Abstract
Activation of a G protein-coupled receptor (GPCR) causes recruitment of multiple intracellular proteins, each of which can activate distinct signaling pathways. This complexity has engendered interest in agonists that preferentially stimulate subsets among the natural signaling pathways (“biased agonists”). We have examined analogues of glucagon-like peptide-1 (GLP-1) containing β-amino acid residues in place of native α residues at selected sites and found that some analogues differ from GLP-1 in terms of their relative abilities to promote G protein activation (as monitored via cAMP production) versus β-arrestin recruitment (as monitored via BRET2 assays). The α→β replacements generally cause modest declines in stimulation of cAMP production and β-arrestin recruitment, but for some replacement sets cAMP production is more strongly affected than is β-arrestin recruitment. The central portion of GLP-1 appears to be critical for achieving bias toward β-arrestin recruitment. These results suggest that backbone modification via α→β residue replacement may be a versatile source of agonists with biased GLP-1R activation profiles.
Graphical Abstract
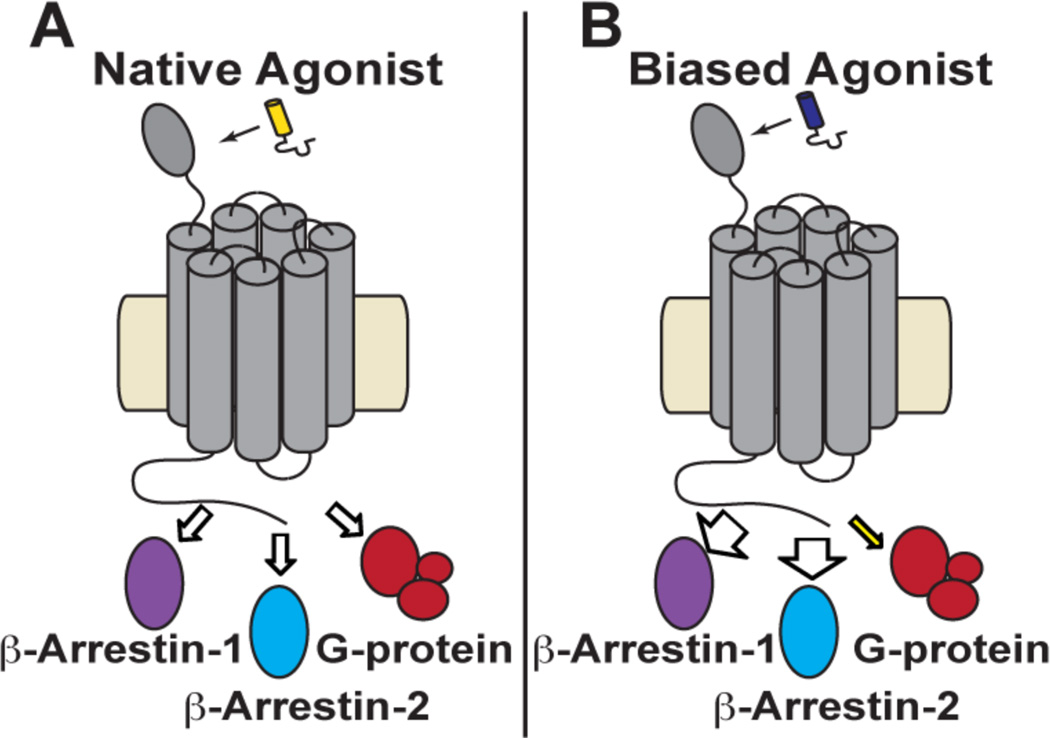
G protein-coupled receptors (GPCRs) play critical physiological roles by transmitting information encoded in extracellular molecules to the cytoplasm. Signal transduction requires interaction of an activated GPCR with intracellular partner proteins, which can include several G proteins (Gαs, Gαi, Gαq, etc.), β-arrestin-1 and β-arrestin-2 (Figure 1A). Each of these effector proteins stimulates one or more signaling pathways. Gαs, for example, leads to the production of the second messenger cAMP, Gαq leads to mobilization of intracellular Ca2+, and β-arrestins mediate receptor internalization and/or serve as scaffolds for G protein-independent signaling.1–3 GPCR structure is dynamic, and each effector protein is thought to bind to a unique receptor conformation.4,5 GPCR ligands that favor engagement of certain effector proteins over others, referred to as “conformationally selective” or “biased” agonists, are believed to favor a subset among the activated GPCR conformations (a hypothetical β-arrestin-biased agonist is shown in Figure 1B).6–8 Mechanistic understanding of ligand bias remains limited, despite extensive study.9–11 Little structural information is available for GPCR complexes of biased agonists,12,13 which hinders both rational design of agonists with pathway selectivity and molecular-level elucidation of the origins of bias.
Figure 1.
Schematic depicting the intracellular proteins, investigated in the current study, that are recruited to a GPCR upon stimulation with (A) the native receptor agonist or with (B) a β–arrestin-biased agonist.
The glucagon-like peptide-1 receptor (GLP-1R) is a member of the B subclass of GPCRs, which are activated by long poly-peptide hormones. GLP-1R is primarily expressed in pancreatic β cells, which secrete insulin to regulate glucose concentration in the bloodstream. This receptor can be engaged by multiple endogenous peptides, including at least six forms of glucagon-like peptide-1. The full-length forms, GLP-1(1–36)NH2 and GLP-1(1–37), are considered weak agonists or antagonists of GLP-1R.14 The mature forms, GLP-1(7–36)NH2 and GLP-1(7–37), are potent agonists; these two forms display indistinguishable activity.14,15 After release into the bloodstream, the mature forms of GLP-1 are rapidly cleaved by dipeptidyl peptidase-4 (DPP-4), to generate GLP-1(9–36)NH2 and GLP-1(9–37), which are low-affinity, low-efficacy agonists of GLP-1R-mediated signaling and can function as antagonists.14 In addition, the peptide hormones glucagon16 and oxyntomodulin16,17 (which is an eight-residue extension of glucagon) can activate GLP-1R (sequences shown in Figure 2).
Figure 2.
Amino acid sequences of each endogenous form of GLP-1, as well as the sequences of related peptides glucagon and oxyntomodulin.
Binding of the mature forms of GLP-1 (referred to below simply as "GLP-1") to the GLP-1R is associated with diverse effects that are critical for human health, but the intracellular signaling networks initiated in this way have not yet been fully elucidated. GLP-1 binding to the GLP-1R activates Gαs and thereby induces a rise in intracellular cAMP, which leads to glucose-stimulated insulin secretion.18 GLP-1 binding recruits also β-arrestin-1 and β-arrestin-2 to the GLP-1R. Recruitment of β-arrestin-1 promotes β cell proliferation19 and protects these cells from apoptosis.20,21 The effects of GLP-1-induced β-arrestin-2 recruitment to the GLP-1R are not yet clear.22 GLP-1R ligands that manifest strong selectivity in terms of effector protein recruitment and activation would be valuable tools for elucidating the role(s) of individual effector proteins.
Synthetic peptide agonists of GLP-1R have been developed to treat type II diabetes by enhancing glucose-stimulated insulin secretion from pancreatic β cells, among other mechanisms. Exendin-4, for example, is a component of the venom of the Gila monster and functions as a potent agonist of this receptor.23 Liraglutide is nearly identical in sequence to GLP-1(7–37) but bears a hexadecanoyl appendage on the side chain of Lys-26.24 This appendage promotes binding to serum albumin, which protects liraglutide from DPP-4 cleavage and dramatically enhances lifetime in the bloodstream.25
Variations in the α-amino acid sequences of GLP-1R agonists lead to differences in the relative and absolute abilities of these agonists to induce recruitment of intracellular effector proteins to the receptor and differences in the stimulation of downstream signaling events. Thus, for example, relative to mature GLP-1, both exendin-4 and oxyntomodulin show a bias toward recruitment of β-arrestin-1 and β-arrestin-2 relative to cAMP generation (which presumably reflects Gαs activation).26 Zhang et al. recently reported discovery of an exendin-4 derivative that displays significant bias in favor of G protein activation, which leads to an altered in vivo efficacy profile relative to exendin-4.27 Furthermore, allosteric GLP-1R ligands can influence the pattern of intracellular protein recruitment induced by a given peptide agonist.28,29 These observations are of great interest from the biomedical perspective, because it has been proposed that therapeutic benefits may be maximized (and deleterious side effects minimized) with drugs that exert specifically tailored activating effects on the GLP-1R (or other GPCRs).6, 30, 31 However, the origins of ligand-dependent variations in signaling outcome are not yet clear at the molecular level, and the rational design of agonists with predetermined bias profiles at a given GPCR is currently not possible.
Here we show that GLP-1 analogues containing β-amino acid residues can display substantial bias in terms of intracellular partner engagement by the GLP-1R. These findings are significant because nearly all known GLP-1 analogues are composed exclusively of α-amino acid residues, although a few examples with unnatural backbone components have been described.32–34 β residue incorporation is simple in the context of conventional solid-phase peptide synthesis, and the findings disclosed here suggest that evaluation of small sets of peptide hormone analogues containing α→β replacements could be a generally productive strategy for discovery of molecules with mechanistic and perhaps ultimately therapeutic utility.
RESULTS
Peptide Design
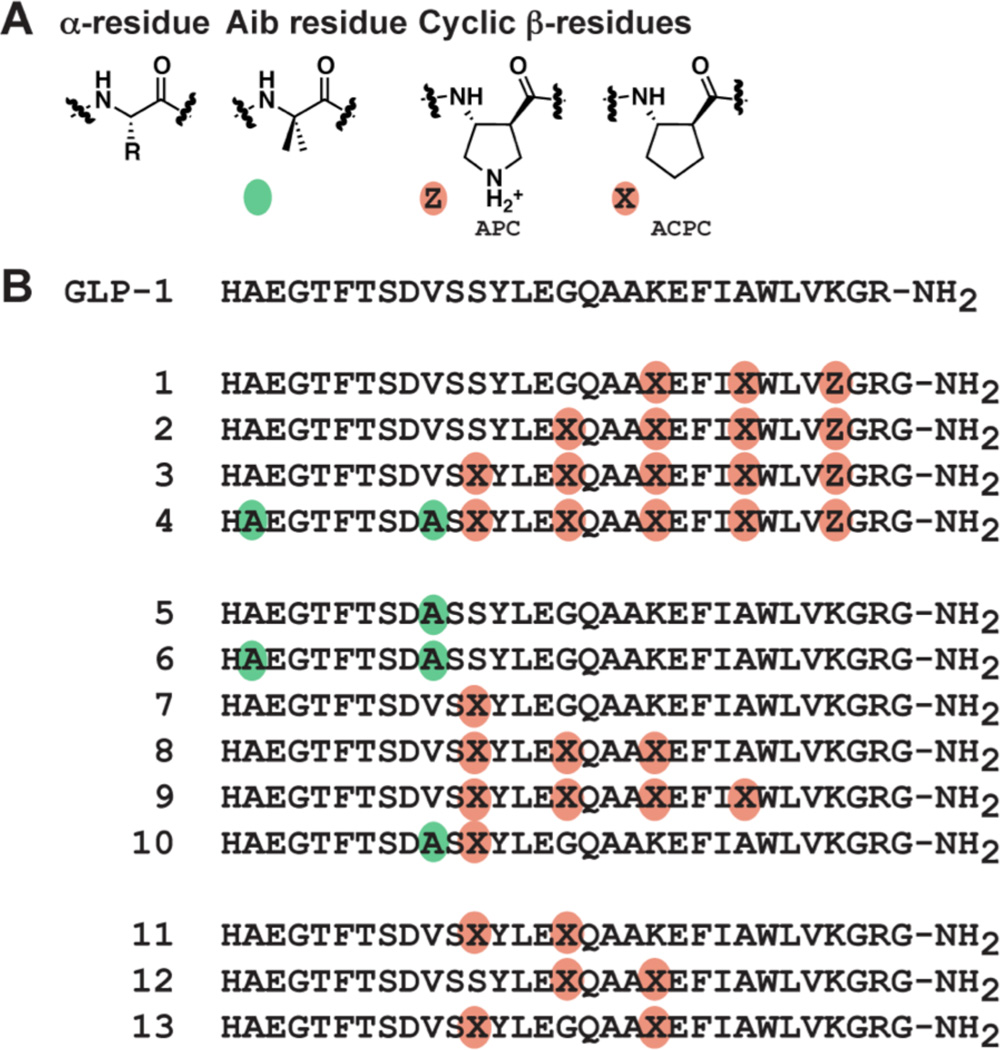
We previously described a small set of GLP-1(7–37)-NH2 analogues, including 1–4 (Figure 3), that contain α→β replacements in the C-terminal region of the hormone and retain agonist activity at the GLP-1R.35 The most heavily modified member of this set, 4, contains two α→Aib (2-aminoisobutyric acid) substitutions as well as five α→β substitutions. The β residue placement in this set conforms to an αααβ pattern. The ring constraint in the β residues employed in 1–4 is known to support adoption of an α-helix-like conformation by α/β-peptides;36 a co-crystal structure of GLP-1 and the GLP-1R extracellular domain shows that the C-terminal portion of the hormone adopts an α-helical conformation in the bound state.37
Figure 3.
(A) α-Amino acid and β-amino acid residues. The colored circles used to indicate non-natural substitutions in sequences below are defined: green circles represent the α residue Aib, and orange circles represent ring-constrained β residues (X = ACPC, Z = APC). (B) GLP-1(7-36)NH2 and α/β-peptide analogues 1 – 13 (based on GLP-1(7-37)NH2). Each peptide has a free N-terminus and a primary amide at the C-terminus.
In the previous study we sought to maximize β residue content in order to minimize the susceptibility of our α/β-peptides to proteolytic degradation. Extension of the αααβ pattern evident in 3 to a sixth site (position 14), however, caused a substantial decline in agonist activity.35 We therefore turned to α→Aib replacements in the N-terminal portion to enhance protease resistance. GLP-1 is rapidly cleaved between Ala8 and Glu9 by dipeptidyl peptidase-4 (DPP-4), but this cleavage can be blocked with maintenance of agonist activity by replacing Ala8 with Aib.38 Aib was therefore placed at position 8 in α/β-peptide 4. The other protease known to degrade GLP-1 in vivo is neprilysin,39 which cleaves at many sites in the C-terminal and central regions of the hormone. We introduced Aib at position 16 in 4 to augment the protection from neprilysin expected from the five α→β substitutions in 3.35
Our initial studies of GLP-1R activation by α/β-peptides35 focused on cAMP accumulation; the work described here expands this evaluation to include recruitment of β-arrestin-1 and β-arrestin-2, as well as a more sensitive alternative assay for cAMP production. In an effort to elucidate the origin of pathway selectivities detected among α/β-peptides 1–4, we expanded this collection to include 5–13 (Figure 3), analogues of GLP-1(7–37)NH2 that contain different subsets among the α→β and/or α→Aib replacements found in 4. Although GLP-1(7–37)NH2 does not occur naturally, this peptide is as potent as the two mature forms of the hormone.40 One of the natural mature forms, GLP-1(7–36)NH2, serves as the reference agonist in these studies; we found that GLP-1(7–36)NH2 and GLP-1(7–37)NH2 behave identically in terms of stimulating cAMP production, β-arrestin-1 recruitment and β-arrestin-2 recruitment (Figure S1).
Evaluation of GLP-1R activation induced by α/β-peptides 1–4
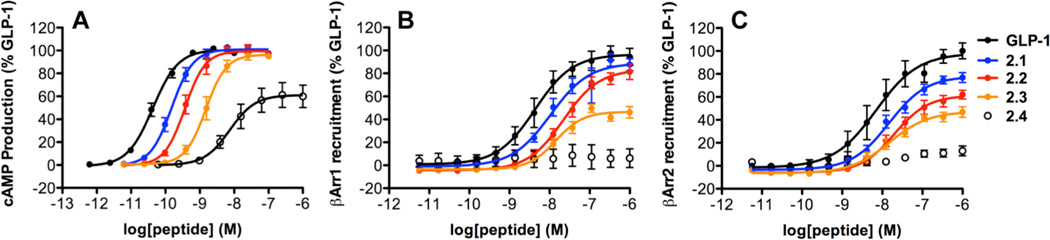
As a prelude to evaluating β-arrestin recruitment induced by α/β-peptide analogues of GLP-1, we reassessed cAMP production via a kinetic, GloSensor assay.41, 42 In contrast to previous results for 1–4,35 which were based on cAMP accumulation and quantification, cAMP generation potency as monitored by the GloSensor assay declines modestly with each additional α→β replacement (Table 1; Figure 4A). However, the two assays are consistent in indicating that α/β-peptides 1–3 match GLP-1 in terms of maximum cAMP generation via the GLP-1R; the maximum activation by 4 is lower than that of GLP-1 or the other α/β-peptides according to the GloSensor assay.
Table 1.
Potency and maximum response measured for GLP-1 and α/β-peptides 1 – 4.
| cAMP | β-Arrestin-1 | β-Arrestin-2 | ||||
|---|---|---|---|---|---|---|
| pEC50 (M) | Max Response (% GLP-1) |
pEC50 (M) | Max Response (% GLP-1) |
pEC50 (M) | Max Response (% GLP-1) |
|
| GLP-1 | −10.3 ± 0.1 | 100 | −8.4 ± 0.2 | 100 | −8.1 ± 0.3 | 100 |
| α/β-Peptide 1 | −9.8 ± 0.1 | 101 ± 1 | −8.0 ± 0.1 | 88 ± 6 | −7.9 ± 0.1 | 78 ± 4 |
| α/β-Peptide 2 | −9.4 ± 0.1 | 100 ± 2 | −7.6 ± 0.1 | 87 ± 6 | −7.7 ± 0.1 | 63 ± 3 |
| α/β-Peptide 3 | −8.8 ± 0.1 | 97 ± 3 | −7.8 ± 0.1 | 48 ± 4 | −7.9 ± 0.1 | 47 ± 4 |
| α/β-Peptide 4 | −8.2 ± 0.1 | 60 ± 5 | – | 6 ± 8 | – | 13 ± 4 |
Values are the mean ± S.E.M of ≥ 3 independent experiments, with duplicate measurements per experiment. Values represent Gαs activation, as measured by the luciferase-based GloSensor cAMP reporter assay, β-arrestin-1 recruitment, as measured using a β-arrestin-1 BRET2 assay and β-arrestin-2 recruitment, as measured using a β-arrestin-2 BRET2 assay. The β-arrestin-2 plasmid contained (R393E, R395E) mutations, which prevent receptor internalization.16 For 4, β-arrestin-1 and β-arrestin-2 maximum responses represent the maximal response at 1 µM peptide.
Figure 4.
GLP-1R activation comparisons among GLP-1(7-36)NH2 and α/β-peptides 1–4 (based on GLP-1(7-37)NH2). Data points are the mean ± S.E.M of ≥ 3 independent experiments, with duplicate measurements for each experiment. Concentration-response curves for peptide-induced activation of GLP-1R as manifested by: (A) Gαs activation, measured by the luciferase-based GloSensor cAMP reporter assay; (B) β-arrestin-1 recruitment, measured using a β-arrestin-1 BRET2 assay; (C) β-arrestin-2 recruitment, measured using a β-arrestin-2 BRET2 assay. The β-arrestin-2 plasmid contains (R393E, R395E) mutations, which prevent receptor internalization.16
Although the new assay for cAMP production indicates that α/β-peptide 4 has significantly lower potency and efficacy relative to GLP-1, previous studies demonstrated that 4 can control blood glucose in vivo, and that this effect is prolonged relative to glucose control achieved with GLP-1.35 In addition, 4 and GLP-1 were indistinguishable as insulin secretagogues in experiments involving mouse pancreatic islets.35 These functional similarities between GLP-1 and 4 may indicate that the resistance to proteolysis engendered by the non-proteinogenic residues in the α/β-peptide compensate for a diminished efficacy in activating the GLP-1R for cAMP production.
Recruitment of β-arrestin-1 and β-arrestin-2 to the GLP-1R induced by α/β-peptides 1–4 was evaluated via previously described bioluminescence resonance energy transfer (BRET) assays.16 Constructs for the GLP1R-RLuc8 and GFP2-β-arrestin-1 or GFP2-β-arrestin-2 (R393E, R395E) fusion proteins were transiently transfected into HEK293FT cells along with a construct for G protein-coupled receptor kinase 5 (GRK5). The (R393E, R395E) variant of β-arrestin-2 prevents loss of BRET signal due to GLP-1R internalization.16 Co-transfection with GRK5 has been shown to enhance the maximum BRET signal in this assay by promoting GLP-1R phosphorylation,16,22 which increases the affinity of both β-arrestins.
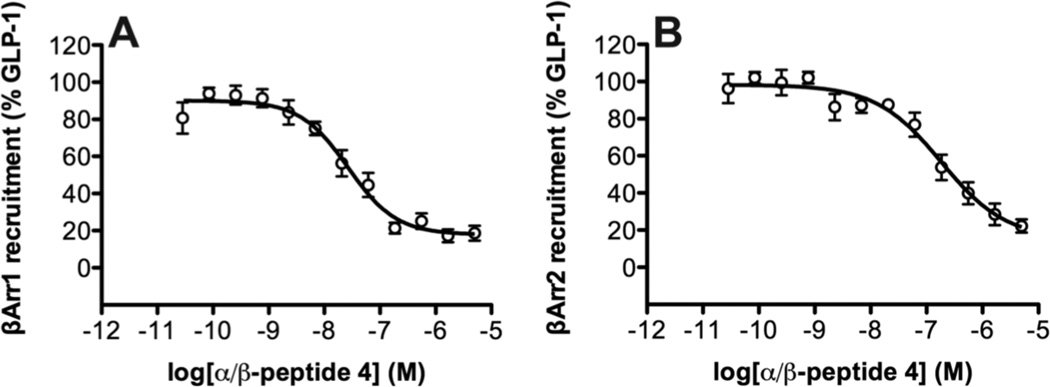
Increasing the number of β residues in the GLP-1 analogues, starting near the C-terminus (1→2→3), causes a progressive decline in the potency and/or maximum level of recruitment for each β-arrestin (Table 1, Figures 4B–C). The most dramatic changes are observed for α/β-peptide 4, which is barely active in these assays. α/β-Peptides 3 and 4 have the same five α→β replacements, but 4 contains two Aib residues not found in 3.
Competition BRET assays, in which 4 was added to cells in the presence of 10 nM GLP-1, show that this α/β-peptide is fully competent to bind to the receptor under these conditions, because 4 serves as an antagonist of GLP-1-induced recruitment of either β-arrestin-1 (pIC50 = −7.6 ± 0.1) or β-arrestin-2 (pIC50= −6.7 ± 0.2) (Figure 5).
Figure 5.
Competition between GLP-1(7-36)NH2 and α/β-peptide 4 for binding to GLP-1R, as detected by BRET2 assays for recruitment of (A) β-arrestin-1 or (B) β-arrestin-2. Data points are the mean ± S.E.M of ≥ 3 independent experiments, with duplicate measurements for each experiment. The concentration of GLP-1(7-36)NH2 was held constant at 10 nM, and the concentration of 4 was varied. The β-arrestin-2 plasmid contains (R393E, R395E) mutations, which prevent receptor internalization.16
Analysis of GLP-1R activation bias by α/β-peptides 1–4
For a receptor such as GLP-1R that can activate multiple intracellular partner proteins, such as G proteins and β-arrestins, the availability of a new agonist leads to an important question: how do the relative efficacies of this new agonist among the possible signaling pathways compare with the relative efficacies of a benchmark agonist? In other words, does the new agonist show a preference pattern among the available signaling pathways that differs from the preference pattern of the benchmark agonist? In the context of the present study, a specific example of this type of question is, "how does the relative efficacy of α/β-peptide 3 for inducing cAMP production (presumably via activation of Gαs) vs. recruiting β-arrestin-2 compare with the relative efficacy of the benchmark agonist GLP-1(7–36)NH2 for these two processes?" If the balance of efficacy along these two signaling axes is substantially different for 3 relative to GLP-1(7–36)NH2, then 3 is identified as a "biased agonist" of the GLP-1R.
We have used the operational model of Black and Leff43,44 to determine whether our α/β-peptide analogues display biased agonism relative to GLP-1(7–36)NH2. This model describes the effect of an agonist on a receptor with two terms, (i) KA, the dissociation constant that characterizes the agonist binding to the receptor conformation induced by association with a given effector protein (e.g., a G protein or a β-arrestin), and (ii) τ, the efficacy of the agonist, a term that accounts for the potency of agonist action on the receptor and assay-dependent factors, including the density of receptors and strength of receptor-effector coupling. KA and τ can be estimated from standard concentration-response data sets. The expression log(τ/KA), referred to as the "transduction coefficient," is a measure of the strength of activation of a particular pathway by a specific agonist. The way a given α/β-peptide differs from benchmark agonist GLP-1(7–36)NH2 can be expressed as a difference in transduction coefficients, Δlog(τ/KA). A "bias factor" for a given α/β-peptide relative to GLP-1(7–36)NH2 can be defined based on comparing two specific outcomes of GLP-1R activation, e.g., cAMP generation vs. β-arrestin-2 recruitment, by calculating ΔΔlog(τ/KA).
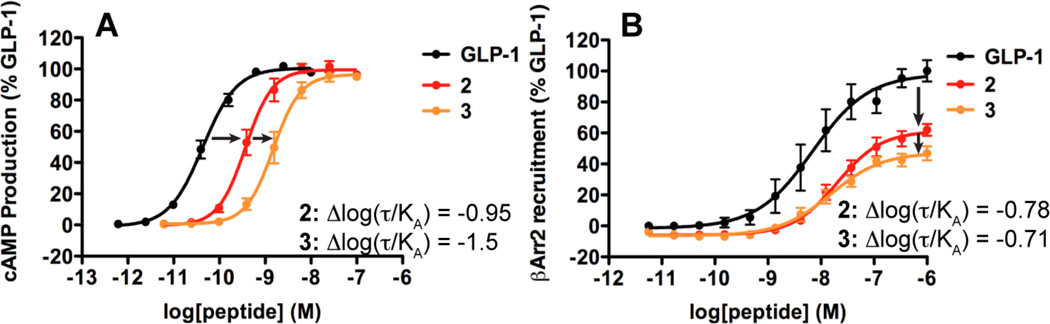
The importance of using the operational model for comparisons among responses involving different effector proteins is illustrated in Figure 6, which focuses on a subset of the data presented in Figure 4. Figure 6A shows that the effect on cAMP production of four or five α→β replacements in GLP-1, to generate 2 or 3, is manifested as a rightward shift in the concentration-response curve. There is very little difference among the maxima in cAMP production for these three compounds. In contrast, Figure 6B shows that the effect on β-arrestin-2 recruitment of the same α→β replacements is manifested primarily as a drop in the maximum recruitment extent. If one focuses on maximum response and pEC50, the parameters provided in Table 1, it is not clear how to compare the impact of the α→β replacements on cAMP production vs. β-arrestin-2 recruitment. However, application of the operational model allows calculation of bias factors for 2 and 3 relative to GLP-1 in both assays, and comparison is now straightforward.
Figure 6.
Illustration of the importance of using the operational model for detecting biased agonism. Concentration-response curves generated for GLP-1 and α/β-peptides 2 and 3 in (A) cAMP production and (B) β-arrestin-2 recruitment. The Δlog(τ/KA) values calculated for α/β-peptide 2 relative to GLP-1 in each response demonstrate that the efficacy of α/β-peptide 2 is similar between cAMP production and β-arrestin-2 recruitment, despite the different changes in the concentration-dependent behavior of this peptide in the two responses. In contrast, the Δlog(τ/KA) values calculated for α/β-peptide 3 relative to GLP-1 in each response demonstrate that the efficacy of α/β-peptide 3 is greater for β-arrestin-2 recruitment than for cAMP production.
Estimation of the ΔΔlog(τ/KA) values reveals parallel losses of efficacy for recruitment of β-arrestin-1 and β-arrestin-2 and for cAMP signaling for α/β-peptides 1 and 2 relative to GLP-1(7–36)NH2 (Table 2). For α/β-peptide 3, however, the loss of cAMP signaling is greater than the loss of β-arrestin-1 or-2 recruitment. Thus, 3 displays a bias for β-arrestin recruitment relative to GLP-1 although the calculated bias factor for 3 is statistically significant only for β-arrestin-2. The lack of statistical significance the for β-arrestin-1 bias factor calculated for 3 may be due to a greater degree of scatter in the for β-arrestin-1 BRET2 data for 1–4 relative to the for β-arrestin-2 BRET2 data for these peptides. Bias factors could not be determined for 4 because no significant recruitment of β-arrestin-1 or -2 could be detected in our assays for this α/β-peptide within the concentration range examined.
Table 2.
Bias factors (ΔΔlog(τ/KA)) calculated for GLP-1 and α/β-peptides 1 – 3.
| ΔΔlog(τ/KA) | ||
|---|---|---|
| β-arrestin-1 vs cAMP |
β-arrestin-2 vs cAMP |
|
| GLP-1 | 0 | 0 |
| α/β-Peptide 1 | 0.1 ± 0.2 | 0.1 ± 0.2 |
| α/β-Peptide 2 | 0.1 ± 0.2 | 0.2 ± 0.3 |
| α/β-Peptide 3 | 0.6 ± 0.2 | 0.8* ± 0.2 |
| α/β-Peptide 4 | N.D. | N.D. |
Bias factors (ΔΔlog(τ/KA)) calculated in terms of β-arrestin-1 recruitment relative to cAMP production, and β-arrestin-2 recruitment relative to cAMP production. These bias factors were derived from experimental data as described in the text. Bias factors could not be calculated for α/β-peptide 4, because the β-arrestin recruitment assays showed no quantifiable activity for this molecule (Figure 2).
Statistically significant difference from GLP-1 using one-way analysis of variance followed by Dunnett’s test (P < 0.05).
Evaluation of peptides 5–13
New analogues of GLP-1(7–37)NH2 were prepared in an effort to determine whether a particular subset among the substitutions in α/β-peptides 3 and 4 relative to GLP-1 plays a dominant role in mediating biased agonism. α-Peptide analogues containing either the Val16→Aib replacement (5) or both Aib replacements (6) were very similar to one another and to GLP-1(7–36)NH2 (Table 3, Figure 6). For the two β-arrestins, both 5 and 6 displayed small declines in maximum recruitment but little change in EC50 relative to GLP-1. For cAMP production, both 5 and 6 were ~3-fold less potent than GLP-1 (EC50), but both α-peptides were indistinguishable from GLP-1 in terms of maximum response. The similarities in activity among 5, 6 and GLP-1(7–36)NH2 suggest that the previously described analogue38 containing only Ala8→Aib should manifest an activity profile comparable to those of 5 and 6.
Table 3.
Potency and maximum response measured for GLP-1 and peptides 5 – 13.
| cAMP | β-Arrestin-1 | β-Arrestin-2 | ||||
|---|---|---|---|---|---|---|
| pEC50 (M) | Max Response (% GLP-1) |
pEC50 (M) | Max Response (% GLP-1) |
pEC50 (M) | Max Response (% GLP-1) |
|
| GLP-1 | −10.3 ± 0.1 | 100 | −8.0 ± 0.1 | 100 | −8.0 ± 0.1 | 100 |
| α-Peptide 5 | −9.8 ± 0.2 | 101 ± 1 | −8.2 ± 0.1 | 97 ± 2 | −8.0 ± 0.1 | 83 ± 2 |
| α-Peptide 6 | −9.8 ± 0.1 | 103 ± 3 | −8.1 ± 0.1 | 86 ± 4 | −7.9 ± 0.1 | 76 ± 4 |
| α/β-Peptide 7 | −10.0 ± 0.1 | 99 ± 1 | −7.9 ± 0.1 | 81 ± 5 | −8.1 ± 0.1 | 84 ± 2 |
| α/β-Peptide 8 | −9.0 ± 0.2 | 96 ± 1 | −7.7 ± 0.2 | 43 ± 3 | −8.0 ± 0.1 | 63 ± 5 |
| α/β-Peptide 9 | −8.4 ± 0.2 | 97 ± 2 | −7.5 ± 0.1 | 41 ± 4 | −8.0 ± 0.1 | 59 ± 2 |
| α/β-Peptide 10 | −9.6± 0.1 | 100 ± 1 | −8.0 ± 0.1 | 30 ± 1 | −8.0 ± 0.1 | 53 ± 5 |
| α/β-Peptide 11 | −9.8 ± 0.1 | 102 ± 1 | −8.0 ± 0.1 | 37 ± 1 | −8.1 ± 0.1 | 52 ± 4 |
| α/β-Peptide 12 | −10.0 ± 0.1 | 103 ± 1 | −7.8 ± 0.1 | 85 ± 3 | −8.0 ± 0.1 | 78 ± 3 |
| α/β-Peptide 13 | −10.1 ± 0.1 | 104 ± 1 | −7.8 ± 0.2 | 80 ± 3 | −8.0 ± 0.1 | 74 ± 4 |
Values are the mean ± S.E.M of ≥ 3 independent experiments, with duplicate measurements for each experiment. Values represent Gαs activation, as measured by the luciferase-based GloSensor cAMP reporter assay, β-arrestin-1 recruitment, as measured using a β-arrestin-1 BRET2 assay and β-arrestin-2 recruitment, as measured using a β-arrestin-2 BRET2 assay. β-Arrestin-2 plasmid contained (R393E, R395E) mutations, which prevent receptor internalization.16
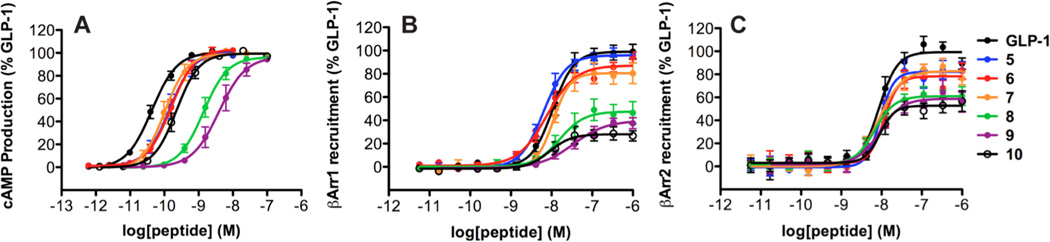
Analogue 7 contains a single α→β replacement at position 18; introduction of a β residue at this position in the original series (2→3) induced an increase in β-arrestin-1 and-2 recruitment bias relative to GLP-1 (Table 2). In contrast, α→β replacement at position 18 alone causes only minimal (and comparable) changes to cAMP production and β-arrestin recruitment (Table 3, Figure 7). Introduction of additional β residues, at positions 22 and 26 (8 and 9), causes decreases in both β-arrestin recruitment and cAMP production relative to GLP-1 (Table 3, Figure 7). Analysis of these data using the operational model (Table 4) indicates that engagement of the GLP-1R by α/β-peptide 8 or 9 causes a preference for recruitment of β-arrestin-1 and -2 over production of cAMP, relative to engagement of the GLP-1R by GLP-1. Thus, α/β-peptides 8 and 9 are β-arrestin-biased agonists of the GLP-1R. α/β-Peptide 10, containing only the Val16→Aib and Ser18→β replacements, displays significant decreases in recruitment of β-arrestin-1 and -2, similar to those seen for 8 and 9; however, the cAMP response for 10 is stronger than that induced by 8 or 9, and application of the operational model indicates that α/β-peptide 10 does not show significant signal bias relative to GLP-1.
Figure 7.
GLP-1R activation comparisons among GLP-1(7-36)NH2 and α/β-peptides 5 – 10 (based on GLP-1(7-37)NH2). Data points are the mean ± S.E.M of ≥ 3 independent experiments, with duplicate measurements for each experiment. Concentration-response curves for peptide-induced activation of GLP-1R as manifested by: (A) Gαs activation, measured by the luciferase-based GloSensor cAMP reporter assay; (B) β-arrestin-1 recruitment, measured using a β-arrestin-1 BRET2 assay; (C) β-arrestin-2 recruitment, measured using a β-arrestin-2 BRET2 assay. The β-arrestin-2 plasmid contains (R393E, R395E) mutations, which prevent receptor internalization.16
Table 4.
Bias factors (ΔΔlog(τ/KA)) calculated for GLP-1 and peptides 5 – 13.
| ΔΔlog(τ/KA) | ||
|---|---|---|
| β-Arrestin-1 vs cAMP |
β-Arrestin-2 vs cAMP |
|
| GLP-1 | 0 | 0 |
| α-Peptide 5 | 0.2 ± 0.1 | 0.2 ± 0.1 |
| α-Peptide 6 | 0.2 ± 0.1 | 0.0 ± 0.1 |
| α/β-Peptide 7 | 0.2 ± 0.1 | 0.2 ± 0.1 |
| α/β-Peptide 8 | 0.6* ± 0.2 | 1.2* ± 0.1 |
| α/β-Peptide 9 | 0.9* ± 0.2 | 1.6 *± 0.1 |
| α/β-Peptide 10 | 0.0 ± 0.1 | 0.3 ± 0.1 |
| α/β-Peptide 11 | 0.1 ± 0.1 | 0.2 ± 0.1 |
| α/β-Peptide 12 | −0.0 ± 0.1 | 0.1 ± 0.1 |
| α/β-Peptide 13 | −0.3 ± 0.2 | −0.1 ± 0.1 |
Bias factors (ΔΔlog(τ/KA)) calculated in terms of β-arrestin-1 recruitment relative to cAMP production, and β-arrestin-2 recruitment relative to cAMP production. These bias factors were derived from experimental data as described in the text.
Statistically significant difference from GLP-1 using one-way analysis of variance followed by Dunnett’s test (P < 0.05).
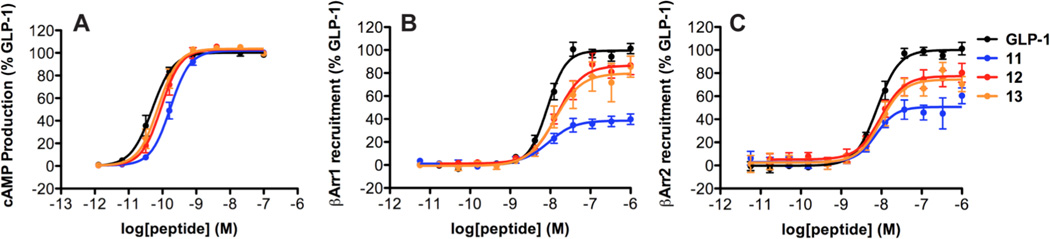
To define the α→β replacement sites that are dominant in terms of the bias toward recruitment of β-arrestin-1 and -2 manifested by α/β-peptides 8 and 9, we prepared and evaluated 11–13 (Table 3, Figure 8). This α/β-peptide set contains each possible pair among the three β residues in 8. Analysis of the results with the operational model indicates that none among 11–13 displays significant signaling bias relative to GLP-1 (Table 4). Thus, it appears that the three α→β replacements in 8 represent a minimum level of modification required to generate a β-arrestin-biased agonist for the GLP-1R.
Figure 8.
GLP-1R activation comparisons among GLP-1(7-36)NH2 and α/β-peptides 11 – 13 (based on GLP-1(7-37)NH2). Data points are the mean ± S.E.M of ≥ 3 independent experiments, with duplicate measurements for each experiment. Concentration-response curves for peptide-induced activation of GLP-1R as manifested by: (A) Gαs activation, measured by the luciferase-based GloSensor cAMP reporter assay; (B) β-arrestin-1 recruitment, measured using a β-arrestin-1 BRET2 assay; (C) β-arrestin-2 recruitment, measured using a β-arrestin-2 BRET2 assay. The β-arrestin-2 plasmid contains (R393E, R395E) mutations, which prevent receptor internalization.16
DISCUSSION
α/β-Peptide 4 contains seven substitutions relative to GLP-1, which, collectively, lead to losses in this analogue’s ability to stimulate cAMP production or induce β-arrestin recruitment, relative to GLP-1. Exploration of subsets among the α→β and α→Aib replacement sites of 4 reveals substitution patterns that have little effect on peptide efficacy in terms of either cAMP production or β-arrestin recruitment (5, 6, 12, and 13), substitution patterns that reduce peptide efficacy in each signaling pathway (1, 2, 7, 10 and 11) and, most interestingly, substitution patterns that differentially affect cAMP production versus β-arrestin recruitment (3, 8 and 9).
Comparing 3, 8 and 9 indicates that α→β replacements in the central region of GLP-1 are the most consequential in terms of engendering a bias toward β-arrestin recruitment relative to cAMP production. While the incorporation of an α →β replacement at position 18 (2→3) is necessary for the induction of selectivity for β-arrestin recruitment, neither the Ser18→β substitution alone (7) nor double substitution at Ser18 and Gly22 (11) is sufficient to induce this selectivity. In addition, implementing only the two outer α→β replacements in 8, at Ser18 and Lys26, to generate 13, does not induce pathway selectivity. These findings are significant because they suggest that G protein activation is more sensitive than is β-arrestin recruitment to changes in a relatively focused region near the middle of a GLP-1R agonist. In addition, our results suggest that this region of the peptide must contain a minimum density of α→β replacements (e.g., all three α →β replacements in 8) in order to induce a bias toward β-arrestin recruitment relative to G protein-mediated signaling.
The importance revealed here of the GLP-1 segment encompassing residues 18–26 in terms of Gαs activation by the GLP-1R, as manifested by cAMP production, can be related to the available structural information for the hormone-GPCR complex. A co-crystal structure of GLP-1(7–37) with the extracellular domain of GLP-1R37 shows that direct contact begins at hormone residue Ala24 and extends toward the hormone C-terminus; thus, interactions of the 18–26 region of GLP-1 with the receptor must largely involve the membrane-embedded domain of the GLP-1R. Photoreactive labeling previously identified a direct interaction between position 20 of GLP-1 and Trp-297 in extracellular loop 2 (ECL2) of the GLP-1R.45 Receptor mutation studies revealed that ECL2 plays an important role in GLP-1-induced signaling (cAMP production, Ca2+ mobilization and ERK phosphorylation).9,11 In addition, recent mutation studies involving ECL3 suggest that several residues in this loop are also critical in mediating signaling induced by GLP-1, exendin-4 or oxyntomodulin.26 The α→β replacements made in the 18–26 region of GLP-1 to generate 8 may disrupt critical interactions between the ligand and the ECLs of the GLP-1R.
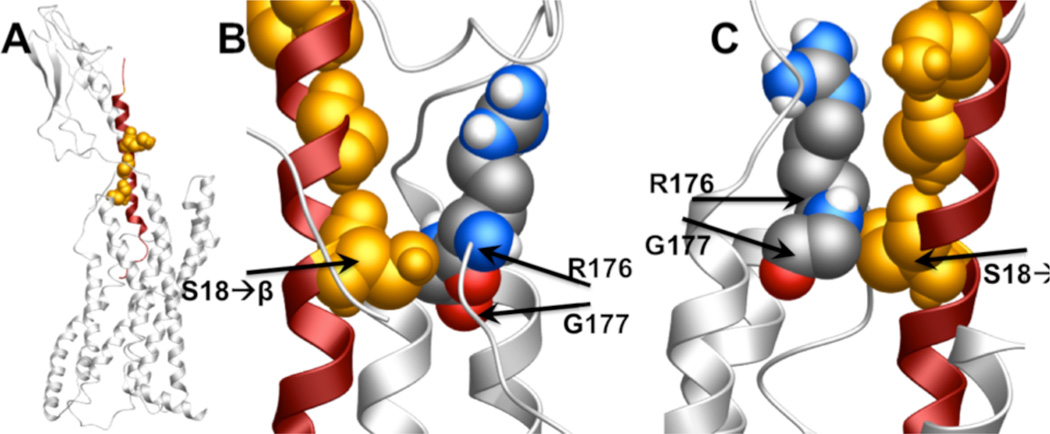
To begin to explore how β-arrestin-biased α/β-peptides might interact with the ECLs of the GLP-1R, we examined a recently reported model of the GLP-1 + GLP-1R complex, and highlighted the positions of GLP-1 that are modified in α/β-peptide 8 to generate Figure 9.46 This image suggests that there may be a close contact between the β residue at position 18 of 8 and residues within ECL3 that were previously found to be critical in mediating ERK1/2 phosphorylation induced by GLP-1, exendin-4 and oxyntomodulin via their interactions with the GLP-1R.26 Oxyntomodulin is biased toward ERK1/2 phosphorylation over cAMP production relative to GLP-1, a selectivity that may arise from the bias of oxyntomodulin toward β-arrestin recruitment.26 Based on this precedent, we speculate that the contacts we propose between the central portion of 8 and ECL3 of the GLP-1R might contribute to the β-arrestin bias displayed by this α/β-peptide.
Figure 9.
Images depicting a hypothetical interaction between the GLP-1R (gray) and α/β-peptide 8 (red), generated from an existing model of the GLP-1R complexed to GLP-1,43 by highlighting in orange the residues in GLP-1 that are modified to β residues in 8. (A) Full view of the GLP-1R + GLP-1 model. (B, C) Close-up views of position 18 of GLP-1 (β residue in 8) in contact with R376 and G377 of ECL3 of GLP-1R. Images were generated using MolSoft ICM Browser software.
As noted above, 8 and related α/β-peptides have lower efficacy relative to GLP-1 in terms of cAMP production and β-arrestin recruitment, but the impact of α→β substitution on cAMP production is larger than the impact on β-arrestin recruitment. The results of the computational modeling raise the possibility that interactions between centrally located β residues on these α/β-peptides and ECL3 on the receptor may explain why efficacy for β-arrestin recruitment suffers less from α→β replacement than does efficacy for cAMP production.
CONCLUSIONS
We have shown that evaluation of a small family of peptide hormone derivatives containing α→β replacements can lead to discovery of agonists with significant signaling bias relative to the native hormone, despite the decline in overall signaling efficacy among the α/β-peptides relative to GLP-1 itself. This type of backbone-modified peptide is readily accessible via conventional solid-phase synthesis but has received relatively little attention from the perspective of exploring new agonists for Class B GPCRs. Promising results have recently been reported for α/β analogues of parathyroid hormone(1–34)47 and vasoactive intestinal peptide.48 Collectively, these recent studies and the work reported here suggest that backbone-modified analogues of peptide hormones may prove to be a rich source of new agonists that can serve as tools for elucidating the physiological consequences of specific GPCR-initiated signaling pathways; these unusual peptides may ultimately lead to therapeutic advances.
Our finding that a central segment of GLP-1 appears to be more critical for G protein-mediated activity than for β-arrestin recruitment at the GLP-1R is of general significance from the perspective of new strategies for discovery of biased agonists of this medically important receptor. In addition, this finding is intriguing in the context of the recent report of a G protein-biased analogue of exendin-427. Exendin-4 is a lizard-derived peptide that serves as a potent GLP-1R agonist and is approved for treatment of type 2 diabetes.23 The new α-peptide was identified via screening of a massive biosynthetic library of exendin-4 variants differing in the N-terminal segment.27 Thus, the bias induced by this combinatorially-derived peptide arises from changes at the N-terminus of exendin-4, an observation that contrasts with and is complemented by our finding that β-arrestin bias can be achieved via modifications in the central segment of GLP-1.
EXPERIMENTAL METHODS
Peptide Synthesis and Purification
Peptides were prepared by microwave-assisted solid phase peptide36 synthesis on ChemMatrix H-PAL amide resin based on Fmoc-protection of the main chain amino groups. Full details on peptide synthesis and purification can be found in the Supporting Information.
cAMP Production Assays
cAMP signaling was assessed in HEK293-derived cells (GS-22A)39 stably expressing the GloSensor cAMP reporter.40 These cells were a gift from Prof. Thomas Gardella at Massachusetts General Hospital. Human GLP-1R was transiently transfected into these cells (10 mg GLP-1R for 10 cm plate of cells) using FuGene HD transfection reagent (3:1 FuGene HD to GLP-1R). On the day of transfection, culture medium was replaced with antibiotic-free DMEM supplemented with 10% FBS, 4 mM L-glutamine and 1 mM sodium pyruvate. GLP-1R and FuGene HD were combined in OptiMEM (1 mL), and this mixture was incubated for 15 minutes before being added to cells. Cells were exposed to transfection reagents for 24 hours before being plated into opaque, clear-bottomed 96-well plates at 30,000 cells per well in antibiotic-free DMEM supplemented with 10% FBS, 4 mM L-glutamine and 1 mM sodium pyruvate. The plate was incubated for 24 hours. For cAMP concentration-response assays, cells were pre-incubated with D-PBS buffer containing D-luciferin (0.5 mM) until a stable background luminescence signal was achieved (30 minutes). Various doses of peptide were then added, and luminescence was measured for 30 minutes on a BioTek Synergy 2 plate reader. The maximal luminescence response (usually observed 16 – 20 minutes after peptide addition) was used to generate concentration-response curves.
Reported EC50 and maximum response values are the average of ≥3 independent experiments. Each experiment involved ≥7 different concentrations of GLP-1 or analogue, with solutions prepared via serial dilution of a stock solution of each peptide (usually 1 µM, which becomes 100 nM in the assay), with each resulting data point representing the average of two replicate wells. Changing pipette tips after each dilution was found to be critical for reproducible concentration-response curves. Both commercially obtained GLP-1 and GLP-1 we had prepared were tested in these assays, with identical results (Figure S2). GLP-1(7–36)NH2 and GLP-1(7–37)NH2 behaved identically in these assays (Figure S1).
Bioluminescence Resonance Energy Transfer Assays
Recruitment of β-arrestin-1 and β-arrestin-2 to the GLP-1R was assessed in HEK293FT cells, using a bioluminescence resonance energy transfer (BRET) assay based on a previous report. GLP-1R-Rluc8, GFP2-β-arrestin-1 or GFP2-β-arrestin-2(R393E, R395E), and GRK5 (which has previously been shown to enhance BRET2 signal in this assay22) were transiently co-transfected using polyethyleneimine (PEI). A 1:1 ratio of PEI:DNA was used, because cell viability was compromised at higher PEI:DNA ratios. On the day of transfection, culture medium was changed to DMEM with no supplements. DNA and PEI were combined in OptiMEM, and the mixture was incubated at room temperature for 20 minutes before being added to cells. After 6 hours, DMEM supplemented with 20% FBS was added to the cells. After 24 hours of transfection, cells were plated into opaque, white-bottomed 96-well plates and incubated for 24 hours.
Prior to BRET experiments, cells were rinsed twice with PBS and then incubated with D-PBS for 1 hr at 37 °C. Various concentrations of peptide were then added and incubated for 20 minutes at 37 °C. Rluc8 substrate DeepBlueC (60 mM in EtOH) was then added and incubated for 20–40 minutes before BRET signal (I515 nm/I410 nm) was measured on a Tecan Infinite 200 or BioTek Synergy 2 plate reader. Incubation with DeepBlueC prior to signal detection reduced signal variability, which presumably arises in the absence of pre-incubation because of variability in time required for DeepBlueC to penetrate cells and become available for oxidation by Rluc8. The BRET signal for GLP-1 usually remained constant over this time, though sometimes the signal for GLP-1 and analogues decreased slightly (to similar extents) over the course of an hour. In some experiments, we observed that the BRET signal for some α/β-peptides (7 and 10) to decrease slightly (~5% decrease relative to GLP-1) over the course of an hour. Thus, the BRET signal for each peptide was measured at the same time in replicate experiments.
Reported EC50 and maximum response values are the average of ≥3 independent experiments. Each experiment consists of ≥7 different concentrations of GLP-1 or analogue, with solutions prepared via serial dilution of the stock solution of each peptide (usually 12 µM, which becomes 1 µM in the assay), with each resulting data point representing the average of two replicate wells.
For competition BRET assays, various concentrations of α/β-peptide 4 were added to cells, and a constant concentration of GLP-1 (130 nM, which becomes 10 nM in the assay) was added immediately after addition of α/β-peptide. All subsequent steps were performed as above.
Full details on molecular biology operations that were employed to access BRET fusion protein constructs and on BRET transfection ratio optimization can be found in the Supporting Information.
Data Analysis
Data were processed using Microscoft Excel and GraphPad Prism 5 software. Concentration-response data for each experiment were normalized to the maximum signal observed for GLP-1(7–36)NH2 for that experiment. The normalized data points from each independent experiment were averaged to determine the average concentration-response behavior for GLP-1 and each analogue as depicted in concentration-response curves. EC50 and maximum response values were extracted from concentration-response data by fitting the concentration-response for each individual experiment to a sigmoidal dose-response model with variable slope, and then calculating the average and SEM for each individual experiment.
To quantify the efficacy of each peptide in the different pathways assayed, each concentration-response curve for each pathway (cAMP production, β-arrestin-1 recruitment and β-arrestin-2 recruitment) was fitted to an operational model of agonism,43,44 as shown in Equation 1. In Equation 1, [A] is the concentration of agonist, and E is the response at each tested [A] (i.e., the independent and dependent variables in the assay). As described in the main text, KA is the dissociation constant that characterizes the agonist binding to the receptor conformation induced by association with a given effector protein. τ is the efficacy of the agonist, a term that accounts for the potency of agonist action on the receptor and assay-dependent factors, including the density of receptors and strength of receptor-effector coupling.
| (Equation 1) |
For each peptide, the efficacy in each signaling pathway was extracted as the log(τ/KA) (mean ± SEM), and then compared to GLP-1 by subtracting the value of log(τ/KA) for GLP-1 from that of the peptide to yield Δlog(τ/KA) for the peptide. The Δlog(τ/KA) values obtained for the peptide relative to GLP-1 were complied to determine the average ± SEM Δlog(τ/KA) for the peptide, The bias factor (differential efficacy of a peptide relative to GLP-1 in two separate pathways) for each peptide was determined by subtracting the Δlog(τ/KA) in the arrestin pathway from the Δlog(τ/KA) for that peptide in the cAMP pathway, yielding ΔΔlog(τ/KA) for each peptide. Error in each ΔΔlog(τ/KA) value was calculated by propogating the error in the log(τ/KA) (SEM obtained from averaging individual Δlog(τ/KA) values) for the peptide and GLP-1 in each pathway (cAMP and either β-arrestin-1 recruitment or β-arrestin-2 recruitment).
To determine whether the ΔΔlog(τ/KA) for a given peptide was statistically significant, ΔΔlog(τ/KA) values were compared using one-way analysis of variance (ANOVA) followed by Dunnett’s post test. P < 0.05 denotes statistical significance.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences (NIGMS) (GM056414, to S.H.G.) and the National Health and Medical Research Council of Australia (NHMRC) (project grants [1061044] and [1065410], and NHMRC program grant [1055134] to P.M.S. and D.W.); P.M.S. is a NHMRC Principal Research Fellow. D.W. is a NHMRC Career Development Fellow. M.V.H. and L.M.J. were supported in part by a Chemical Biology Interface Training Grant from NIGMS (T32 GM008505). Support for this research was provided by the University of Wisconsin–Madison, Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation. The authors gratefully acknowledge Prof. Thomas Gardella for sharing cells containing the GloSensor cAMP plasmid, Jakob Lerche Hansen and Rasmus Jorgensen for sharing GLP1R-Rluc8, GRK5 and GFP2-β-Arrestin-2 plasmids, and Prof. Alan Attie and group members Dr. Mark Keller, Dr. Sushant Bhatnagar, Dr. Lindsay Wrighton and Donnie Stapleton for technical expertise and helpful discussion.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Additional details for experimental protocols, Figures S1-S19 (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): L.M.J. and S.H.G. are inventors on a patent application covering GLP-1 analogues described here; S.H.G. is a cofounder of Longevity Biotech, Inc., which is pursuing biomedical applications of α/β-peptides.
REFERENCES
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Nat.Rev. Mol. Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Shenoy SK, Lefkowitz RJ. Sci. STKE. 2005;308:cm10. doi: 10.1126/stke.2005/308/cm10. [DOI] [PubMed] [Google Scholar]
- 3.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Annu. Rev. Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 4.Kenakin T. Trends Pharm. Sci. 2003;24:346–354. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 5.Kobilka BK, Deupi X. Trends Pharm. Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Kenakin T, Christopoulos A. Nat. Rev. Drug Discov. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 7.Rajagopal S, Rajagopal K, Lefkowitz RJ. Nat. Rev. Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Annu. Rev. Pharmacol. Toxicol. 2012;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koole C, Wootten D, Simms J, Savage EE, Miller LJ, Christopoulos A, Sexton PM. J. Biol. Chem. 2012;287:3659–3673. doi: 10.1074/jbc.M111.309369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wootten D, Simms J, Miller LJ, Christopoulos A, Sexton PM. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5211–5216. doi: 10.1073/pnas.1221585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koole C, Wootten D, Simms J, Miller LJ, Christopoulos A, Sexton PM. J. Biol. Chem. 2012;287:3642–3658. doi: 10.1074/jbc.M111.309328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warne T, Edwards PC, Leslie AG, Tate CG. Structure. 2012;20:841–849. doi: 10.1016/j.str.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla AK, Singh G, Ghosh E. Trends Biochem. Sci. 2014;39:594–602. doi: 10.1016/j.tibs.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Baggio LL, Drucker DJ. Gastroenterology. 2007;132:2131–2157. doi: 10.1053/j.gastro.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 15.Hansen L, Deacon CD, Orskov C, Holst JJ. Endocrinology. 1999;140:5356–5363. doi: 10.1210/endo.140.11.7143. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen R, Kubale V, Vrecl M, Schwartz TW, Elling CE. J Pharmacol. Exp. Ther. 2007;322:148–154. doi: 10.1124/jpet.107.120006. [DOI] [PubMed] [Google Scholar]
- 17.Baggio LL, Huang Q, Brown TJ, Drucker DJ. Gastroenterology. 2004;127:546–558. doi: 10.1053/j.gastro.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 18.Willard FS, Sloop KW. Exp. Diabetes Res. 2012;2012:470851. doi: 10.1155/2012/470851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talbot J, Joly E, Prentki M, Buteau J. Mol. Cell. Endocrinol. 2012;364:65–70. doi: 10.1016/j.mce.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Quoyer J, Longuet C, Broca C, Linck N, Costes S, Varin E, Bockaert J, Bertrand G, Dalle S. J. Biol. Chem. 2010;285:1989–2002. doi: 10.1074/jbc.M109.067207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonoda N, Imamura T, Yoshizaki T, Babendure JL, Lu JC, Olefsky JM. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6614–6619. doi: 10.1073/pnas.0710402105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorgensen R, Martini L, Schwartz TW, Elling CE. Mol. Endocrinol. 2005;19:812–823. doi: 10.1210/me.2004-0312. [DOI] [PubMed] [Google Scholar]
- 23.Robles GI, Singh-Franco Drug Des. Dev. Ther. 2009;3:219–240. doi: 10.2147/dddt.s3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudsen SM, Nielsen PF, Huusfeldt PO, Johansen NL, Madsen K, Pederson FZ, Thorgerson H, Wilken M, Agerso H. J. Med. Chem. 2000;43:1664–1669. doi: 10.1021/jm9909645. [DOI] [PubMed] [Google Scholar]
- 25.Steensgaard DB, Thomsen JK, Olsen HB, Knudsen SM. Diabetes. 2008;51(Suppl. 1):A164. [Google Scholar]
- 26.Wootten D, Reynolds CA, Smith KJ, Mobarec JC, Koole C, Savage EE, Pabreja K, Simms J, Sridhar R, Furness SGB, Lie M, Thompson PE, Miller LJ, Christopoulos A, Sexton PM. Cell. 2016;165:1632–1643. doi: 10.1016/j.cell.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Sturchler E, Zhu J, Nieto A, Cistrone PA, Xie J, He L, Yea K, Jones T, Turn R, Di Stefano PS, Griffin PR, Dawson PE, McDonald PH, Lerner RA. Nat. Comm. 2015;6:8918. doi: 10.1038/ncomms9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wootten D, Savage EE, Willard FS, Bueno AB, Sloop KW, Christopoulos A, Sexton PM. Mol. Pharmacol. 2013;83:822–834. doi: 10.1124/mol.112.084525. [DOI] [PubMed] [Google Scholar]
- 29.Koole C, Wootten D, Simms J, Valant C, Sridhar R, Woodman OL, Miller LJ, Summers RJ, Christopoulos A, Sexton PM. Mol. Pharmacol. 2010;78:456–465. doi: 10.1124/mol.110.065664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Appleton KM, Luttrell LM. J. Recept. Signal Transduct. Res. 2013;33:153–161. doi: 10.3109/10799893.2013.769004. [DOI] [PubMed] [Google Scholar]
- 31.Whalen EJ, Rajagopal S, Lefkowitz RJ. Trends Mol. Med. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denton EV, Craig CJ, Pongratz RL, Appelbaum JS, Doerner AE, Narayanan A, Shulman GI, Cline GW, Schepartz A. Org. Lett. 2013;15:5318–5321. doi: 10.1021/ol402568j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briochhagen J, Podewin T, Meyer-Brig H, von Olhen Y, Johnston NR, Jones BJ, Bloom SR, Rutter GA, Hoffman-Roder A, Hodson DJ, Trauner D. Angew. Chem. Int. Ed. 2015;54:15565–15569. doi: 10.1002/anie.201506384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai X, Niu Y, Zhu J, Yang AQ, Wu YF, Ye XS. Bioorg. Med. Chem. 2016;24:1163–1170. doi: 10.1016/j.bmc.2016.01.036. [DOI] [PubMed] [Google Scholar]
- 35.Johnson LM, Barrick S, Hager MV, McFedries A, Homan EA, Rabaglia ME, Keller MP, Attie AD, Saghatelian A, Bisello A, Gellman SH. J. Am. Chem. Soc. 2014;136:12848–12851. doi: 10.1021/ja507168t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horne SW, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Proc. Natl. Acad. Sci. USA. 2009;106:14751–14756. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Underwood CR, Garibay P, Knudsen LB, Hastrup S, Peters GH, Rudolph R, Reedtz-Runge S. J. Biol. Chem. 2010;285:723–730. doi: 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deacon CF, Madsen K, Wiberg FC, Jacobsen O, Holst JJ. Diabetologia. 1998;41:271–278. doi: 10.1007/s001250050903. [DOI] [PubMed] [Google Scholar]
- 39.Hupe-Sodmann K, McGregor GP, Bridgenbaugh R, Goke R, Goke B, Thole H, Zummermann B, Voigt K. Regul. Pept. 1995;58:149–156. doi: 10.1016/0167-0115(95)00063-h. [DOI] [PubMed] [Google Scholar]
- 40.de Menthiere CS, Chavanieu A, Grassy G, Dalle S, Salazar G, Kervran A, Pfieiffer B, Renard P, Delagrange P, Manachez D, Bakes D, Ktorza A, Calas B. Eur. J. Med. Chem. 2004;39:473–480. doi: 10.1016/j.ejmech.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Carter PH, Dean T, Bhayana B, Khatri A, Rajur R, Gardella TJ. Mol. Endocrinol. 2015;29:307–321. doi: 10.1210/me.2014-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Binkowski BF, Butler BL, Stecha PF, Eggers CT, Otto P, Zimmerman K, Vidugiris G, Wood MG, Encell LP, Fan F, Wood KV. ACS Chem. Biol. 2011;6:1193–1197. doi: 10.1021/cb200248h. [DOI] [PubMed] [Google Scholar]
- 43.Black JW, Leff P. Proc. R. Soc. Lond. 1983;220:121. doi: 10.1098/rspb.1983.0093. [DOI] [PubMed] [Google Scholar]
- 44.Kenakin T, Watson C, Muniz-Medina V, Christopoulos A, Novick S. ACS Chem. Neurosci. 2012;3:193–203. doi: 10.1021/cn200111m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller LJ, Chen Q, Lam PC, Pinon DI, Sexton PM, Abagyan R, Dong M. J. Biol. Chem. 2011;286:15895–15907. doi: 10.1074/jbc.M110.217901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wootten D, Reynolds CA, Koole C, Smith KJ, Mobarec JC, Simms J, Quan T, Coudrat T, Furness SGB, Miller LJ, Christopoulos A, Sexton PM. Mol. Pharmacol. 2016;89:335–347. doi: 10.1124/mol.115.101246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheloha RW, Maeda A, Dean T, Gardella TJ, Gellman SH. Nat. Biotechnol. 2014;32:653–655. doi: 10.1038/nbt.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olson KE, Kosloski-Bilek LM, Anderson KM, Diggs BJ, Clark BE, Gledhill JM, Jr, Shandler SJ, Mosley RL, Gendelman HE. J. Neurosci. 2015;35:16463–16470. doi: 10.1523/JNEUROSCI.2131-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.