancer. Clin Epigenetics. 2016;8:17. doi: 10.1186/s13148-016-0184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khanna A. DNA damage in cancer therapeutics: a boon or a curse? Cancer Res. 2015;75:2133–8. doi: 10.1158/0008-5472.CAN-14-3247. [DOI] [PubMed] [Google Scholar]
- 5.O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60:547–60. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 6.Gavande NS, VanderVere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS, et al. DNA repair targeted therapy: the past or future of cancer treatment? Pharmacol Ther. 2016;160:65–83. doi: 10.1016/j.pharmthera.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao D, Herman JG, Guo M. The clinical value of aberrant epigenetic changes of DNA damage repair genes in human cancer. Oncotarget. 2016;7:37331–46. doi: 10.18632/oncotarget.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 10.Ralhan R, Kaur J, Kreienberg R, Wiesmüller L. Links between DNA double strand break repair and breast cancer: accumulating evidence from both familial and nonfamilial cases. Cancer Lett. 2007;248:1–17. doi: 10.1016/j.canlet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–87. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 13.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hitrik A, Abboud-Jarrous G, Orlovetskie N, Serruya R, Jarrous N. Targeted inhibition of WRN helicase by external guide sequence and RNase P RNA. Biochim Biophys Acta. 2016;1859:572–80. doi: 10.1016/j.bbagrm.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Samadder P, Aithal R, Belan O, Krejci L. Cancer TARGETases: DSB repair as a pharmacological target. Pharmacol Ther. 2016;161:111–31. doi: 10.1016/j.pharmthera.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Pandey M, Kumar S, Goldsmith G, Srivastava M, Elango S, Shameem M, et al. Identification and characterization of novel ligase I inhibitors [published online ahead of print June 14, 2016] Mol Carcinog. doi: 10.1002/mc.22516. [DOI] [PubMed] [Google Scholar]
- 17.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23(Suppl 2):S60–4. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 18.Gruver AM, Portier BP, Tubbs RR. Molecular pathology of breast cancer: the journey from traditional practice toward embracing the complexity of a molecular classification. Arch Pathol Lab Med. 2011;135:544–57. doi: 10.5858/2010-0734-RAIR.1. [DOI] [PubMed] [Google Scholar]
- 19.Kimbung S, Loman N, Hedenfalk I. Clinical and molecular complexity of breast cancer metastases. Semin Cancer Biol. 2015;35:85–95. doi: 10.1016/j.semcancer.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Jekimovs C, Bolderson E, Suraweera A, Adams M, O’Byrne KJ, Richard DJ. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: the good, the bad, and the promising. Front Oncol. 2014;4:86. doi: 10.3389/fonc.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez GR, Loureiro AP, Marques SA, Miyamoto S, Yamaguchi LF, Onuki J, et al. Oxidative and alkylating damage in DNA. Mutat Res. 2003;544:115–27. doi: 10.1016/j.mrrev.2003.05.005. [DOI] [PubMed] [Google Scholar]
- 22.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–85. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 23.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Li Y, Lu X. Regulators in the DNA damage response. Arch Biochem Biophys. 2016;594:18–25. doi: 10.1016/j.abb.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 25.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–95. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleibl Z, Kristensen VN. Women at high risk of breast cancer: Molecular characteristics, clinical presentation and management. Breast. 2016;28:136–44. doi: 10.1016/j.breast.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Menghi F, Inaki K, Woo X, Kumar PA, Grzeda KR, Malhotra A, et al. The tandem duplicator phenotype as a distinct genomic configuration in cancer. Proc Natl Acad Sci U S A. 2016;113:E2373–82. doi: 10.1073/pnas.1520010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–81. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 29.Bogdanova N, Helbig S, Dörk T. Hereditary breast cancer: ever more pieces to the polygenic puzzle. Hered Cancer Clin Pract. 2013;11:12. doi: 10.1186/1897-4287-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–81. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 31.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 32.Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–54. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeggo PA, Löbrich M. How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. Biochem J. 2015;471:1–11. doi: 10.1042/BJ20150582. [DOI] [PubMed] [Google Scholar]
- 35.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 36.Yuan J, Chen J. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J Biol Chem. 2009;284:31746–52. doi: 10.1074/jbc.M109.023424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yimit A, Kim T, Anand RP, Meister S, Ou J, Haber JE, et al. MTE1 functions with MPH1 in double-strand break repair. Genetics. 2016;203:147–57. doi: 10.1534/genetics.115.185454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–38. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sfeir A, Symington LS. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci. 2015;40:701–14. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma S, Javadekar SM, Pandey M, Srivastava M, Kumari R, Raghavan SC. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 2015;6:e1697. doi: 10.1038/cddis.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan K, Cramer-Morales K, McElroy DL, Ostrov DA, Haas K, Childers W, et al. Identification of a small molecule inhibitor of RAD52 by structure-based selection. PLoS One. 2016;11:e0147230. doi: 10.1371/journal.pone.0147230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajamanickam S, Panneerdoss S, Gorthi A, Timilsina S, Onyeagucha B, Kovalskyy D, et al. Inhibition of FoxM1-mediated DNA repair by imipramine blue suppresses breast cancer growth and metastasis. Clin Cancer Res. 2016;22:3524–36. doi: 10.1158/1078-0432.CCR-15-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrs M, Korabecny J, Nepovimova E, Jun D, Hodny Z, Kuca K. Small molecules targeting ataxia telangiectasia and Rad3-related (ATR) kinase: an emerging way to enhance existing cancer Therapy. Curr Cancer Drug Targets. 2016;16:200–8. doi: 10.2174/156800961603160206122927. [DOI] [PubMed] [Google Scholar]
- 44.Vecchio D, Frosina G. Targeting the ataxia telangiectasia mutated protein in cancer therapy. Curr Drug Targets. 2016;17:139–53. doi: 10.2174/1389450115666141110154621. [DOI] [PubMed] [Google Scholar]
- 45.Olcina MM, O’Dell S, Hammond EM. Targeting chromatin to improve radiation response. Br J Radiol. 2015;88:20140649. doi: 10.1259/bjr.20140649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mei Z, Su T, Ye J, Yang C, Zhang S, Xie C. The miR-15 family enhances the radiosensitivity of breast cancer cells by targeting G2 checkpoints. Radiat Res. 2015;183:196–207. doi: 10.1667/RR13784.1. [DOI] [PubMed] [Google Scholar]
- 47.Harris RS. Molecular mechanism and clinical impact of APOBEC3B-catalyzed mutagenesis in breast cancer. Breast Cancer Res. 2015;17:8. doi: 10.1186/s13058-014-0498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faversani A, Vaira V, Moro GP, Tosi D, Lopergolo A, Schultz DC, et al. Survivin family proteins as novel molecular determinants of doxorubicin resistance in organotypic human breast tumors. Breast Cancer Res. 2014;16:R55. doi: 10.1186/bcr3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gasparini P, Lovat F, Fassan M, Casadei L, Cascione L, Jacob NK, et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci U S A. 2014;111:4536–41. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang F, Mazin AV. A small molecule inhibitor of human RAD51 potentiates breast cancer cell killing by therapeutic agents in mouse xenografts. PLoS One. 2014;9:e100993. doi: 10.1371/journal.pone.0100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi YE, Pan Y, Park E, Konstantinopoulos P, De S, D’Andrea A, et al. MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. Elife. 2014;3:e02445. doi: 10.7554/eLife.02445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu R, Huang Y, Mai J, Zhang G, Guo X, Xia X, et al. Multistage vectored siRNA targeting ataxia-telangiectasia mutated for breast cancer therapy. Small. 2013;9:1799–808. doi: 10.1002/smll.201201510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett CN, Tomlinson CC, Michalowski AM, Chu IM, Luger D, Mittereder LR, et al. Cross-species genomic and functional analyses identify a combination therapy using a CHK1 inhibitor and a ribonucleotide reductase inhibitor to treat triple-negative breast cancer. Breast Cancer Res. 2012;14:R109. doi: 10.1186/bcr3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flores-Pérez A, Rafaelli LE, Ramírez-Torres N, Aréchaga-Ocampo E, Frías S, Sánchez S, et al. RAD50 targeting impairs DNA damage response and sensitizes human breast cancer cells to cisplatin therapy. Cancer Biol Ther. 2014;15:777–88. doi: 10.4161/cbt.28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heravi M, Kumala S, Rachid Z, Jean-Claude BJ, Radzioch D, Muanza TM. ZRBA1, a mixed EGFR/DNA targeting molecule, potentiates radiation response through delayed DNA damage repair process in a triple negative breast cancer model. Int J Radiat Oncol Biol Phys. 2015;92:399–406. doi: 10.1016/j.ijrobp.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 56.Zheng Z, Ng WL, Zhang X, Olson JJ, Hao C, Curran WJ, et al. RNAi-mediated targeting of noncoding and coding sequences in DNA repair gene messages efficiently radiosensitizes human tumor cells. Cancer Res. 2012;72:1221–8. doi: 10.1158/0008-5472.CAN-11-2785. [DOI] [PubMed] [Google Scholar]
- 57.Fleyshman D, Cheney P, Ströse A, Mudambi S, Safina A, Commane M, et al. ARTIK-52 induces replication-dependent DNA damage and p53 activation exclusively in cells of prostate and breast cancer origin. Cell Cycle. 2016;15:455–70. doi: 10.1080/15384101.2015.1127478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nestal de Moraes G, Bella L, Zona S, Burton MJ, Lam EW. Insights into a critical role of the FOXO3a-FOXM1 axis in DNA damage response and genotoxic drug resistance. Curr Drug Targets. 2016;17:164–77. doi: 10.2174/1389450115666141122211549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438–51. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy CG, Dickler MN. The role of CDK4/6 inhibition in breast cancer. Oncologist. 2015;20:483–90. doi: 10.1634/theoncologist.2014-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albarakati N, Abdel-Fatah TM, Doherty R, Russell R, Agarwal D, Moseley P, et al. Targeting BRCA1-BER deficient breast cancer by ATM or DNA-PKcs blockade either alone or in combination with cisplatin for personalized therapy. Mol Oncol. 2015;9:204–17. doi: 10.1016/j.molonc.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.El-Awady RA, Semreen MH, Saber-Ayad MM, Cyprian F, Menon V, Al-Tel TH. Modulation of DNA damage response and induction of apoptosis mediates synergism between doxorubicin and a new imidazopyridine derivative in breast and lung cancer cells. DNA Repair (Amst) 2016;37:1–11. doi: 10.1016/j.dnarep.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 64.Pennisi R, Ascenzi P, di Masi A. Hsp90: a new player in DNA repair? Biomolecules. 2015;5:2589–618. doi: 10.3390/biom5042589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fukuda T, Wu W, Okada M, Maeda I, Kojima Y, Hayami R, et al. Class I histone deacetylase inhibitors inhibit the retention of BRCA1 and 53BP1 at the site of DNA damage. Cancer Sci. 2015;106:1050–6. doi: 10.1111/cas.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tonsing-Carter E, Bailey BJ, Saadatzadeh MR, Ding J, Wang H, Sinn AL, et al. Potentiation of carboplatin-mediated DNA damage by the Mdm2 modulator nutlin-3a in a humanized orthotopic breast-to-lung metastatic model. Mol Cancer Ther. 2015;14:2850–63. doi: 10.1158/1535-7163.MCT-15-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng E, Hanna A, Samant RS, Shevde LA. The impact of hedgehog signaling pathway on DNA repair mechanisms in human cancer. Cancers (Basel) 2015;7:1333–48. doi: 10.3390/cancers7030839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai TH, Ewald B, Zecevic A, Liu C, Sulda M, Papaioannou D, et al. HDAC inhibition induces microRNA-182, which targets RAD51 and impairs HR repair to sensitize cells to sapacitabine in acute myelogenous leukemia. Clin Cancer Res. 2016;22:3537–49. doi: 10.1158/1078-0432.CCR-15-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schöffski P, Blay JY, De Greve J, Brain E, Machiels JP, Soria JC, et al. Multicentric parallel phase II trial of the polo-like kinase 1 inhibitor BI 2536 in patients with advanced head and neck cancer, breast cancer, ovarian cancer, soft tissue sarcoma and melanoma. The first protocol of the European Organization for Research and Treatment of Cancer (EORTC) Network Of Core Institutes (NOCI) Eur J Cancer. 2010;46:2206–15. doi: 10.1016/j.ejca.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 70.Sanhaji M, Louwen F, Zimmer B, Kreis NN, Roth S, Yuan J. Polo-like kinase 1 inhibitors, mitotic stress and the tumor suppressor p53. Cell Cycle. 2013;12:1340–51. doi: 10.4161/cc.24573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livraghi L, Garber JE. PARP inhibitors in the management of breast cancer: current data and future prospects. BMC Med. 2015;13:188. doi: 10.1186/s12916-015-0425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dizdar O, Arslan C, Altundag K. Advances in PARP inhibitors for the treatment of breast cancer. Expert Opin Pharmacother. 2015;16:2751–8. doi: 10.1517/14656566.2015.1100168. [DOI] [PubMed] [Google Scholar]
- 73.Adams SF. PARP-inhibition and CTLA-4 blockade in BRCA-deficient ovarian cancer. Bethesda, MD: ClinicalTrials.gov identifier, New Mexico Cancer Care Alliance; 2016. [Google Scholar]
- 74.Lee JM. ClinicalTrials.gov. Bethesda, MD: National Institutes of Health; 2016. A phase II single arm pilot study of the Chk1/2 inhibitor (LY2606368) in BRCA1/2 mutation associated breast or ovarian cancer, triple negative breast cancer, high grade serous ovarian cancer, and metastatic castrate-resistant prostate cancer. [Google Scholar]
- 75.Isakoff S. ClinicalTrials.gov. Bethesda, MD: National Institutes of Health; 2016. ABT-888 and temozolomide for metastatic breast cancer and BRCA1/2 breast cancer ABT-888 and temozolomide to test PARP inhibitor ABT-188 to enhance the potential of chemotherapy therapy like temozolomide to induce cell death. [Google Scholar]
- 76.LoRusso PM. ClinicalTrials.gov. Bethesda, MD: National Institutes of Health; 2016. Veliparib and atezolizumab either alone or in combination in treating patients with stage iii-iv triple negative breast cancer. [Google Scholar]