Abstract
Background
Alcohol is known to affect two epigenetic phenomena, DNA methylation and DNA hydroxymethylation, and iron is a cofactor of ten-eleven translocation (TET) enzymes that catalyze the conversion from methylcytosine to hydroxymethylcytosine. In the present study we aimed to determine the effects of alcohol on DNA hydroxymethylation and further effects of iron on alcohol associated epigenetic changes.
Methods
Twenty-four male Sprague-Dawley rats were fed either Lieber-DeCarli alcohol diet (36% calories from ethanol) or Lieber-DeCarli control diet along with or without iron supplementation (0.6% carbonyl iron) for 8 weeks. Hepatic non-heme iron concentrations were measured by colorimetric assays. Protein levels of hepatic ferritin and transferrin receptor were determined by Western blotting. Methylcytosine, hydroxymethylcytosine and unmodified cytosine in DNA were simultaneously measured by liquid chromatography/mass spectrometry method.
Results
Iron supplementation significantly increased hepatic non-heme iron contents (P < 0.05) but alcohol alone did not. However, both alcohol and iron significantly increased hepatic ferritin levels and decreased hepatic transferrin receptor levels (P < 0.05). Alcohol reduced hepatic DNA hydroxymethylation (0.21% ± 0.04% vs. 0.33% ± 0.04%, P = 0.01) compared to control, while iron supplementation to alcohol diet did not change DNA hydroxymethylation. There was no significant difference in methylcytosine levels, while unmodified cytosine levels were significantly increased in alcohol-fed groups compared to control (95.61% ± 0.08% vs. 95.26% ± 0.12%, P = 0.03), suggesting that alcohol further increases the conversion from hydroxymethylcytosine to unmodified cytosine.
Conclusions
Chronic alcohol consumption alters global DNA hydroxymethylation in the liver but iron supplementation reverses the epigenetic effect of alcohol.
Keywords: Alcohols, Iron, DNA hydroxymethylation, DNA methylation, Epigenetics
INTRODUCTION
DNA methylation, methylated cytosine in CpG dinucleotides, has been most extensively studied due to its influence on gene transcription. This characteristic DNA methylation is involved in many physiologic and pathologic processes and is an important etiologic mechanism for the development and progression of cancer. DNA hydroxymethylation is another epigenetic modification of cytosines within DNA, and has recently been the topic of epigenetic studies. Hydroxymethylated cytosines in CpG residues function as an active DNA demethylation step, but also have unique gene regulatory functions that are somewhat different from DNA methylation. Furthermore the distribution of DNA hydroxymethylation is more variable in each tissue than that of DNA methylation.1
Our previous animal study demonstrated that chronic alcohol consumption at 18% of energy significantly reduced global levels of hepatic DNA hydroxymethylation in young mice but not old ones.2 This pattern of reduced global hydroxymethylcytosine is similarly seen in cancerous tissue and, therefore, it is plausible that this alcohol consumption may create a tumorigenic environment in the liver, thereby predisposing the tissue to cancer.3–6 By better understanding how chronic alcohol consumption may affect global DNA hydroxymethylation we may gain insight into the early stages of this tumorigenic environment.
The process of hydroxylation of methylcytosine is catalyzed by ten-eleven translocation (TET) enzymes, which has been regarded as the first step of active demethylation. In many cancers TET gene mutations and aberrant DNA hydroxymethylation have been found, suggesting that the alteration of DNA hydroxymethylation is critical to carcinogenesis.7 Interestingly, the conversion of methylcytosine to hydroxymethylcytosine is dependent on iron,8 a cofactor of TET enzymes, which is an essential nutrient for an array of key biological processes including oxygen transport, cellular respiration through electron transport, DNA replication, DNA repair, and free radical production.
Because alcohol often interferes with iron metabolism,9–12 we wanted to investigate whether the influence of alcohol consumption on the hydroxylation could be modulated by iron supplementation. In the present study we attempted to validate the epigenetic effect of alcohol on DNA hydroxymethylation in a different animal model at the different dietary alcohol level and further to demonstrate the effect of iron on alcohol associated epigenetic change.
MATERIALS AND METHODS
1. Animal study and diets
Twenty four 8 week old male Sprague-Dawley rats (SLC Inc., Hamamatsu, Japan) were fed one of four different diets: 1) control group, Lieber-DeCarli control diet (0% calorie from ethanol); 2) alcohol group, Lieber-DeCarli alcohol diet (36% calories from ethanol); 3) iron group, Lieber-DeCarli control diet (0% calorie from ethanol) with iron supplementation (0.6% carbonyl iron); and 4) iron + alcohol group, Lieber-DeCarli alcohol diet (36% calories from ethanol) with iron supplementation (0.6% carbonyl iron) (n = 6 per each group).13,14 We chose the dose, 0.6% iron, based on the results from a previous study,14 which showed a significant interaction between iron and alcohol on liver damage. The alcohol feeding protocol with Lieber-DeCarli alcohol diet is a standard method that has been extensively used as an animal model of alcohol consumption.13,15 In particular, the Lieber-DeCarli alcohol diet provides sufficient amount of all essential nutrients in a liquid formula, and successfully induces alcoholic liver disease within 4 to 8 weeks of feeding.
After one-week acclimation on a chow diet, all animals were fed a Lieber-DeCarli liquid diet (Dyets, Inc., Bethlehem, PA, USA) without ethanol for five days. Ethanol was gradually introduced over a 10-day period before providing animals with the final concentration of 6.2% (vol/vol) (36% of total calories as ethanol). In the control diet, ethanol was replaced by an isocaloric amount of maltodextrin. Rats were killed after 8 weeks of pair feeding and harvested liver tissues were stored at −80°C. This study was reviewed and approved by the Institutional Animal Care and Use Committee of Kyung Hee University (KHUASP(SE)-09-002).
2. Measurement of hepatic non-heme iron contents and protein levels of ferritin and transferrin receptor
Liver non-heme iron content was measured by colorimetric assay.16 Briefly, 0.1 g liver tissues were digested in 2 mL acidic solution (3 mole/L HCl and 10% trichloroacetic acid) for 20 hours at 65°C. Digested samples were incubated with chromogen reagent containing 0.1% bathophenathrolinesulfonate and 1% thioglycolic acid for 10 minutes at room temperature, and the absorbance at 535 nm was measured by spectrophotometer (Bio-Tek Instruments Inc., Winooski, VT, USA). The protein expressions of hepatic ferritin, which stores iron, and transferrin receptor, which imports iron into the cell, were measured using the western blotting according to a standard procedure. Tissue level of ferritin is known to be positively associated with iron, whereas tissue levels of transferrin receptor are negatively associated with that of iron.17,18 Liver tissue is dissolved in the lysis buffer containing 1 mmol/L phenylmethanesulfonylfluoride (25 mmol/L Tris-HCl, 1% NP-40, 1% sodium deoxycholate, 150 mmol/L NaCl, 1% SDS) and added protease inhibitors. After centrifuge protein levels of supernatants were determined by PierceTM BCA protein assay kit protein assay kit (Thermo Fisher Scientific, Rockford, IL, USA) and protein extracts were separated on the 10% SDS PAGE gel. Primary antibodies used in this study were ferritin (The Binding Site Group Ltd., Birmingham, UK), transferrin receptor (Thermo Fisher Scientific, Bremen, Germany) and beta-actin (Santa Cruz Biotechnology, Dallas, TX, USA) as a loading control. Band densities were quantified by using a chemi-doc imaging system (Clinx Science Instruments Co., Shanghai, China).
3. Measurement of methylcytosine, hydroxymethylcytosine and unmodified cytosine
Extraction of DNA was conducted using the conventional phenol/chloroform/isoamyl alcohol method. Hydrolysis of DNA was performed as previously described.19 In brief DNA was hydrolyzed to nucleosides using nuclease P1, phosphodiesterase I and alkaline phosphatase (all from Sigma, St. Louis, MO, USA). Thereafter, internal standards were added to samples. The isotope-labeled internal standard for deoxycytidine was [15N3]-2′-deoxycytidine, while that of 5-methyl-deoxycytidine and 5-hydroxymethyl-deoxycytidine were (methyl-d3, ring-6-d1)-5′-methyl-2′-deoxycytidine (both from Cambridge Isotopes Laboratories, Inc., Andover, MA, USA). The DNA hydrolysates were separated by an Agilent 1100 high-performance liquid chromatograph (Agilent Technologies, Palo Alto, CA, USA) and masses were detected through a 3200 Q Trap MS-MS system (Applied Biosystem, Concord, ON, Canada). Using the known masses of isotope-labeled internal standards added to each sample and the area of each peak the absolute amount of unmodified cytosine, 5′-methylcytosine and 5′-hydroxymethylcytosine per 1 μg of DNA can be calculated. Thereafter each amount was expressed as a percentage of total cytosine as we previously described.20
4. Statistics
Differences in percent methylation, hydroxymethylation and unmodified cytosine were determined through a two-way ANOVA followed by a Tukey-Kramer adjustment for multiple comparisons. Hepatic non-heme iron contents and protein levels of ferritin and transferrin receptor were compared by one-way ANOVA followed by Duncan’s multiple range tests. Differences were considered statistically significant at a P < 0.05. Data were analyzed using SAS ver. 9.3 (SAS Institute, Cary, NC, USA).
RESULTS
1. Hepatic non-heme iron levels and protein expression of hepatic ferritin and transferrin receptor
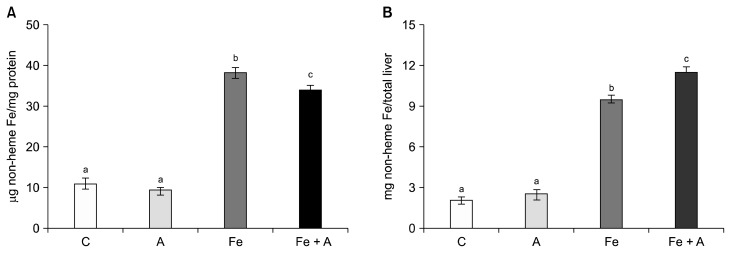
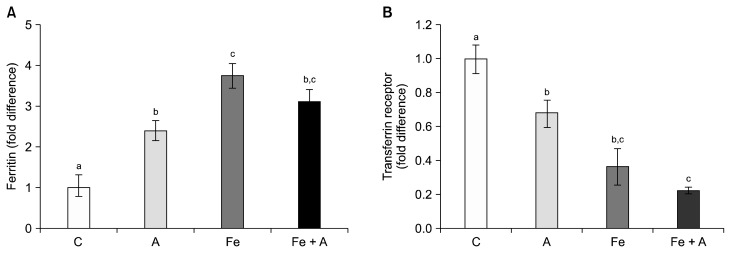
Hepatic non-heme iron contents, expressed either μg/mg protein (Fig. 1A) or mg/total liver (Fig. 1B), were significantly increased in the iron supplemented groups regardless of alcohol consumption, while alcohol consumption alone did not increase hepatic non-heme contents. On the other hand, all three diet groups showed significantly increased ferritin levels compared to the control group (Fig. 2A) and decreased transferrin receptor levels (Fig. 2B) (P < 0.05), indicating that chronic alcohol consumption and iron supplementation, both individually and combined, modify the proteins of which gene expression is responsive to cellular iron contents.
Figure 1.
Hepatic non-heme iron concentrations in rats fed 4 different diets. Iron supplementation increased hepatic non-heme iron contents, expressed either (A) μg non-heme Fe/mg protein or (B) mg non-heme Fe/total liver, but not alcohol consumption only. C, Lieber-DeCarli control diet (0% calorie from ethanol); A, Lieber-DeCarli alcohol diet (36% calories from ethanol); Fe, Lieber-DeCarli control diet (0% calorie from ethanol) with iron supplementation (0.6% carbonyl iron); Fe + A, Lieber-DeCarli alcohol diet (36% calories from ethanol) with iron supplementation (0.6% carbonyl iron). a–cBars with different alphabet were significant at P < 0.05 (Mean ± SE for n = 6 per group).
Figure 2.
Protein expression of hepatic ferritin and transferrin receptor. Compared to the control group, three diet groups showed significantly increased (A) ferritin levels and (B) decreased transferrin receptor levels (P < 0.05), indicating that both alcohol consumption and iron supplementation increases iron storage in the liver. C, Lieber-DeCarli control diet (0% calorie from ethanol); A, Lieber-DeCarli alcohol diet (36% calories from ethanol); Fe, Lieber-DeCarli control diet (0% calorie from ethanol) with iron supplementation (0.6% carbonyl iron); Fe + A, Lieber-DeCarli alcohol diet (36% calories from ethanol) with iron supplementation (0.6% carbonyl iron). a–cBars with different alphabet were significant at P < 0.05. Values are averages and SE (n = 6 per group).
2. DNA methylation and hydroxymethylation as well as unmodified cytosine
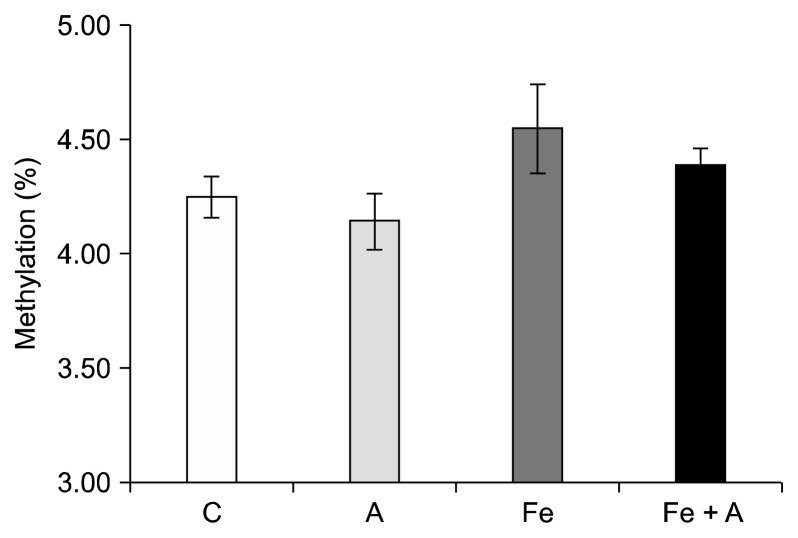
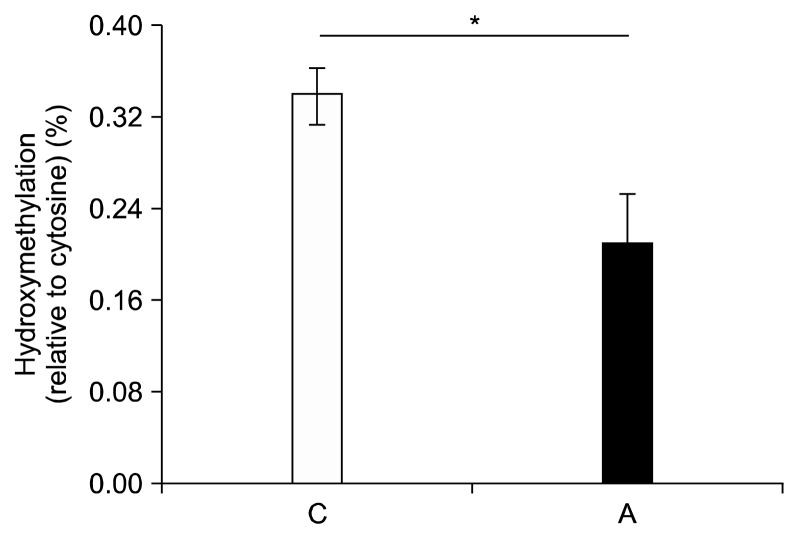
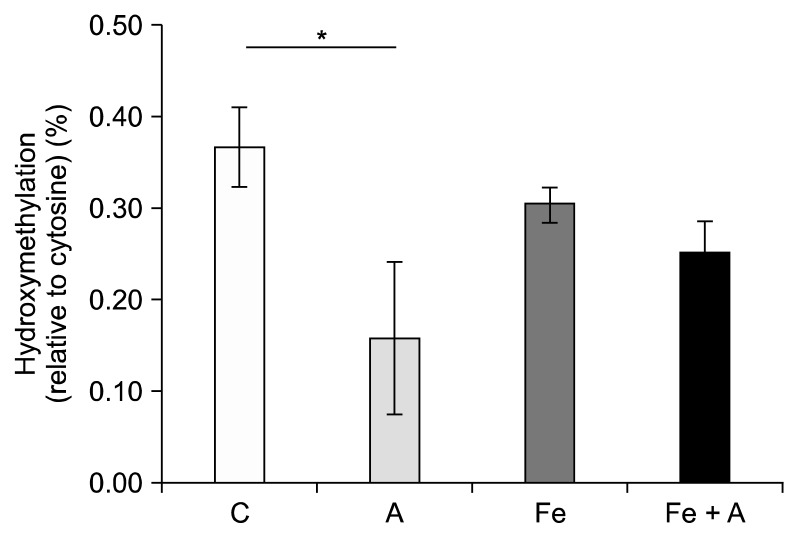
There were no significant changes in DNA methylation among any of the diet groups (Fig. 3); however, significant changes occurred in the percent of cytosines that were hydroxymethylated. When determining the effect of alcohol alone on percent hydroxymethylation regardless of whether the rats were also fed an iron supplemented diet, we found a significant decrease in the livers of rats fed an alcohol-containing diet (Fig. 4) (0.21% ± 0.04% vs. 0.33% ± 0.04%, P = 0.01; Tukey-Kramer Adjustment). Furthermore, when the data is broken up into four groups to include the effect of iron and alcohol, a significant decrease in DNA hydroxymethylation was seen in rats fed the alcohol diet relative to the control group (0.30% ± 0.02% vs. 0.37% ± 0.04%, P = 0.03) (Fig. 5). Rats that were fed the iron + alcohol diet, had a percent hydroxymethylation that was comparable to both the rats in the control or iron diet group.
Figure 3.
Influence of chronic alcohol consumption and iron on methylcytosine levels. There was no significant difference in methyl-cytosine levels among 4 groups (P = NS). Values are averages and SE (n = 6 per group). C, Lieber-DeCarli control diet (0% calorie from ethanol); A, Lieber-DeCarli alcohol diet (36% calories from ethanol); Fe, Lieber-DeCarli control diet (0% calorie from ethanol) with iron supplementation (0.6% carbonyl iron); Fe + A, Lieber-DeCarli alcohol diet (36% calories from ethanol) with iron supplementation (0.6% carbonyl iron).
Figure 4.
Influence of alcohol on DNA hydroxymethylation regardless of iron supplementation. DNA hydroxymethylation was significant decreased in the livers of rats fed an alcohol-containing diet compared to control (P = 0.01). Values are averages and SE (n = 12 per group). C, Lieber-DeCarli control diet (0% calorie from ethanol); A, Lieber-DeCarli alcohol diet (36% calories from ethanol). *Statistically significant at a P < 0.05.
Figure 5.
Influence of iron supplementation on DNA hydroxymethylation reduced by chronic alcohol consumption. A significant decrease in DNA hydroxymethylation was seen in rats fed the alcohol diet without iron relative to the control group (P = 0.03). When rats were fed a diet containing both alcohol and iron, the percent hydroxymethylation was equivalent to the rats fed a control diet with or without iron. Values are averages and SE (n = 6 per group). C, Lieber-DeCarli control diet (0% calorie from ethanol); A, Lieber-DeCarli alcohol diet (36% calories from ethanol); Fe, Lieber-DeCarli control diet (0% calorie from ethanol) with iron supplementation (0.6% carbonyl iron); Fe + A, Lieber-DeCarli alcohol diet (36% calories from ethanol) with iron supplementation (0.6% carbonyl iron). *Statistically significant at a P < 0.05.
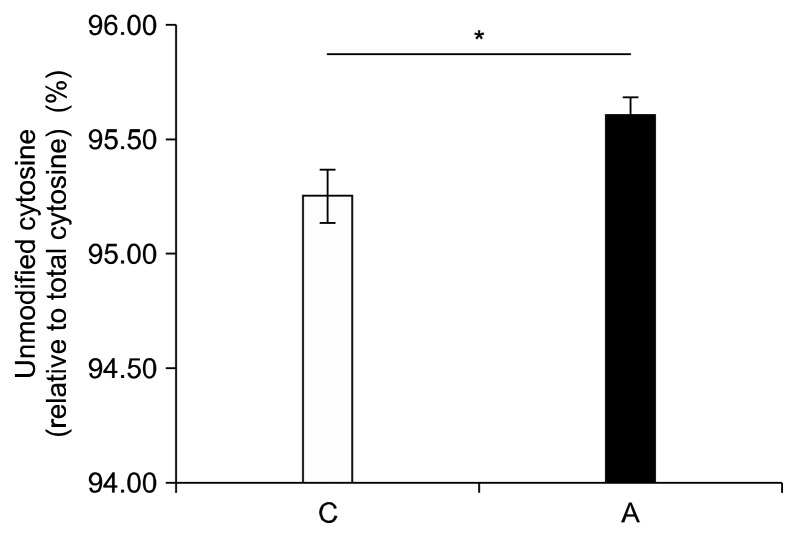
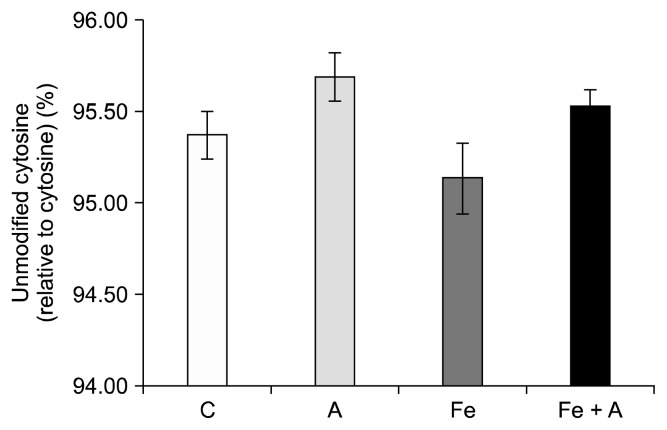
Alcohol increased the percentage of cytosines that were unmodified, regardless of iron supplementation (95.61% ± 0.08% vs. 95.26% ± 0.12%, P = 0.03) (Fig. 6). Alcohol tended to increase the percentage of cytosine that was unmodified in both the control and the iron supplemented groups, though neither of these results reached significance (Fig. 7).
Figure 6.
Influence of chronic alcohol consumption on unmodified cytosine levels. The level of unmodified cytosine level was significantly increased in the livers of rats fed an alcohol-containing diet compared to control regardless of iron supplementation (P = 0.03). Values are averages and SE (n = 12 per group). C, Lieber-DeCarli control diet (0% calorie from ethanol); A, Lieber-DeCarli alcohol diet (36% calories from ethanol). *Statistically significant at a P < 0.05.
Figure 7.
Influence of chronic alcohol consumption and iron on unmodified cytosine levels. There was no significant difference in unmodified cytosine among 4 groups (P = NS). Values are averages and SE (n = 6 per group). C, Lieber-DeCarli control diet (0% calorie from ethanol); A, Lieber-DeCarli alcohol diet (36% calories from ethanol); Fe, Lieber-DeCarli control diet (0% calorie from ethanol) with iron supplementation (0.6% carbonyl iron); Fe + A, Lieber-DeCarli alcohol diet (36% calories from ethanol) with iron supplementation (0.6% carbonyl iron).
DISCUSSION
The chronic consumption of alcohol significantly decreased hepatic DNA hydroxymethylcytosine. An interesting finding here is that the supplementation of iron into a diet containing alcohol seems to reverse the reduction of hydroxymethylcytosine. Because iron is a cofactor for the TET enzyme catalysis of methylcytosine to hydroxymethylcytosine, this increase in iron loading in the liver would seemingly increase the enzymatic reaction.21,22 There is, however, the possibility that the binding constant of iron to the TET enzymes is low enough that the iron-TET binding was saturated in the absence of alcohol, because the level of DNA hydroxymethylation in the liver of rats fed an iron containing diet without alcohol is not significantly different from that of control diet group.
We also show here that rats fed a diet containing alcohol had a significant increase in the portion of cytosines that were unmodified. Previous studies have reported a significant decrease in DNA methylation in alcohol models, and this increase in unmodified cytosine may coincide with those reports, even though the decrease in methylcytosine here does not reach significance.23–25 It appears that alcohol reduces DNA hydroxymethylation by increasing the degradation process of hydroxymethylcytosine to cytosine. In fact TET enzymes not only catalyze the reaction from methylcytosine to hydroxymethylcytosine but also catalyze the reactions from hydroxymethylcytosine to formylcytosine as well as from formylcytosine to carboxycytosine, which is finally converted to unmodified cytosine.26
Ferritin, the major iron storage protein composed of multimeric H and L subunits, is transcriptionally and post-transcriptionally up-regulated under iron loading.27 In this study, alcohol consumption was associated with a decrease in DNA hydroxymethylation, and the supplementation of iron in the diet blocked this alcohol-associated decrease in DNA hydroxymethylation. Conversely, both the alcohol group and the iron + alcohol group had increased ferritin protein levels. Thus our data suggest that iron itself, rather than iron-bound ferritin, modulates the degree of DNA hydroxymethylation. Future study is needed to confirm this hypothesis.
Taken together, the results presented here validate our previous finding that chronic alcohol consumption decreases the percentage of total cytosine that is hydroxymethylated in hepatic DNA. Even though our previous study took place in mice, the amount of alcohol fed to the mice was lower (18% of total calories were derived from ethanol, compared to the 36% of total calories here), and the duration of the study was shorter (5 weeks vs. 8 weeks), the end results were similar. Interestingly, we only saw a decrease in hydroxymethylated DNA in the young mice fed alcohol in our previous study. The mice were 5.5 months old at the end of the study, which is a similar age as the rats here (4 months old), which aided in the validation of our previous findings. While this data is intriguing, without gene specific techniques, such as a microarray, it is unknown whether this decrease in hydroxymethylcytosine is occurring at genes that may promote tumorigenesis. It is interesting that a similar decrease in hydroxymethylation is shown in cancer, particularly in alcohol associated hepatocellular carcinoma.5,28–30 Through microarray and sequencing technologies, the role of hydroxymethylcytosine in the normal and diseased cell states can be further delineated. Other limitations of the study include the lack of TET enzyme activities, which might further explain the molecular mechanisms by which iron reverses the epigenetic effect of alcohol.
ACKNOWLEDGMENTS
This work was supported in part by the Korean Food Research Institute (S.W.C.) and the Basic Science Research Program of the National Research Foundation (NRF-2010-0011226 to J.C.).
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Aspects Med. 2013;34:753–64. doi: 10.1016/j.mam.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tammen SA, Dolnikowski GG, Ausman LM, Liu Z, Sauer J, Friso S, et al. Aging and alcohol interact to alter hepatic DNA hydroxymethylation. Alcohol Clin Exp Res. 2014;38:2178–85. doi: 10.1111/acer.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 2012;103:670–6. doi: 10.1111/j.1349-7006.2012.02213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin. 2013;6:10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen ML, Shen F, Huang W, Qi JH, Wang Y, Feng YQ, et al. Quantification of 5-methylcytosine and 5-hydroxymethylcytosine in genomic DNA from hepatocellular carcinoma tissues by capillary hydrophilic-interaction liquid chromatography/quadrupole TOF mass spectrometry. Clin Chem. 2013;59:824–32. doi: 10.1373/clinchem.2012.193938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–46. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroeze LI, van der Reijden BA, Jansen JH. 5-Hydroxymethylcytosine: An epigenetic mark frequently deregulated in cancer. Biochim Biophys Acta. 2015;1855:144–54. doi: 10.1016/j.bbcan.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Ponnaluri VK, Maciejewski JP, Mukherji M. A mechanistic overview of TET-mediated 5-methylcytosine oxidation. Biochem Biophys Res Commun. 2013;436:115–20. doi: 10.1016/j.bbrc.2013.05.077. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki Y, Saito H, Suzuki M, Hosoki Y, Sakurai S, Fujimoto Y, et al. Up-regulation of transferrin receptor expression in hepatocytes by habitual alcohol drinking is implicated in hepatic iron overload in alcoholic liver disease. Alcohol Clin Exp Res. 2002;26(8 Suppl):26S–31S. doi: 10.1111/j.1530-0277.2002.tb02698.x. [DOI] [PubMed] [Google Scholar]
- 10.Charlton RW, Jacobs P, Seftel H, Bothwell TH. Effect of alcohol on iron absorption. Br Med J. 1964;2:1427–9. doi: 10.1136/bmj.2.5422.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison-Findik DD, Schafer D, Klein E, Timchenko NA, Kulaksiz H, Clemens D, et al. Alcohol metabolism-mediated oxidative stress down-regulates hepcidin transcription and leads to increased duodenal iron transporter expression. J Biol Chem. 2006;281:22974–82. doi: 10.1074/jbc.M602098200. [DOI] [PubMed] [Google Scholar]
- 12.Whitfield JB, Zhu G, Heath AC, Powell LW, Martin NG. Effects of alcohol consumption on indices of iron stores and of iron stores on alcohol intake markers. Alcohol Clin Exp Res. 2001;25:1037–45. doi: 10.1111/j.1530-0277.2001.tb02314.x. [DOI] [PubMed] [Google Scholar]
- 13.Lieber CS, DeCarli LM, Sorrell MF. Experimental methods of ethanol administration. Hepatology. 1989;10:501–10. doi: 10.1002/hep.1840100417. [DOI] [PubMed] [Google Scholar]
- 14.Olynyk J, Hall P, Reed W, Williams P, Kerr R, Mackinnon M. A long-term study of the interaction between iron and alcohol in an animal model of iron overload. J Hepatol. 1995;22:671–6. doi: 10.1016/0168-8278(95)80222-3. [DOI] [PubMed] [Google Scholar]
- 15.Bertola A, Mathews S, Ki SH, Wang H, Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–37. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torrance JD, Bothwell TH. A simple technique for measuring storage iron concentrations in formalinised liver samples. S Afr J Med Sci. 1968;33:9–11. [PubMed] [Google Scholar]
- 17.Cairo G, Tacchini L, Pogliaghi G, Anzon E, Tomasi A, Bernelli-Zazzera A. Induction of ferritin synthesis by oxidative stress. Transcriptional and post-transcriptional regulation by expansion of the “free” iron pool. J Biol Chem. 1995;270:700–3. doi: 10.1074/jbc.270.2.700. [DOI] [PubMed] [Google Scholar]
- 18.Dongiovanni P, Lanti C, Gatti S, Rametta R, Recalcati S, Maggioni M, et al. High fat diet subverts hepatocellular iron uptake determining dysmetabolic iron overload. PLoS One. 2015;10:e0116855. doi: 10.1371/journal.pone.0116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem. 2002;74:4526–31. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 20.Tammen SA, Dolnikowski GG, Ausman LM, Liu Z, Kim KC, Friso S, et al. Aging alters hepatic DNA hydroxymethylation, as measured by liquid chromatography/mass spectrometry. J Cancer Prev. 2014;19:301–8. doi: 10.15430/JCP.2014.19.4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi SW, Stickel F, Baik HW, Kim YI, Seitz HK, Mason JB. Chronic alcohol consumption induces genomic but not p53-specific DNA hypomethylation in rat colon. J Nutr. 1999;129:1945–50. doi: 10.1093/jn/129.11.1945. [DOI] [PubMed] [Google Scholar]
- 24.van Engeland M, Weijenberg MP, Roemen GM, Brink M, de Bruïne AP, Goldbohm RA, et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 2003;63:3133–7. [PubMed] [Google Scholar]
- 25.Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15:395–8. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 26.Scourzic L, Mouly E, Bernard OA. TET proteins and the control of cytosine demethylation in cancer. Genome Med. 2015;7:9. doi: 10.1186/s13073-015-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 28.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–5. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Liu L, Chen X, Shen J, Shan J, Xu Y, et al. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One. 2013;8:e62828. doi: 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udali S, Guarini P, Moruzzi S, Ruzzenente A, Tammen SA, Guglielmi A, et al. Global DNA methylation and hydroxymethylation differ in hepatocellular carcinoma and cholangiocarcinoma and relate to survival rate. Hepatology. 2015;62:496–504. doi: 10.1002/hep.27823. [DOI] [PubMed] [Google Scholar]