Abstract
Aging is a universal process that causes deterioration in biological functions of an organism over its lifetime. There are many risk factors that are thought to contribute to aging rate, with disruption of metabolic homeostasis being one of the main factors that accelerates aging. Previously, we identified a new function for the putative G-protein-coupled receptor, Bride of sevenless (BOSS), in energy metabolism. Since maintaining metabolic homeostasis is a critical factor in aging, we investigated whether BOSS plays a role in the aging process. Here, we show that BOSS affects lifespan regulation. boss null mutants exhibit shortened lifespans, and their locomotor performance and gut lipase activity—two age-sensitive markers—are diminished and similar to those of aged control flies. Reactive oxygen species (ROS) production is also elevated in boss null mutants, and their ROS defense system is impaired. The accumulation of protein adducts (advanced lipoxidation end products [ALEs] and advanced glycation end products [AGEs]) caused by oxidative stress are elevated in boss mutant flies. Furthermore, boss mutant flies are sensitive to oxidative stress challenges, leading to shortened lives under oxidative stress conditions. Expression of superoxide dismutase 2 (SOD2), which is located in mitochondria and normally regulates ROS removal, was decreased in boss mutant flies. Systemic overexpression of SOD2 rescued boss mutant phenotypes. Finally, we observed that mitochondrial mass was greater in boss mutant flies. These results suggest that BOSS affects lifespan by modulating the expression of a set of genes related to oxidative stress resistance and mitochondrial homeostasis.
Introduction
Aging is a complex phenomenon with a multifactorial etiology. Several theories have been proposed to explain how endogenous and exogenous factors affect aging. Reduced dietary caloric intake is known to extend lifespan in a wide range of organisms including mammals and worms [1, 2]. Caloric restriction induces changes in metabolic response. Loss of fat mass is the most prominent metabolic change in response to restriction, and lipid homeostasis seems to be an important factor in regulating lifespan [3]. Concomitantly, caloric restriction reduces ROS generation [4]. Thus, the regulation of oxidant production and the ability of organisms to respond to oxidative stress are intricately linked to aging and lifespan [5, 6].
According to the free-radical theory of aging, ROS are predominantly byproducts of mitochondrial metabolism and are thought to be a risk factor for aging [7]. Thus, the regulation of mitochondrial mass and activity is very important. How, though, are mitochondrial mass and activity regulated? Although mitochondria possess their own genome (mtDNA), the majority of mitochondrial proteins are encoded by the nuclear genome, translated in the cytoplasm, and imported into mitochondria [8]. Therefore, mitochondrial mass and activity must be coordinated with nutrient availability. Indeed, starvation or exercise increases mitochondrial mass [9].
Here, we describe the functions of BOSS in the aging process. BOSS was originally identified as a ligand of the receptor SEVENLESS and required for the development of the R7 photoreceptor neuron in the compound eye of Drosophila melanogaster [10]. Then we previously discovered that BOSS possesses a critical metabolic function: maintaining energy homeostasis [11, 12]. Insulin signaling is disrupted in boss mutant flies. Furthermore, boss mutant flies are lean but hyperphagia. Because changes in metabolism are tightly linked with aging, we speculated that BOSS might play a role in aging. Thus, we examined the lifespan of boss mutant flies and various aging phenotypes of boss mutant flies. We discovered that boss mutants have a short lifespan. Interestingly, we found that aging processes were accelerated in boss mutant flies. In young boss mutant flies, locomotor activity and gut lipase activity were reduced. Furthermore, oxidative stress (ALEs and AGEs) and oxidative damage were enhanced in boss mutants. However, overexpression of SOD2 rescued boss mutant flies from oxidative damage and from exhibiting an aged phenotype. Our findings suggest that BOSS may play a central role in lifespan control through its involvement in mitochondrial function.
Results
boss mutant flies have shortened lifespan and mobility impairment
To determine the role of BOSS in aging, we first examined the lifespan of boss mutant flies (Fig 1A). Both female and male boss mutant flies had shorter lifespans compared to control flies, indicating that aging in boss mutants might be accelerated.
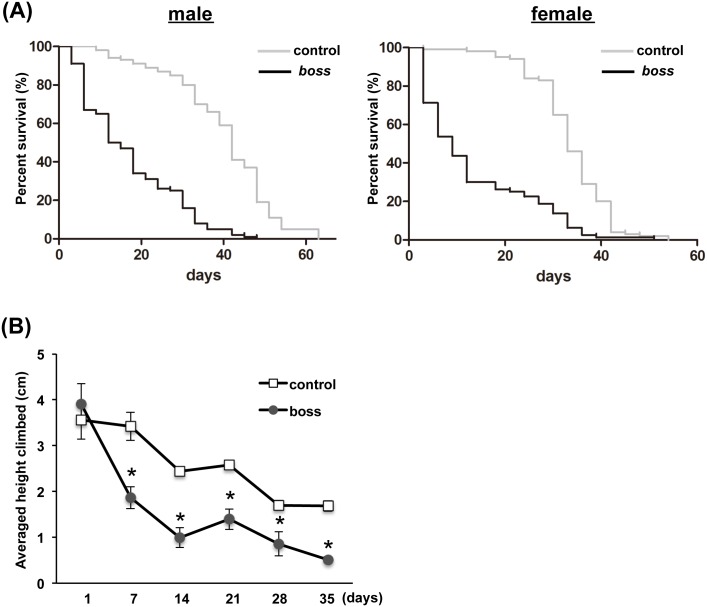
Fig 1. Aging is accelerated in boss mutant flies.
(A) Lifespan for male and female boss mutant flies. boss mutant flies have shorter lifespan than control flies. Survival was presented by Kaplan Meier curves of control and boss mutant flies. n = 100 for each genotypes. Median survival for control male = 40days, boss mutant males = 13.5 days, control female = 33days, boss mutant females = 9days (Log-rank test, p<0.0001). (B) Analysis of vertical distance climbed in the rapid iterative negative geotaxis assay of boss mutant and control (w) male flies (n = 3 groups; each group contains 10 flies per genotype per time point). Data are means ± SEM, and differences are significant by t-test (*P<0.05).
To test the accelerated aging hypothesis, we used a behavioral task sensitive to age. Negative geotaxis is a natural escape response of flies to climb vertically, and it is an ability that declines with age [13, 14]. We compared age-related locomotor performance of mutant and control flies using the rapid iterative negative geotaxis assay [15], which objectively measures climbing performance by measuring negative geotaxis. We assessed performance of boss mutant and control flies of different ages: 1, 7, 14, 21, 28, and 35 days old. Young 1-day-old boss mutant flies displayed the same climbing performance as control flies, confirming normal locomotor development. However, performance declined more rapidly in boss mutant flies with age compared to control flies, with differences evident as young as 7 days old (Fig 1B). Additionally, the performance decline in control flies paralleled the survival rate, while in boss mutant flies the initial decline significantly exceeded the observed decline in survival rate. This hyperbolic decline characteristic in boss mutant flies can be interpreted as an increased population sickness already at a young age. Together, these results suggest that BOSS affects the lifespan of flies.
Young boss mutant flies exhibit age-related decline of gut function
Next, we assessed gut lipase activity in boss mutant flies. Recently, it was reported that gut lipase activity decreases during aging [16, 17]. Gastric lipases digest lipids in food, which are then absorbed in the gut. Therefore, a reduction in gastric lipase activity decreases gut lipid storage. We found that gut lipid storage is reduced in young (10-day-old) boss mutant flies, similar to that in aged control flies (Fig 2A).
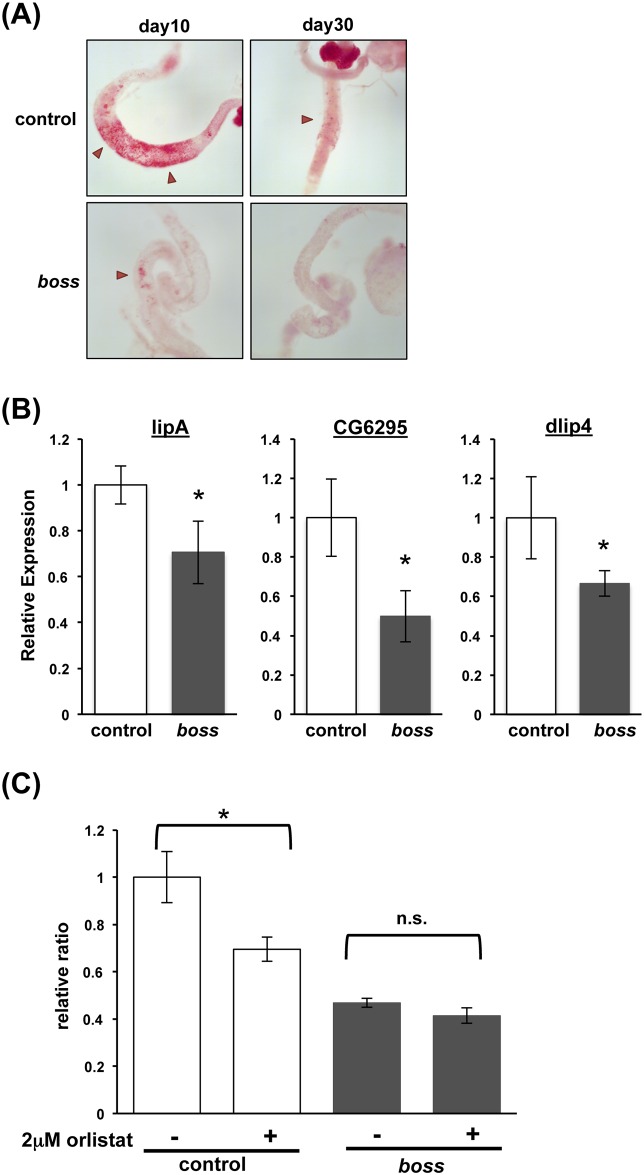
Fig 2. Gut lipase activity declines with aging in boss mutant flies.
(A) Oil-Red-O staining of neutral lipids in the guts of young (10-days) and old (30-days) control and boss mutant female flies. Anterior midgut region stores lipid (arrowhead). (B) Expression of gut lipases (lipA/margo, CG6295, and dlip4) in the intestines of young (day 10) flies, as assessed by qRT-PCR (n = 3, 5 guts per replicate). (C) boss mutant flies are resistant to orlistat treatment. Young adult male boss mutant flies and control flies were fed a diet with or without orlistat (2.0 μM) for 5 days. Afterward TAG levels were determined (n = 5, 10 flies per replicate) and normalized for total protein. Levels are presented as normalized to levels for wild-type flies. Data are mean relative ratios ± SEM, and differences are significant by t-test (*P<0.05).
The expression of gastric lipases was also decreased in boss mutant flies. This is consistent with the decreased Oil-red-O staining in mutant flies (Fig 2A). Transcription of gastric lipases (lipA/margo, CG6295, and dlip4) was also significantly downregulated in boss mutant flies, as confirmed by qRT-PCR (Fig 2B).
To confirm and extend these results, we inhibited gastric lipases with orlistat (tetrahydrolipstatin). Orlistat is widely used as an over-the-counter weight-loss drug [18, 19]. It acts inside the intestine as a competitive inhibitor of pancreatic and gastric lipases, preventing their interaction with dietary triacylglycerides (TAG) and thus blocking fatty acid release and dietary lipid uptake. In control flies exposed to orlistat, TAG levels were significantly reduced (Fig 2C). Orlistat, however, failed to affect TAG levels in boss mutant flies, indicating that endogenous gastric lipase activity is very low. These data show that gastric lipase activity is decreased in boss mutant flies due to downregulation of gastric lipase expression.
Age-related chronic FOXO activation in enterocytes leads to sustained repression of gut lipase expression and disruption of lipid homeostasis [17]. We measured the expression of FOXO target genes (Drosophila insulin receptor [dInR] and Thor/4E-BP) [20], and found that dInR mRNA expression was significantly increased while Thor mRNA expression exhibited a slight, but statistically non-significant increase. This suggests that FOXO activation in the gut of boss mutant flies is enhanced (S1 Fig). Taken together, these data indicate that tissue homeostasis is disrupted in the gut of boss mutant flies similar to that in aged control flies.
Oxidative stress is enhanced in boss mutant flies
Oxidative damage is a major risk factor for age-related diseases and aging. ROS, such as hydrogen peroxide (H2O2) and superoxide anions (O2-), are highly reactive molecules produced by incomplete reduction of oxygen, and their production causes oxidative damage [21]. Oxidative damage contributes to the onset of aging and to age-related diseases such as diabetes, obesity, and neurodegenerative disorders [21]. This prompted us to ask the question: Does oxidative damage accumulate more in boss mutant flies?
Since elevated ROS levels promote the oxidation of lipids and glucose, resulting in the accelerated formation of ALEs and AGEs [22, 23], we examined ALEs and AGEs levels to know oxidative levels in control and boss mutant flies. First, we measured the level of 4-hydroxy-2-nonenal (4-HNE), a major type of ALE. Under oxidative-stress conditions, lipid peroxidation of ω6-polyunsaturated fatty acids leads to the production of 4-HNE [24]. We found that 4-HNE levels were higher in boss mutant flies than in control flies at late stage (Fig 3A).
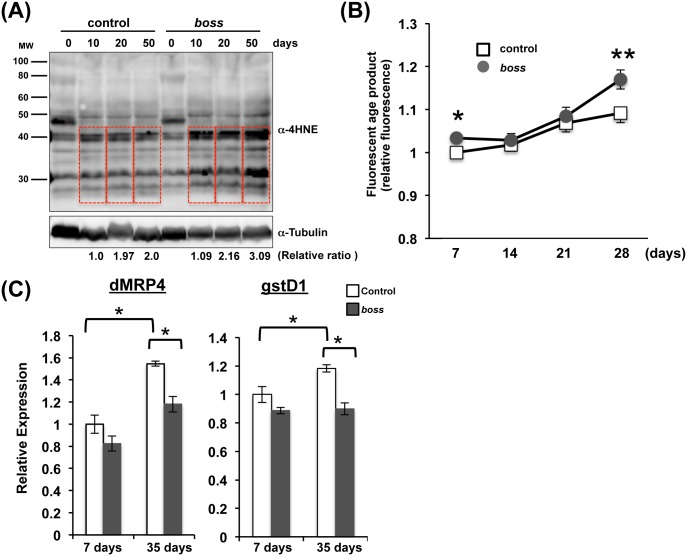
Fig 3. Oxidative stress is enhanced in boss mutant flies.
(A) ALEs were detected by Western blot and antibodies against 4-hydroxy-2-nonenal (4HNE). Ages (0~50-days old) are shown at the top of the blot. (B) Accumulation of fluorescent AGEs was measured in control and boss mutant flies (n = 3, 20 flies per replicate). Data are mean relative fluorescence ± SEM, and differences are significant by t-test (*P<0.05, **P<0.01). (C) Expression of ROS-inducible genes (dMRP4 and gstD1) in young (7-days) and old (35- days) flies, assessed by qRT-PCR (n = 3, 10 flies per replicate). Data are mean relative expression ± SEM, and differences are significant by t-test (*P<0.05).
Next, we examined AGE levels during aging. In Drosophila, AGEs are also used as biomarkers of age-related damage [25]. AGEs fluoresce in a nonenzymatic reaction between reducing sugars and amine residues on proteins, lipoproteins, and nucleic acids. AGEs also accumulate during aging and correlate strongly with mortality rate in flies. We quantified fluorescent AGE levels in sets of young and old male flies during aging (7- to 28-days old). Fluorescent AGE levels were higher in boss mutant flies than in control flies at late stage (Fig 3B), indicating that oxidative stress was enhanced in boss mutant flies.
The elevated fluorescent AGE levels in boss mutant flies prompted us to examine the expression of marker genes (dMRP4 and gstD1) that mediate oxidative stress resistance (Fig 3C) [26, 27]. These genes are induced in adult flies during normal aging and when flies are subjected to oxidative stress, for example, by exposure to hyperoxic conditions or to paraquat. Both dMRP4 and gstD1 mRNAs were elevated in old control flies (35-days old) compared to young control flies (7-days old). However, dMRP4 and gstD1 mRNA expression was not elevated in old boss mutant flies, indicating that boss mutant flies are more sensitive to oxidative stress.
boss mutant flies are sensitive to oxidative stress
Since oxidative stress is enhanced in boss mutant flies and expression of genes involved in resistance to it is reduced, we speculated that boss mutant flies might be more susceptible to conditions that increase oxidative stress. If this were the case, oxidative damage would accumulate faster as these flies age. To examine this hypothesis, we first measured boss mutant flies’ survival curve when subjected to oxidative stress.
boss mutant and control flies were given food containing either paraquat (2 mM) or H2O2 (0.5%). Under these conditions, boss mutant flies died more quickly than control flies (Fig 4A), providing more evidence that boss mutant flies are more sensitive to oxidative stress.
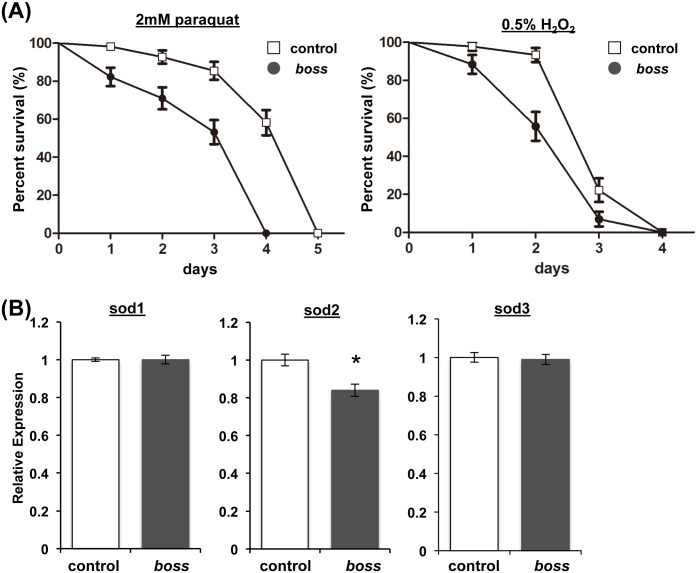
Fig 4. boss mutant flies are sensitive to oxidative stress.
(A) Young adult male boss mutant and control flies were fed a diet containing either 2.0 mM paraquat or 0.5% H2O2, and survival rate of the groups was measured (n = 30 flies for each genotypes). Log-rank test, p<0.0001. Error bars represent S.E. (B) Expression of superoxide dismutases (SOD1-3) in young (10-days) flies as measured by qRT-PCR. Data are means ± SEM (t-test, *P<0.05).
One reason to explain this increased susceptibility to oxidative stress is that levels of ROS in the mutant flies may be dysregulated. Cellular levels of ROS are controlled by antioxidant enzymes. Superoxide dismutase (SOD) is one of several major enzymes that regulate the removal of ROS. Similar to other metazoans, Drosophila has three SOD isoforms (SOD1-3) [28, 29]. SOD1 localizes to the cytoplasm, SOD2 to the in the mitochondrial matrix, and SOD3 to the extracellular space. Loss of either SOD1 or SOD2 causes early lethality in flies and mice [30, 31], whereas ubiquitous overexpression of either SOD1 or SOD2 extends lifespan [32–36]. This is in line with our findings that boss mutant flies have increased oxidative stress levels and increased sensitivity to oxidative stress.
We next measured the expression levels of SODs in boss mutant flies to test this idea. Quantitative RT-PCR revealed that expression of sod2 mRNA was decreased in boss mutant flies, while expression of sod1 and sod3 mRNA was comparable to that in control flies (Fig 4B).
The mitochondrion is the major organelle that produces ROS, and SOD2 is required to prevent increased ROS levels. Consistent with our findings and hypothesis is that sod2 mutant flies have a shortened lifespan [37]. We speculated that decreased SOD2 level in boss mutant flies is one cause of their accelerated aging.
SOD2 expression rescues sensitivity to oxidative stress of boss mutant flies
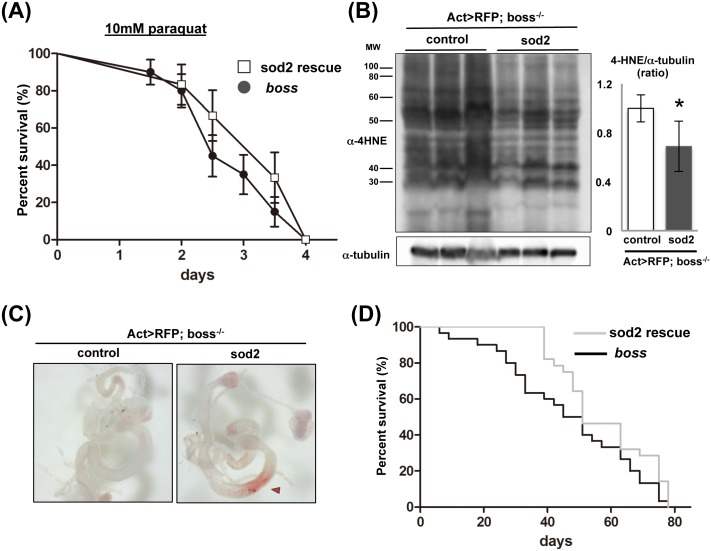
To assess the role of SOD2 in boss mutant flies, we generated SOD2-overexpressing boss mutant flies by crossing Act-Gal4 flies with UAS SOD2 flies. This resulted in Act-GAL4/UAS-SOD2 flies with a boss mutant background (S2 Fig). First, we examined whether these flies are sensitive to oxidative stress by exposing them to paraquat (10 mM) in their feed, as before (Fig 5A). Under this condition, SOD2-overexpressing boss mutant flies lived longer than boss mutant flies, supporting our hypothesis that decreased SOD2 levels in boss mutant flies is one cause of their accelerated aging under oxidative stress-producing conditions.
Fig 5. SOD2 overexpression rescues aging phenotype of boss mutant flies.
(A) Survival curves of SOD2-overexpressing boss mutant flies (Act-Gal4, UAS-RFP/ UAS-SOD2; boss-/-) and boss mutant flies (Act-Gal4, UAS-RFP/ +; boss-/-) treated with 10 mM paraquat. SOD2 overexpression prolonged the survival of boss mutant flies, shifting the survival curve to the right of the population of SOD2-overexpressing boss mutant flies (n = 30 for each genotypes). Error bars represent S.E. (B) ALEs were detected by Western blot and anti-4HNE antibody. SOD2 overexpression reduced the staining intensity for ALEs (n = 3, 10 flies per replicate). (C) Oil-Red-O staining for neutral lipids in the guts of young (10-days) SOD2-overexpressing control and boss mutant female flies. Anterior midgut region stores lipids (arrowhead). (D) Lifespan for male SOD2-overexpressing boss mutant and boss mutant flies. SOD2 overexpression prolonged the survival of boss mutant flies. Survival was presented by Kaplan Meier curves of SOD2-overexpressing boss mutant and boss mutant flies. n = 50 for each genotypes. Median survival for SOD2-overexpressing boss mutant = 51 days, boss mutant males = 48 days (Log-rank test, p = 0.044).
Next, we assessed oxidative stress levels in SOD2-overexpressing boss mutant flies by measuring 4-HNE levels (Fig 5B). SOD2 overexpression reduced 4-HNE levels. These results demonstrate that sensitivity to oxidative stress was rescued by SOD2 overexpression in boss mutant flies.
We also examined gut lipid storage in SOD2-overexpressing boss mutant flies (Fig 5C). The decreased Oil-red-O staining we observed earlier in young boss mutant flies (cf. Fig 2A) was recovered in SOD2-overexpressing boss mutant flies. This suggests that SOD2 expression can rescue gastric lipase activity of boss mutant flies. This is also in line with the observation that SOD2 overexpression increases the lifespan of boss mutant flies under normal conditions (Fig 5D). From these SOD2 rescue experiments, we conclude that decreased SOD2 expression in boss mutant flies contributes to their shortened lifespan.
Regulation of mitochondrial activity is disrupted in boss mutant flies
We previously reported that energy state is reduced in boss mutant flies [11, 12]. Reducing cellular energy levels by starvation or exercise increases mitochondrial activity, and hence production of the energy source, ATP[9,38]. This led us to speculate that mitochondrial mass increases in boss mutant flies in order to meet energy demand.
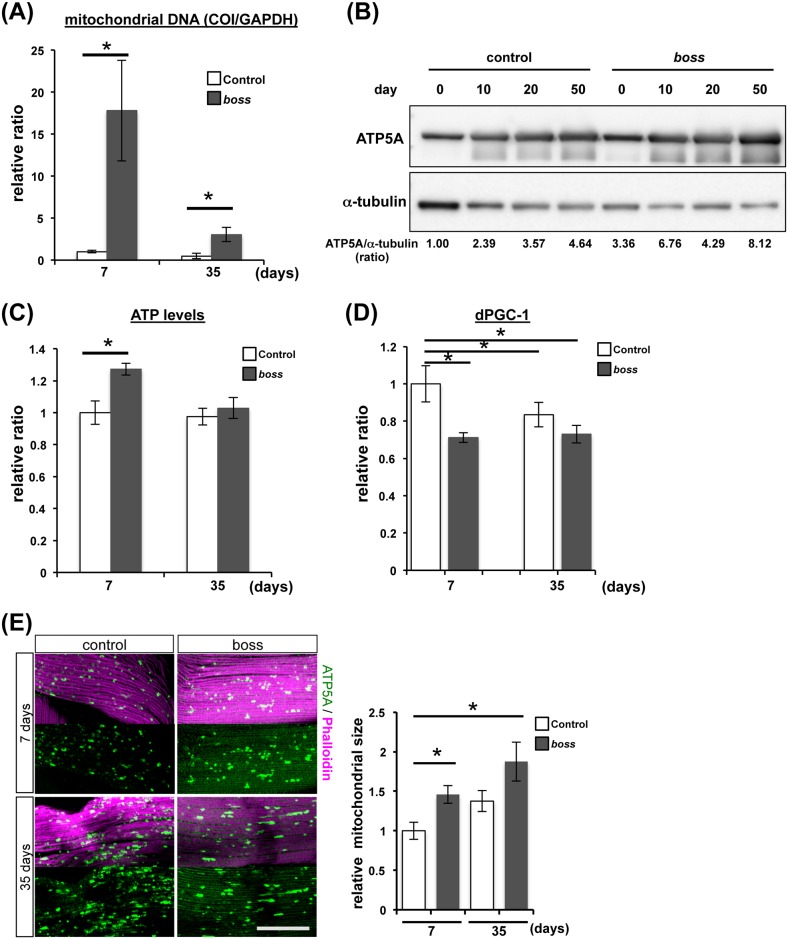
To determine mitochondrial mass as a proxy measure, we measured the amount of mitochondrial DNA (mtDNA) relative to the amount of nuclear DNA (nDNA) in young (7-days old) and old (35-days old) flies. The amount of mtDNA increased in boss mutant flies (Fig 6A). We also examined the expression of ATP5A, a mitochondrial marker, in order to monitor the amount of mitochondria in flies. ATP5A levels were higher in boss mutant flies compared to control flies (Fig 6B), which is consistent with our results on mtDNA quantification (c.f. Fig 6A).
Fig 6. Mitochondrial mass is increased in boss mutant flies.
(A) Quantification of mitochondrial DNA (mtDNA) in control and boss mutant flies as determined by qPCR (n = 3, 5 flies per replicate). mtDNA of both young (7-days) and old (35-days) boss mutant flies was greater than in control flies. Data are means ± SEM (t-test, *P<0.05). (B) The amount of mitochondria was determined using Western blotting and anti-ATP5A antibody. ATP5 levels were increased in boss mutant flies, suggesting that the total mass of mitochondria increased. Values at bottom of columns are quantitation of ATP5A densitometry analysis (ATP5A/tubulin ratio). (C) ATP production levels were measured in young (7-days) and old (35-days) flies (n = 3, 5 flies per replicate). Data are means ± SEM (t-test, *P<0.05). (D) Expression of pgc1 mRNA was measured in young (7-days) and old (35 days) flies by qRT-PCR (n = 3, 10 flies per replicate). pgc-1 mRNA levels were significantly decreased in young boss mutant flies, indicating indicates mitochondrial biogenesis is reduced. Data are means ± SEM (t-test, *P<0.05). (E) Representative confocal images showing indirect flight muscles stained with phalloidin (magenta) and anti-ATP5 antibody (green). One representative optical section is shown from each phenotype. Scale bar, 50μm. Comparison of the mitochondrial size of both young (7-days) and old (35-days) of control and boss mutant flies are shown to the right. Data are means ± SEM (t-test, *P<0.05).
Next, we measured ATP levels in control and boss mutant flies, since mitochondria is at the core of cellular energy metabolism, being the site of most ATP generation but also responsible for generation of substantial amounts of superoxide caused by electron leakage from the oxidative phosphorylation pathway [7, 38]. ATP concentrations were increased in young boss mutant flies, indicating that mitochondrial activity is upregulated in boss mutant flies; ATP concentrations were the same in old control and boss mutant flies (Fig 6C). The increased mitochondrial mass without increased ATP production indicated that unfunctional mitochondria are accumulated in old boss mutant flies.
Mitochondria are energy-converting organelles that produce most of the ATP required for cellular functions and integrity, and they produce ROS as byproducts of electron transport during the generation of ATP. ROS cause mtDNA mutations to accumulate, which may be responsible for a decline in mitochondrial energy production. Thus, cells have developed mechanisms—or mitochondria “quality control system”—to eliminate damaged mitochondria [21].
The increased mitochondrial mass in boss mutant flies may be due to dysregulation of this mitochondrial quality control system. To test this possibility, we examined the expression of a key molecule in this system called peroxisome proliferator-activated receptor gamma co-activator 1 (PGC-1), a transcription coactivator. PGC-1 regulates gene expression for mitochondrial biogenesis and maintains the structural integrity of mitochondria [39, 40]. PGC-1 mRNA levels in control and boss mutant flies at young and old time points were compared (Fig 6D). We compared PGC-1 mRNA levels in young and old control and boss mutant flies, and found that PGC-1 mRNA levels were significantly decreased in young boss mutant flies. This indicates that mitochondrial biogenesis is reduced (Fig 6D). Mitochondrial mass was increased throughout the lifespan of boss mutant flies. However, increased ATP levels were present only in young flies, strongly indicating that dysfunctional mitochondria accumulate as boss mutant flies age. In Drosophila, size of mitochondria is increased during aging[41]. This is consistent with our finding that mitochondria in the flight muscles of young (7-days old) and old (35-days old) boss mutant flies were much larger than those in comparably aged control flies (Fig 6E). Taken together, the age-related increase in mtDNA and mitochondrial size with a concomitant decrease in ATP levels might point towards reduced mtDNA quality in aging boss mutants.
Hsp22 expression is not increased in boss mutant flies during aging and oxidative stress stimulation
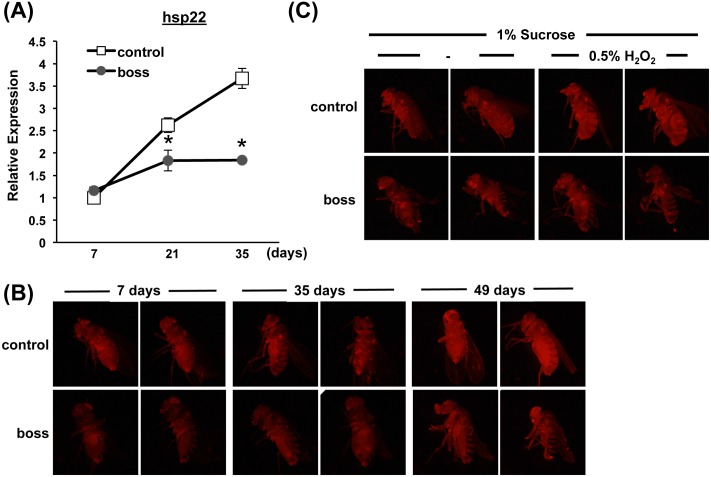
Finally, we examined hsp22 mRNA expression levels. Hsp22 belongs to the sHSP family, localizes in the mitochondrial matrix, and is involved in aging processes, and imparts resistance to oxidative stress [42, 43]. HSP22 expression is upregulated during aging [44], in a long-lived fly line [45], and in response to oxidative stress [27]. Corroborating a previous report [44], hsp22 expression progressively increased with aging in control flies (Fig 7A). However, while hsp22 expression was indistinguishable in young (7-days old) control and boss mutant flies, expression diverged with aging between the two groups. In boss mutant flies aged 21and 35 days, hsp22 expression was significantly lower in the older flies.
Fig 7. hsp22 expression is not induced in boss mutant flies.
(A) Expression of hsp22 mRNA was measured in 7-, 21-and 35-days-old control and boss mutant flies by using qRT-PCR (n = 3, 10 flies per replicate). Data are means ± SEM (t-test, *P<0.05). (B) DsRed fluorescence was observed in control and boss mutant flies bearing the Hsp22-DsRed reporter. DsRed fluorescence progressively increased during aging in control but not in boss mutant flies. (C) Flies bearing the Hsp22-DsRed reporter were transferred to a diet containing 0.5% H2O2 for 24 h, and then DsRed fluorescence was observed. Under this oxidative stressful condition, boss mutant flies still failed to exhibit increased hsp22 expression, suggesting dysfunctional mitochondria and oxidative stress dysregulation.
We also examined hsp22 levels a second way, using hsp22 reporter (hsp22-dsRED) [44]. The promoter sequence of hsp22 gene is fused to DsRED; thus, DsRED expression level reflects hsp22 expression level. hsp22-DsRED expression increased with age in wild-type flies but not in boss mutant flies (Fig 7B). Oxidative stress stimulation with H2O2 also clearly increased Hsp22-dsRED expression in wild-type flies but slightly in boss mutant flies (Fig 7C), supporting the notion that boss mutant flies apparently cannot respond adequately to oxidative stress, and thus fail to maintain mitochondrial integrity.
Discussion
Aging is associated with a decline of function at the organismal level. It begins with cellular deterioration, which ultimately leads to the disruption of tissue homeostasis. The present study indicates that BOSS influences on aging. We showed that boss mutant flies have a shortened lifespan (Fig 1), which is caused by deterioration of gut homeostasis and by elevation of oxidative stress. Even in young boss mutant flies, gastric lipase expression and activity are already reduced (Fig 2). Enhanced oxidative stress was also observed in boss mutant flies (Fig 3), and this was caused by an impaired ROS defense system (Figs 4–6). Our genetic analysis of SOD2 expression rescue showed that reducing oxidative stress could rescue all aged phenotypes of the boss mutant (Fig 5).
We previously reported that energy metabolism is disrupted in boss mutant flies, causing lean phenotype [11, 12]. The gut is one of the key tissues for aging regulation [17]. Hyper-proliferation of intestinal stem cells (ISCs) occurs in the aged gut, resulting in loss of tissue homeostasis, elevated oxidative stress, and chronic JNK activation [16, 46, 47]. In boss mutant flies, we found that oxidative stress is increased (Fig 3) and gut homeostasis is impaired (Fig 2). Moreover, the expression and activity of gastric lipases were decreased in boss mutant flies, producing a similar phenotype to that observed in aged control flies. Previously, we demonstrated that BOSS is expressed in enteroendocrine cells, and knockdown of boss in these cells lead to the lean phenotype [12]. Thus, it would be interesting to know whether BOSS expressed in enteroendocrine cells and/or in other tissues (such as neurons and the fat body) affect gut homeostasis.
A number of recent studies indicate that regulation of ROS levels seems to be very important for aging. ROS is concomitantly generated during ATP production and causes oxidative damage and cellular/tissue dysfunction if it accumulates [4]. Excessive ROS levels have a harmful effect on cellular physiology [31, 37, 48], whereas physiological levels of ROS are likely essential for the maintenance of cellular homeostasis [49]. This is called mitohormesis or adaptive cytoprotective responses to low levels of oxidative stress in the mitochondria [50, 51]. Thus, disrupted mitohormesis affects disease onset, progression, and aging.
Mitochondria are energy-converting organelles that produce most of the ATP required for cellular functions and integrity, and they produce ROS as byproducts of electron transport during the generation of ATP. ROS cause mtDNA mutations to accumulate, which may be responsible for a decline in mitochondrial energy production. Thus, cells have developed mechanisms—or mitochondria “quality control system”—to eliminate damaged mitochondria [21]. In the present study, ATP production was increased in young boss mutant flies, and mitochondrial mass was increased throughout their life (Fig 6). This indicates that dysregulation of mitochondrial homeostasis lead to loss of tissue homeostasis. Indeed, we observed that the expression of hsp22—which localizes in mitochondria and activates the ROS defense system (sod2, dMRP4, and gstD1)—was decreased in boss mutants (Fig 7). Aged phenotypes were also rescued by sod2 overexpression (Fig 5). Finally, we found that PGC-1 expression was decreased in boss mutant flies (Fig 6D).
Since PGC-1 and Hsp22 increase mitochondrial biogenesis and extend lifespan in Drosophila [43, 47], we speculate that the acceleration of aging seen in the boss mutant flies is likely caused by disruption of mitochondrial homeostasis. We previously found that boss mutants are hyperphagic but paradoxically are lean, indicating that nutrient absorption might be disrupted [12]. If this were the case, more mitochondria would be required to generate more ATP. Thus, we speculate that the number of mitochondria and their activity might be increased in boss mutant flies. However, concomitant generation of ROS might lead to cellular damage and loss of metabolic homeostasis, ultimately leading to accelerated aging in boss mutant flies.
Our data strongly indicate that increased ROS levels influence the aging process, and that normal mitochondrial function and structure are also damaged in the boss mutant. Very recently, it was proposed that mitochondria affect the aging process not only by producing ROS but also via other mitochondrial signaling pathways. Thus, changes in mitochondrial dynamics and the electron transport chain could have an impact on aging [7, 52]. It would be interesting and important, then, in the future to examine the molecular mechanism of how BOSS localized in cell membranes regulates mitochondrial function and structure.
BOSS is an evolutionarily conserved protein [53]. The expression of GPRC5B, a mammalian BOSS homologue, is similar to that of BOSS [54–56]. GPRC5B is broadly expressed in brain, gut, and adipose tissues. Recently, aberrant expression of GPRC5B was identified to be an obesity risk factor [57]. Considering that the regulatory mechanisms of energy metabolism and mitochondrial quality control are highly conserved between Drosophila and mammals [58, 59], specific analysis of Drosophila BOSS will open the door for understanding the broader physiological functions of the highly conserved membrane protein BOSS/GPRC5B.
Materials and Methods
Fly stocks and genetics
The following fly strains were used: w (WT, control); boss1 (null mutant, homozygous viable), Act-Gal4>UAS-RFP (Kyoto Stock Center, DGGR#4414), UAS-Sod2 (Bloomington Drosophila Stock Center, #24494); hsp22-RED (a kind gift from Dr. John Tower).
Husbandry
Flies were grown on standard food (10% glucose/4% yeast/4% corn meal/1% agar) at a temperature of 25°C under a 12h-12h light-dark regime.
Lifespan analysis
Female or male flies were collected at day 0 and maintained at a density of 10 or 20 flies per vial. Flies were transferred to new vials every 3 days, and dead ones were noted in order to calculate survival curves.
Rapid iterative negative geotaxis assay
Fly climbing speed was assessed using the RING assay, as described previously [15]. 10 flies (n = 3 groups) were transfer into each vial without anesthetizing. The vials were tapped and all flies were knocked down to the bottom of each vial. After 3 seconds a picture was taken to record the climb height. The procedure was repeated 5 times at 1min intervals and the average height climbed of each vial was scored.
Quantitative real-time PCR (qRT-PCR)
Guts of female flies, aged 5 days, were used for RNA samples to measure gastric lipase expression (Fig 2B). To measure SOD1~3 expression (Fig 4B and S2 Fig) were collected from whole flies. Samples were immediately frozen in liquid nitrogen for later analysis. Total RNA was extracted using Trizol Reagent (Invitrogen). Total RNA samples (1 μg per reaction) were reverse transcribed using oligo-dT and random primers and Superscript RT-III (Invitrogen). The generated cDNA was used for real-time RT-PCR (qPCR Mastermix Plus for SYBERGreen; Applied Biosystems). Rp49 gene was used as reference gene. Two-step RT-PCR was performed with the following condition: denature temperature was 95°C and anneal and extend temperature was 60°C. Three separate samples were collected from each condition, and triplicate measurements were made. Primers (S1 Table) were designed using the Universal Probe Library Assay Design Center (Roche Applied Science).
Western blot analysis
Ten male flies were used for each condition. The antibodies used in Western blot analysis were as follows: anti-4HNE (1:1000, JalCA); anti-ATP5A (1:100,000, Abcam); and anti-tubulin (1:10,000, Sigma). HRP-conjugated anti-rabbit or anti-mouse IgG antibodies (Cell Signaling Technology) were used as secondary antibodies. Blots were visualized using Chemi-Lumi One Super reagent (Nacali Tesque Inc.). To quantify 4-HNE and ATP5A levels, staining intensity of each lane was measured using ImageJ (http://imagej.nih.gov/ij/). Measured area is indicated in the box.
Oil-Red-O staining
Guts from female adult flies, aged 10 and 30 days, were fixed in 4% paraformaldehyde for 25 min at room temperature, washed with distilled water, and incubated in 100% propylene glycol for 5 min. Specimens were then incubated at 60°C in Oil-Red-O stain (0.5% Oil Red O in propylene glycol), washed twice with propylene glycol, washed three times with PBS, and mounted onto glass slides in 20% glycerol/PBS for imaging.
TAG assay
Orlistat treatment procedures involved growing flies on standard food with or without 2 μM orlistat for 5 days. TAG levels were then assayed. For the TAG assay, male flies aged 3–5 days were homogenized in PBS. Supernatant was collected after heat inactivation at 70°C for 5 min, and centrifugation at 13000 rpm for 5 min. TAG was measured using a triglyceride assay kit (Sigma). Data were normalized to total protein.
Oxidative stress assay
To induce oxidative stress, we fed male flies aged 5 days with standard food with 0.5% H2O2 or 2 mM paraquat (Fig 4A) or 10mM paraquat (Fig 5A). The number of living flies were noted every day and the survival rate was calculated.
Fluorescent advanced glycation end products (AGEs)
Fluorescent AGEs were measured as previously described [22]. Twenty male flies were homogenized in 900 μl of 10 mM EDTA-Na2.H2O in PBS. Homogenate was transferred to a microcentrifuge tube containing 100 μl of a solution containing 10 mg trypsin/10 mM EDTA-Na2.H2O in PBS. Following incubation for 24 h at 37°C, the digested homogenate was centrifuged at 11,000g for 5 min. The supernatant was spin-filtered through a 0.22 μm cellulose membrane (Millipore). Aliquots of the filtrate (200 μl) were transferred to 96-well plates, and fluorescence was measured at excitation and emission wavelengths of 365 nm and 440 nm, respectively. Fluorescence values for analysis were taken as the mean fluorescence from triplicate wells.
Mitochondrial DNA measurement
Mitochondrial DNA content was determined by the ratio of the gene for cytochrome oxidase subunit I [57] to a nuclear gene GAPDH (S1 Table). Total DNA from the whole flies (n = 3, 20 flies per replicate) were prepared by homogenization in 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.1% Triton X-100 and 10μg/ml protease K. Following 60 min incubation at 37°C, Protease K was heat inactivated at 95°C for 5 min. Mitochondrial DNA was quantified relative to nuclear DNA by the ratio of cytochrome oxidase subunit I [57] to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in quantitative real-time PCRs. DNA copy numbers were measured by using real-time PCR as described above.
ATP quantification
ATP levels were detected according to the procedures detailed in Tennessen et al. [60]. Five 10-day-old adult flies were used for this assay in the present experiment. Cellular ATP content was quantified with a luciferin/luciferase-based assay using an ATP Determination Kit (Sigma-Aldrich), and the data were normalized to the protein content.
Immunohistochemistry
7-day-old and 35-day old adult flies were used for the flight muscles staining with anti-ATP5A (1:10,000, Abcam). To measure mitochondrial dimensions, the particle analysis function of ImageJ was used.
Supporting Information
Expression of thor and dInR mRNAs was measured in young (7-days old) flies by qRT-PCR (n = 3, 10 flies per replicate).
(TIF)
Expression of sod2 mRNA was measured in young (7-days old) flies by qRT-PCR (n = 3, 10 flies per replicate). Data are means ± SEM (*P<0.05).
(TIF)
(TIF)
Acknowledgments
We would like to thank the Bloomington Drosophila Stock Center (Indiana University), the Vienna RNAi Center (VDRC), the Developmental Studies Hybridoma Bank (University of Iowa), and FLYBASE for providing fly lines, antibodies, and information useful for these studies. This work was supported in part by grants from the RIKEN Brain Science Institute (to Y. H.) and by the 2013 Danone Institute of Japan Foundation Research Grant (to A. K.). We thank T. Shimizu for technical assistance. We are grateful to the Support Unit for Bio-material Analysis, RIKEN Brain Science Institute Research Resources Center, for help with the nucleotide sequencing analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the RIKEN Brain Science Institute to YH, and from the 2013 Danone Institute of Japan Foundation Research Grant to AK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Clancy DJ, Gems D, Hafen E, Leevers SJ, Partridge L. Dietary restriction in long-lived dwarf flies. Science. 2002;296(5566):319 10.1126/science.1069366 [DOI] [PubMed] [Google Scholar]
- 2.Tissenbaum HA, Guarente L. Model organisms as a guide to mammalian aging. Dev Cell. 2002;2(1):9–19. [DOI] [PubMed] [Google Scholar]
- 3.Hansen M, Flatt T, Aguilaniu H. Reproduction, fat metabolism, and life span: what is the connection? Cell metabolism. 2013;17(1):10–9. 10.1016/j.cmet.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120(4):483–95. 10.1016/j.cell.2005.02.001 [DOI] [PubMed] [Google Scholar]
- 5.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–47. 10.1038/35041687 [DOI] [PubMed] [Google Scholar]
- 6.Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15(2):247–54. [DOI] [PubMed] [Google Scholar]
- 7.Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest. 2013;123(3):951–7. 10.1172/JCI64125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88(2):611–38. 10.1152/physrev.00025.2007 [DOI] [PubMed] [Google Scholar]
- 9.Guarente L. Mitochondria—a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132(2):171–6. 10.1016/j.cell.2008.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinke R, Zipursky SL. Cell-cell interaction in the Drosophila retina: the bride of sevenless gene is required in photoreceptor cell R8 for R7 cell development. Cell. 1988;55(2):321–30. [DOI] [PubMed] [Google Scholar]
- 11.Kohyama-Koganeya A, Kim YJ, Miura M, Hirabayashi Y. A Drosophila orphan G protein-coupled receptor BOSS functions as a glucose-responding receptor: loss of boss causes abnormal energy metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(40):15328–33. 10.1073/pnas.0807833105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohyama-Koganeya A, Kurosawa M, Hirabayashi Y. Differential Effects of Tissue-Specific Deletion of BOSS on Feeding Behaviors and Energy Metabolism. PloS one. 2015;10(7):e0133083 10.1371/journal.pone.0133083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gargano JW, Martin I, Bhandari P, Grotewiel MS. Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Experimental gerontology. 2005;40(5):386–95. 10.1016/j.exger.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 14.Rhodenizer D, Martin I, Bhandari P, Pletcher SD, Grotewiel M. Genetic and environmental factors impact age-related impairment of negative geotaxis in Drosophila by altering age-dependent climbing speed. Experimental gerontology. 2008;43(8):739–48. 10.1016/j.exger.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nichols CD, Becnel J, Pandey UB. Methods to assay Drosophila behavior. Journal of visualized experiments: JoVE. 2012;(61). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010;6(10):e1001159 10.1371/journal.pgen.1001159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karpac J, Biteau B, Jasper H. Misregulation of an adaptive metabolic response contributes to the age-related disruption of lipid homeostasis in Drosophila. Cell Rep. 2013;4(6):1250–61. 10.1016/j.celrep.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heck AM, Yanovski JA, Calis KA. Orlistat, a new lipase inhibitor for the management of obesity. Pharmacotherapy. 2000;20(3):270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell metabolism. 2009;10(6):481–90. 10.1016/j.cmet.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell metabolism. 2008;7(1):21–32. 10.1016/j.cmet.2007.11.010 [DOI] [PubMed] [Google Scholar]
- 21.Fischer F, Hamann A, Osiewacz HD. Mitochondrial quality control: an integrated network of pathways. Trends in biochemical sciences. 2012;37(7):284–92. 10.1016/j.tibs.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 22.Ayala A, Munoz MF, Arguelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014:360438 10.1155/2014/360438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott C, Jacobs K, Haucke E, Navarrete Santos A, Grune T, Simm A. Role of advanced glycation end products in cellular signaling. Redox Biol. 2014;2:411–29. 10.1016/j.redox.2013.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyd O, Weng P, Sun X, Alberico T, Laslo M, Obenland DM, et al. Nectarine promotes longevity in Drosophila melanogaster. Free radical biology & medicine. 2011;50(11):1669–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson J, Lambert AJ, Portero-Otin M, Pamplona R, Magwere T, Miwa S, et al. Biomarkers of aging in Drosophila. Aging Cell. 2010;9(4):466–77. 10.1111/j.1474-9726.2010.00573.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landis G, Shen J, Tower J. Gene expression changes in response to aging compared to heat stress, oxidative stress and ionizing radiation in Drosophila melanogaster. Aging (Albany NY). 2012;4(11):768–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Lu-Bo Y, Haddad GG. A Drosophila ABC transporter regulates lifespan. PLoS Genet. 2014;10(12):e1004844 10.1371/journal.pgen.1004844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mech Ageing Dev. 2005;126(3):365–79. 10.1016/j.mad.2004.08.012 [DOI] [PubMed] [Google Scholar]
- 29.Jung I, Kim TY, Kim-Ha J. Identification of Drosophila SOD3 and its protective role against phototoxic damage to cells. FEBS Lett. 2011;585(12):1973–8. 10.1016/j.febslet.2011.05.033 [DOI] [PubMed] [Google Scholar]
- 30.Muller FL, Song W, Jang YC, Liu Y, Sabia M, Richardson A, et al. Denervation-induced skeletal muscle atrophy is associated with increased mitochondrial ROS production. Am J Physiol Regul Integr Comp Physiol. 2007;293(3):R1159–68. 10.1152/ajpregu.00767.2006 [DOI] [PubMed] [Google Scholar]
- 31.Oka S, Hirai J, Yasukawa T, Nakahara Y, Inoue YH. A correlation of reactive oxygen species accumulation by depletion of superoxide dismutases with age-dependent impairment in the nervous system and muscles of Drosophila adults. Biogerontology. 2015. [DOI] [PubMed] [Google Scholar]
- 32.Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263(5150):1128–30. [DOI] [PubMed] [Google Scholar]
- 33.Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19(2):171–4. 10.1038/534 [DOI] [PubMed] [Google Scholar]
- 34.Sun J, Tower J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol Cell Biol. 1999;19(1):216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orr WC, Mockett RJ, Benes JJ, Sohal RS. Effects of overexpression of copper-zinc and manganese superoxide dismutases, catalase, and thioredoxin reductase genes on longevity in Drosophila melanogaster. J Biol Chem. 2003;278(29):26418–22. 10.1074/jbc.M303095200 [DOI] [PubMed] [Google Scholar]
- 36.Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161(2):661–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Celotto AM, Liu Z, Vandemark AP, Palladino MJ. A novel Drosophila SOD2 mutant demonstrates a role for mitochondrial ROS in neurodevelopment and disease. Brain Behav. 2012;2(4):424–34. 10.1002/brb3.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neretti N, Wang PY, Brodsky AS, Nyguyen HH, White KP, Rogina B, et al. Long-lived Indy induces reduced mitochondrial reactive oxygen species production and oxidative damage. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(7):2277–82. 10.1073/pnas.0812484106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119(1):121–35. 10.1016/j.cell.2004.09.013 [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell metabolism. 2005;1(6):361–70. 10.1016/j.cmet.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 41.Demontis F, Piccirillo R, Goldberg AL, Perrimon N. Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Disease models & mechanisms. 2013;6(6):1339–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrow G, Inaguma Y, Kato K, Tanguay RM. The small heat shock protein Hsp22 of Drosophila melanogaster is a mitochondrial protein displaying oligomeric organization. J Biol Chem. 2000;275(40):31204–10. 10.1074/jbc.M002960200 [DOI] [PubMed] [Google Scholar]
- 43.Morrow G, Tanguay RM. Drosophila melanogaster Hsp22: a mitochondrial small heat shock protein influencing the aging process. Front Genet. 2015;6:1026 10.3389/fgene.2015.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Tower J. Expression of hsp22 and hsp70 transgenes is partially predictive of drosophila survival under normal and stress conditions. J Gerontol A Biol Sci Med Sci. 2009;64(8):828–38. 10.1093/gerona/glp054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Sun H, Lu J, Li X, Chen X, Tao D, et al. Lifespan extension and elevated hsp gene expression in Drosophila caused by histone deacetylase inhibitors. J Exp Biol. 2005;208(Pt 4):697–705. 10.1242/jeb.01439 [DOI] [PubMed] [Google Scholar]
- 46.Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3(4):442–55. 10.1016/j.stem.2008.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, et al. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell metabolism. 2011;14(5):623–34. 10.1016/j.cmet.2011.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cocheme HM, Quin C, McQuaker SJ, Cabreiro F, Logan A, Prime TA, et al. Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell metabolism. 2011;13(3):340–50. 10.1016/j.cmet.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Copeland JM, Cho J, Lo T Jr., Hur JH, Bahadorani S, Arabyan T, et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Current biology: CB. 2009;19(19):1591–8. 10.1016/j.cub.2009.08.016 [DOI] [PubMed] [Google Scholar]
- 50.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–85. 10.1146/annurev.genet.40.110405.090613 [DOI] [PubMed] [Google Scholar]
- 51.Yun J, Finkel T. Mitohormesis. Cell metabolism. 2014;19(5):757–66. 10.1016/j.cmet.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ziegler DV, Wiley CD, Velarde MC. Mitochondrial effectors of cellular senescence: beyond the free radical theory of aging. Aging Cell. 2015;14(1):1–7. 10.1111/acel.12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Metpally RP, Sowdhamini R. Cross genome phylogenetic analysis of human and Drosophila G protein-coupled receptors: application to functional annotation of orphan receptors. BMC genomics. 2005;6:106 10.1186/1471-2164-6-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YJ, Sano T, Nabetani T, Asano Y, Hirabayashi Y. GPRC5B activates obesity-associated inflammatory signaling in adipocytes. Sci Signal. 2012;5(251):ra85 10.1126/scisignal.2003149 [DOI] [PubMed] [Google Scholar]
- 55.Robbins MJ, Michalovich D, Hill J, Calver AR, Medhurst AD, Gloger I, et al. Molecular cloning and characterization of two novel retinoic acid-inducible orphan G-protein-coupled receptors (GPRC5B and GPRC5C). Genomics. 2000;67(1):8–18. 10.1006/geno.2000.6226 [DOI] [PubMed] [Google Scholar]
- 56.Sano T, Kim YJ, Oshima E, Shimizu C, Kiyonari H, Abe T, et al. Comparative characterization of GPRC5B and GPRC5CLacZ knockin mice; behavioral abnormalities in GPRC5B-deficient mice. Biochem Biophys Res Commun. 2011;412(3):460–5. 10.1016/j.bbrc.2011.07.118 [DOI] [PubMed] [Google Scholar]
- 57.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–48. 10.1038/ng.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith WW, Thomas J, Liu J, Li T, Moran TH. From fat fruit fly to human obesity. Physiol Behav. 2014;136:15–21. 10.1016/j.physbeh.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23(2):64–71. 10.1016/j.tcb.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tennessen JM, Barry WE, Cox J, Thummel CS. Methods for studying metabolism in Drosophila. Methods. 2014;68(1):105–15. 10.1016/j.ymeth.2014.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of thor and dInR mRNAs was measured in young (7-days old) flies by qRT-PCR (n = 3, 10 flies per replicate).
(TIF)
Expression of sod2 mRNA was measured in young (7-days old) flies by qRT-PCR (n = 3, 10 flies per replicate). Data are means ± SEM (*P<0.05).
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.