Abstract
Arabidopsis is emerging as a model system to study the biochemistry, biological functions, and evolution of plant terpene secondary metabolism. It was previously shown that the Arabidopsis genome contains over 30 genes potentially encoding terpene synthases (TPSs). Here we report the characterization of a monoterpene synthase encoded by two identical, closely linked genes, At3g25820 and At3g25830. Transcripts of these genes were detected almost exclusively in roots. An At3g25820/At3g25830 cDNA was expressed in Escherichia coli, and the protein thus produced was shown to catalyze the formation of 10 volatile monoterpenes from geranyl diphosphate, with 1,8-cineole predominating. This protein was therefore designated AtTPS-Cin. The purified recombinant AtTPS-Cin displayed similar biochemical properties to other known monoterpene synthases, except for a relatively low Km value for geranyl diphosphate of 0.2 μm. At3g25820/At3g25830 promoter activity, measured with a β-glucuronidase (GUS) reporter gene, was primarily found in the epidermis, cortex, and stele of mature primary and lateral roots, but not in the root meristem or the elongation zone. Although the products of AtTPS-Cin were not detected by direct extraction of plant tissue, the recent report of 1,8-cineole as an Arabidopsis root volatile (Steeghs M, Bais HP, de Gouw J, Goldan P, Kuster W, Northway M, Fall R, Vivanco JM [2004] Plant Physiol 135: 47–58) suggests that the enzyme products may be released into the rhizosphere rather than accumulated. Among Arabidopsis TPSs, AtTPS-Cin is most similar to the TPS encoded by At3g25810, a closely linked gene previously shown to be exclusively expressed in flowers. At3g25810 TPS catalyzes the formation of a set of monoterpenes that is very similar to those produced by AtTPS-Cin, but its major products are myrcene and (E)-β-ocimene, and it does not form 1,8-cineole. These data demonstrate that divergence of organ expression pattern and product specificity are ongoing processes within the Arabidopsis TPS family.
Terpenes comprise a very large class of plant secondary metabolites that serve a variety of different functions in basic and specialized metabolism. The majority of these compounds are likely to operate as direct defense agents because they often exhibit toxic and deterrent effects against plant pathogens and herbivores (Langenheim, 1994; Gershenzon and Kreis, 1999). However, since all monoterpenes, many sesquiterpenes, and a few diterpenes have high vapor pressure at ambient temperature and volatilize easily, these compounds could also act at a distance from the plant, serving as attractants or deterrents for pollinators and herbivores (Dobson, 1994; Dudareva and Pichersky, 2000; Pichersky and Gershenzon, 2002), or as signals released after damage by herbivores to attract their predators and parasitoids (Pare and Tumlinson, 1999; Dicke and Van Loon, 2000; Kessler and Baldwin, 2001). In fact, volatile monoterpenes and sesquiterpenes have frequently been shown to be emitted from flowers and other aerial parts of the plant, and their release is often developmentally regulated or induced by damage (Chen et al., 2003b; Dudareva et al., 2003; Arimura et al., 2004). Curiously, volatile monoterpenes and sesquiterpenes have also been reported to be synthesized and accumulated in roots and rhizomes of various plant species (Wichtmann and Stahl-Biskup, 1987; Bos et al., 2002; Kovacevic et al., 2002). As a consequence of their antimicrobial and antiherbivore activity, these substances may serve as direct defenses of below-ground tissue when they accumulate. However, if volatile terpenes were released from roots, they could also play interesting roles in interactions with other rhizosphere organisms analogous to roles of terpenes emitted from aerial parts.
Monoterpenes contain a 10-carbon backbone and are all synthesized from the common precursor geranyl diphosphate (GPP) by the action of various monoterpene synthases (Wise and Croteau, 1999). In a similar way, sesquiterpenes (C15) and diterpenes (C20) are synthesized from farnesyl diphosphate (FPP) and geranylgeranyl diphosphate, respectively, by sesquiterpene and diterpene synthases (Bohlmann et al., 1998; Davis and Croteau, 2000). One characteristic feature of most terpene synthases (TPSs) is the formation of multiple products from a single substrate (Steele et al., 1998). All plant TPSs are related to each other, and thus form a large family of proteins (Martin et al., 2004). Comparative sequence analyses have shown that, within the same plant genome, TPSs making different products may be more similar to each other than TPSs making the same products in different species, suggesting that convergent evolution is common in this family of proteins (Bohlmann et al., 1998; Aubourg et al., 2002).
The Arabidopsis genome contains more than 30 genes that encode proteins with sequence similarity to the TPS family (Aubourg et al., 2002). Within this group, phylogenetic analysis identified a subset of seven genes (but only six sequences, since two genes in the database have identical sequences) that are most closely related to each other and to monoterpene synthases from other species (Aubourg et al., 2002). Heterologous expression of cDNAs of four of these genes, At3g25810, At1g61680, At4g16740, and At2g24210 in Escherichia coli, followed by enzyme assays, have shown that they indeed encode monoterpene synthases (Bohlmann et al., 2000; Chen et al., 2003b; Fäldt et al., 2003). In the plant, expression of these four genes occurs exclusively in the aerial parts of ecotype Columbia (Col), with some of these genes expressed exclusively in the flowers (Chen et al., 2003b). However, preliminary data indicated that a fifth TPS gene in this subfamily, corresponding to the two identical sequences of At3g25820 and At3g25830, is primarily expressed in root tissue (Chen et al., 2003b). Here we demonstrate that these genes both encode a TPS whose principal product is the volatile monoterpene 1,8-cineole. We further show that these genes are expressed in specific types of root cells, but never in the root tip, and that their expression seems to change during the course of root growth. Although we could not measure 1,8-cineole accumulation in the root tissue of Arabidopsis, it was very recently found that this oxygenated monoterpene is emitted from Arabidopsis roots (Steeghs et al., 2004), suggesting that 1,8-cineole is released directly into the rhizosphere without accumulation.
RESULTS
The Arabidopsis cDNAs of At3g25820 and At3g25830 Are Identical and Have High Sequence Similarity to Other Monoterpene Synthases
The Arabidopsis genome database shows three tandemly linked TPS genes, At3g25810, At3g25820, and At3g25830 on chromosome 3 (Fig. 1). Gene At3g25810 was previously found to be expressed exclusively in flowers and to encode a TPS that catalyzes the formation mostly of myrcene and (E)-β-ocimene (Chen et al., 2003b). The two other genes are identical in sequence (Aubourg et al., 2002). Furthermore, the annotation of this region by The Arabidopsis Information Resource (TAIR; http://Arabidopsis.org) shows two identical blocks of 8,262 nucleotides. Each block encompasses not only a TPS gene but also part of an adjacent sequence of the pseudo non-long-terminal-repeat retroelements At3g25815 and At3g25826, respectively (Fig. 1). The duplication of these gene elements was confirmed by genomic PCR analysis (data not shown). To determine the catalytic activity of the enzyme encoded by the identical TPS genes At3g25820 and At3g25830, we first isolated a full-length cDNA by reverse transcription (RT)-PCR from isolated root mRNA. Because the sequences of these two genes, including the promoter region, are identical, it is not possible to determine which gene was the source of the transcript we converted into cDNA in our RT-PCR procedure, and the cDNA will therefore be referred to henceforth as At3g25820/At3g25830 cDNA. This cDNA encodes a protein of 600 amino acids, with 78% sequence identity to the TPS encoded by At3g25810, and 27% to 63% identity to the other identified Arabidopsis monoterpene synthases (Fig. 2).
Figure 1.
Organization of the three monoterpene synthase genes At3g25810, At3g25820, and At3g25830 on Arabidopsis chromosome 3. The location of the two identical blocks of sequences is indicated.
Figure 2.
Alignment of deduced amino acid sequences of the five presently identified monoterpene synthases of Arabidopsis and a 1,8-cineole synthase from S. officinalis. AtTPS-Cin, encoded by the identical genes At3g25820 and At3g25830, is shown on top. At5g25810 encodes a monoterpene synthase whose main products are myrcene and (E)-β-ocimene, and it has 78% sequence identity to AtTPS-Cin. At2g24210 is a myrcene/(E)-β-ocimene synthase (shown here is the protein of ecotype C24; accession no. AF178535), with 63% sequence identity to AtTPS-Cin. At4g16740 encodes an (E)-β-ocimene synthase (protein of ecotype C24; accession no. AY151086), with 52% sequence identity to AtTPS-Cin. At1g61680 is a linalool synthase, with 27% sequence identity to AtTPS-Cin. S. officinalis cineole synthase (accession no. AF051899) shares 45% sequence identity with AtTPS-Cin. Residues shaded in black indicate conserved identical residues in at least four out of six sequences shown. Dashes indicate gaps inserted for optimal alignment. The horizontal line marks the putative N-terminal transit peptide. The arrowhead indicates the truncation site of the expressed recombinant protein encoded by At3g25820/At3g25830. The conserved RRX8W and DDXXD motifs are marked with frames. The dashed horizontal line indicates a region of 74 amino acids which, when swapped from S. officinalis cineole synthase into the corresponding region of S. officinalis sabinene synthase, enabled the formation of 1,8-cineole (Peters and Croteau, 2003).
The At3g25820/At3g25830-Encoded Protein Expressed in E. coli Converts GPP to 1,8-Cineole as the Principal Product
Because sequence analysis indicated that the protein encoded by At3g25820 and At3g25830 contains a transit peptide (Aubourg et al., 2002), a truncated cDNA coding for an open reading frame that started with an added Met codon immediately upstream of the two Arg codons (positions 47 and 48) of the conserved RRX8W motif was inserted into the E. coli expression vector pCRT7/CT-TOPO (Invitrogen, Carlsbad, CA). The removal of the first 46 codons was based on previous observations that the N terminus of the mature part of many TPSs begins at or slightly upstream of the double-Arg motif, and that a truncated TPS that includes the two Args at its N terminus shows maximal activity (Williams et al., 1998).
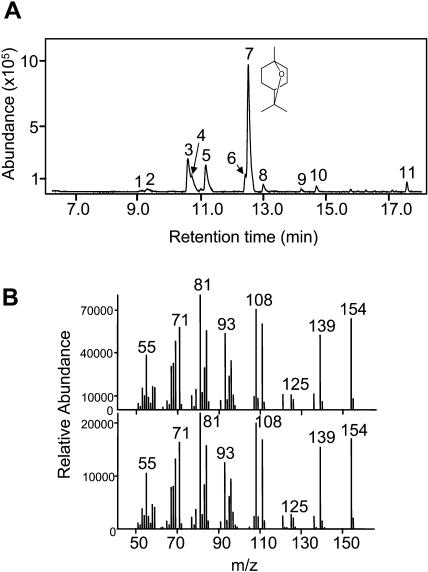
The crude protein extracts from bacterial cultures expressing the truncated At3g25820/At3g25830-encoded protein were incubated with GPP or FPP in assay buffer as previously described for other TPSs (Bohlmann et al., 2000; Chen et al., 2003b; Fäldt et al., 2003), and the compounds produced in these reactions were analyzed by gas chromatography-mass spectrometry (GC-MS). While the enzyme was essentially inactive with FPP, it catalyzed the formation of multiple monoterpenes using GPP as substrate (Fig. 3A). The major enzymatic product was identified as 1,8-cineole, which accounted for 52% of the total reaction products (Fig. 3, A and B). Besides 1,8-cineole, nine minor products were identified, including (+)-α-thujene (0.6%), (−)-(1S)-α-pinene (1.9%), sabinene [14.5%; (−)-(1S):(+)-(1R), 3:1], (−)-(1S)-β-pinene (7.8%), myrcene (13.3%), (−)-(4S)-limonene (4.0%), (E)-β-ocimene (2.7%), terpinolene (0.8%), and α-terpineol [2.4%; (−)-(4S):(+)-(4R), 4.6:1] (Fig. 3A). Because 1,8-cineole was the major product of this enzyme, we designate it as a 1,8-cineole synthase (AtTPS-Cin). To test if the product profile of the multiproduct AtTPS-Cin enzyme was affected by the nature of the recombinant expression system, the cDNA was also expressed in the pET101/D-TOPO vector (Invitrogen), yielding a similar product profile with 1,8-cineole accounting for 62% of total product, followed by sabinene (18%) and α-terpineol (11%).
Figure 3.
Identification of the monoterpene products obtained with GPP as substrate in a reaction catalyzed by AtTPS-Cin synthesized in E. coli following expression of the truncated cDNA of At3g25820/At3g25830. A, Gas chromatographic separation of products formed in the in vitro reaction. Peak 1, (+)-α-thujene; peak 2, (−)-(1S,5S)-α-pinene; peak 3, sabinene [75% (−)-(1S), 25% (+)-(1R)]; peak 4, (−)-(1S,5S)-β-pinene; peak 5, myrcene; peak 6, (−)-(4S)-limonene; peak 7, 1,8-cineole; peak 8, (E)-β-ocimene; peak 9, terpinolene; peak 10, linalool (racemic, also produced in assay of control E. coli cultures transformed with the empty vector); peak 11, α-terpineol [82% (−)-(4S), 18% (+)-(4R)]. B, Mass spectra of peak 7 (top) and an authentic 1,8-cineole standard (bottom).
Biochemical Properties of At3g25820/At3g25830-Encoded AtTPS-Cin
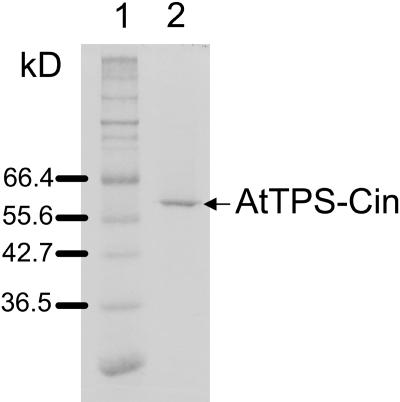
The E. coli-expressed mature AtTPS-Cin was purified and its biochemical properties determined. The crude enzyme preparation was subjected to a combination of DEAE and Mono-Q ion-exchange chromatography to yield a nearly homogeneous protein with a molecular mass of approximately 60 kD (Fig. 4). The enzyme showed a pH optimum at 8.0. In the absence of a divalent metal ion, AtTPS-Cin was inactive, but activity was restored by provision of either Mg2+ or Mn2+. The maximum activity levels with either ion were nearly identical, but with Mg2+, 40 mm were required to achieve these levels, while with Mn2+, the enzyme was fully active at a concentration of 1 mm. The specific activity of Arabidopsis AtTPS-Cin with GPP was 10.9 ± 0.4 pkat mg−1 protein, giving a kcat of 0.001 s−1. The enzyme had an apparent Km value for GPP of 0.2 ± 0.03 μm.
Figure 4.
Purification of AtTPS-Cin from an E. coli expression system. An aliquot (1 μg) of AtTPS-Cin from the Mono-Q fraction with the highest activity was separated on a 10% (w/v) SDS-polyacrylamide gel (lane 2). The gel was stained with Coomassie Brilliant Blue. The positions of protein molecular mass markers are indicated in lane 1.
At3g25820/At3g25830 Are Expressed in Specific Tissues of the Roots
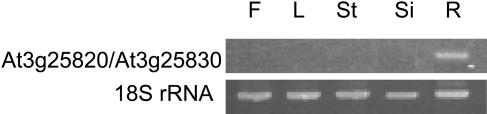
In a preliminary RT-PCR characterization of the entire family of TPS genes in Arabidopsis (ecotype Col), we reported that At3g25820/At3g25830 transcripts were found in roots and at much lower levels in siliques also, but not in leaves, flowers, or stems under normal growing conditions (Chen et al., 2003b). Since the promoter regions of both genes are completely identical, it is reasonable to assume that both genes are transcribed at equal rates. Additional RT-PCR experiments showed that in 6-week-old bolted Col plants, At3g25820/At3g25830 were expressed exclusively in roots (Fig. 5). A very similar expression pattern of At3g25820/At3g25830 was observed using gene-specific longmer-oligonucleotide microarrays with the Landsberg erecta ecotype (Ler), but, in addition to roots, comparable expression of At3g25820/At3g25830 was detected in flowers of Ler (J. Ehlting and J. Bohlmann, unpublished data). Gene expression in the aerial parts of prebolting Col plants could not be induced by treatments including the application of jasmonic acid, the 12-oxo-phytodienoic acid/coronatine analog 6-ethyl indanoyl-l-Ile, which has been shown to act as a strong elicitor of induced terpene volatile emission in plants (Schuler et al., 2001), the fungal peptaibol elicitor alamethicin (Engelberth et al., 2001), and damage by Plutella xylostella larvae (data not shown).
Figure 5.
RT-PCR analysis of At3g25820/At3g25830 gene expression. RNA was extracted from roots (R), flowers (F), leaves (L), stems (St), and siliques (Si) of mature 6-week-old plants. PCR reactions with primers for 18S rRNA were performed to judge the quantitative equality of the RNA templates used for various samples.
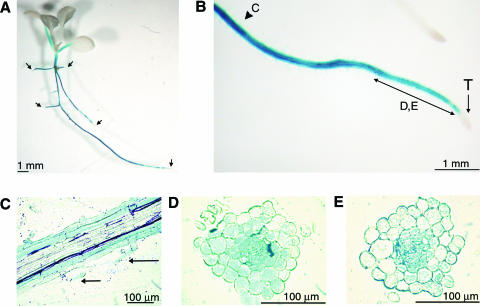
We looked at the expression of At3g25820/At3g25830 in roots and seedlings in more detail by examining the staining patterns of transgenic Arabidopsis Col plants carrying an At3g25820/At3g25830 promoter::β-glucuronidase (GUS) fusion construct. Five to 10 seedlings of four independent T2 lines showed consistently strong GUS staining in the mature primary and lateral roots. However, GUS activity was never observed in a 0.2- to 0.5-cm-long zone situated at the tip of the root, which includes the apical meristem and elongation zone (Fig. 6, A and B). The staining pattern of intact roots in comparison to that of transverse and lateral root sections suggested a developmental change in the location of At3g25820/At3g25830 promoter activity. Whereas younger root sections often showed GUS staining primarily in the vascular system, in older root zones, strong GUS activity was exclusively or additionally observed in the cortex and epidermal cell layer (Fig. 6, B–E). Staining was found with less consistency in root hairs (Fig. 6, B and C). In addition to the roots, GUS staining also occurred in the hypocotyl and, to a lesser degree, in the petioles of the seedling, but the rest of the aerial parts were not stained (Fig. 6A). GUS activity was not affected by the addition of Suc or kanamycin to the medium. In mature plants, GUS staining was also observed in roots, but not in stems or flowers (data not shown). Control lines carrying the promoterless GUS insertion cassette did not show any GUS activity.
Figure 6.
Histochemical analysis of GUS activity of transgenic Arabidopsis (ecotype Col) seedlings transformed with an At3g25820/At3g25830 promoter::uidA gene fusion construct. A, GUS staining was observed in the hypocotyl, petioles of cotyledons, and primary and lateral roots, except for root tips as indicated by arrows. B, GUS staining of different growth zones of a primary root. C to E, Growth zones of which detailed sections were obtained as shown in C to E, respectively. C, GUS activity in a longitudinal section of an older root growth zone. Staining occurs in the cortex and the epidermis, including root hairs, marked with arrows. D and E, GUS activity in transversal sections of younger root growth zones occurs primarily in the vascular tissue with a transition to cortex and epidermal cells. T, Root tip.
The products of AtTPS-Cin were sought in planta by solvent extraction and solid-phase microextraction of ground roots of Arabidopsis plants grown in soil or under hydroponic conditions. However, neither 1,8-cineole nor any of the other enzyme products were detectable by GC-MS analysis above the level of 10 to 20 ng g−1 fresh weight. Two AtTPS-Cin products, myrcene and (−)-limonene, have previously been found in headspace collections of aerial parts of Arabidopsis (ecotype Col; Chen et al., 2003b). However, these are unlikely to represent the activity of AtTPS-Cin since At3g25820/At3g25830 expression is almost completely absent in aerial parts (Fig. 5), and myrcene and (−)-limonene are also known to be products of other Arabidopsis TPSs, including the enzyme encoded by At3g25810 (Bohlmann et al., 2000; Chen et al., 2003b; Fäldt et al., 2003).
DISCUSSION
At3g25820 and At3g25830 Encode a Root-Specific AtTPS-Cin
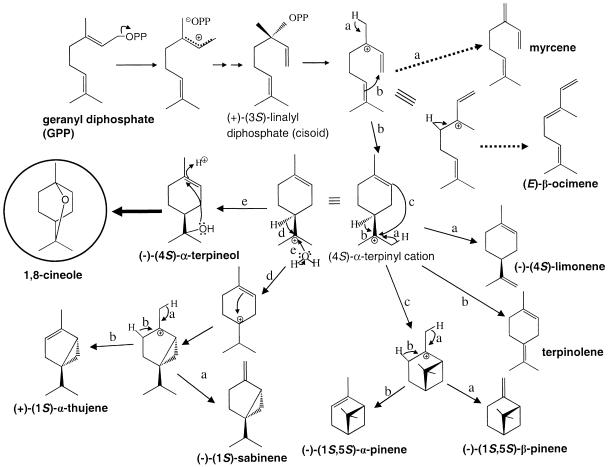
While much research has focused on terpenoid biosynthesis in aerial parts of plants, little is known about the formation of terpenes in below-ground organs. Here we show that the tandem Arabidopsis genes At3g25820 and At3g25830, with identical open reading frames and promoter regions, encode a root-localized monoterpene synthase that produces the cyclic ether 1,8-cineole as its principal product when expressed in E. coli. The enzyme also catalyzes the formation of nine other monoterpenes (eight olefins and one alcohol) as minor products, and has properties generally similar to those of other monoterpene synthases. An exception is the kcat value, which is approximately one order of magnitude lower than the turnover rates usually reported for plant monoterpene and sesquiterpene synthases (Cane, 1999; Wise and Croteau, 1999). However, the apparent Km value for the substrate GPP (0.2 μm) is also lower than apparent Km values for most other enzymes of this class, including cineole synthase from Salvia officinalis (Croteau et al., 1994; Wise et al., 1998). The product distribution of cineole synthase from S. officinalis includes the olefins α-pinene, β-pinene, sabinene, and limonene and hence shows some similarity to the product profile of the corresponding Arabidopsis enzyme. However, AtTPS-Cin forms twice as many olefins as the S. officinalis enzyme and the reaction products primarily belong to the opposite antipodal series (Fig. 7). This suggests that the enzyme reaction catalyzed by AtTPS-Cin involves the cyclization of a bound (3S)-linalyl diphosphate intermediate into a (4S)-α-terpinyl cation with similar stereochemistry to that determined for (−)-(4S)-limonene synthase from spearmint (Davis and Croteau, 2000).
Figure 7.
Comparison of the products formed by AtTPS-Cin and the monoterpene synthase encoded by the closely related gene At3g25810, and proposed mechanisms of the reactions involved. GPP is ionized in the presence of a divalent metal ion. All compounds are produced by both enzymes, except 1,8-cineole, which is synthesized by AtTPS-Cin only. The major products formed by At3g25810 [myrcene, (E)-β-ocimene] are shown with bold broken arrows. Formation of mono- and bicyclic reaction products involves the isomerization of GPP to a linalyl diphosphate intermediate. Acyclic products might be formed via carbocations derived from either GPP or linalyl diphosphate. OPP, Diphosphate moiety.
The nearly exclusive root-specific expression of At3g25820 and At3g25830 was revealed by RT-PCR and promoter::GUS fusion analysis. To our knowledge, this is the first report of a functionally characterized root-specific monoterpene synthase gene. Among sesquiterpene synthases, a gene encoding an epi-aristolochene synthase from Nicotiana attenuata was shown to be expressed in roots (Bohlmann et al., 2002), while a number of the uncharacterized root-expressed TPS genes of Arabidopsis are likely to encode diterpene synthases (Aubourg et al., 2002; Chen et al., 2003b).
1,8-Cineole and Other At3g25820/At3g25830-Encoded Enzyme Products Are Not Accumulated in Plants, but Are Likely Released from the Roots as Volatiles
Our inability to detect 1,8-cineole and other products of AtTPS-Cin in Arabidopsis roots is puzzling given the high apparent level of At3g25820/At3g25830 gene expression in this organ. Due to the presence of a potential plastidic transit peptide, AtTPS-Cin is most likely located in root plastids that have been shown, in other plant species, to contain an active methylerythritol phosphate pathway for formation of the C5 building blocks of terpenoids (Hans et al., 2004). Cineole biosynthesis rates in Arabidopsis roots might be limited by the level of enzymatically active AtTPS-Cin protein or the availability of the substrate GPP in root plastids, although AtTPS-Cin showed a very low apparent Km value for GPP in vitro.
On the other hand, our findings are consistent with previous studies on Arabidopsis aerial parts where, despite high expression of several TPS genes (Chen et al., 2003b), only trace amounts of monoterpenes and sesquiterpenes were found to accumulate (A. Farooq and D. Tholl, unpublished data). Instead, monoterpenes and sesquiterpenes were found to be released from aerial parts, especially flowers, at readily detectable levels (Chen et al., 2003b). Conceivably, monoterpenes formed in Arabidopsis roots might also be released as volatiles. In fact, 1,8-cineole was just recently detected as a volatile compound emitted from hairy-root cultures of Arabidopsis using a very sensitive method employing proton transfer reaction mass spectrometry (Steeghs et al., 2004).
The volatilization of 1,8-cineole and other AtTPS-Cin products from Arabidopsis roots would be consistent with the overall staining pattern observed in the At3g25820/At3g25830 promoter::GUS fusion transformants. Expression is strongest in the outer cell layers, the cortex, and the epidermis of mature roots, from which these lipophilic compounds could readily diffuse or be released directly into the atmosphere. In younger roots, GUS staining was weaker and localized in the vascular system, while staining was completely absent from the root tip. These observations are supported by recent cell type-specific microarray expression profiling of Arabidopsis roots, which showed that transcripts of At3g25820/At3g25830 were detected primarily in fully elongated epidermal and lateral root cap cells (localized expression domain group 6; Birnbaum et al., 2003).
The lack of accumulation of 1,8-cineole and other AtTPS-Cin products in the plant may also be attributed to their rapid conversion to other metabolites. However, except for limonene and sabinene, bona fide metabolites of AtTPS-Cin monoterpenes have only rarely been reported from plant species (Wise and Croteau, 1999).
AtTPS-Cin Products May Function as Defensive or Allelopathic Agents
1,8-Cineole and several other AtTPS-Cin products have long been known to be active against plant pests. For example, 1,8-cineole is toxic and deterrent to certain insect and mammalian herbivores (Tripathi et al., 2001; Wiggins et al., 2003), and also has antimicrobial activity (Hammer et al., 2003; Pina-Vaz et al., 2004). Indeed, in Arabidopsis hairy-root cultures, infection with compatible pathogens, the bacterium Pseudomonas syringae strain DC3000 or the fungus Alternaria brassicola, triggered increased emission of 1,8-cineole (Steeghs et al., 2004) as an expected response if this compound serves as a defense against microbes. 1,8-Cineole also exhibits allelopathic activity, reducing the germination and growth of a variety of plant species (Fischer, 1991; Romagni et al., 2000; Singh et al., 2002). Thus, the products of AtTPS-Cin may play a role in protecting Arabidopsis from its enemies and increase its ability to compete below-ground with neighboring plants for limited water and nutrient supplies. However, the effectiveness of these monoterpenes is hard to assess without knowing more about the rate of their release from intact roots growing in soil. Comparison of wild-type Arabidopsis with lines overexpressing At3g25820/At3g25830 or knocked out in these genes in bioassays with pathogens or competitors should help establish the biological role(s) of AtTPS-Cin in the plant.
In evaluating the biological significance of AtTPS-Cin products, it will be important to keep in mind that these compounds do not appear to accumulate in plant tissue, but are instead released from the roots. A large variety of primary and secondary metabolites are known to be exuded from the roots of many plant species (Walker et al., 2003a), including Arabidopsis (Narasimhan et al., 2003; Walker et al., 2003b), though terpenes are poorly represented in these observations and have never been reported to exude from Arabidopsis. Exudation of metabolites may be critical for their biological function (Bais et al., 2004). For example, allelopathic agents must contact seeds or roots of other plants to inhibit their growth (Bais et al., 2003). Exudation of antiherbivore defenses may allow plants to deter subterranean herbivores in the rhizosphere even before they initiate feeding. With respect to microorganisms, exudation of specific metabolites may permit plants to block the growth of pathogenic microorganisms in the immediate vicinity of roots while promoting growth of beneficial microorganisms involved in symbiotic or protective interactions.
Besides being of direct benefit to plants, exudates could also serve as signals for a diverse group of rhizosphere organisms. For example, a group of sesquiterpene lactones that are released in low amounts from roots of several different plant species, including sorghum, maize, and rice, trigger the germination of the parasitic weeds Orobanche and Striga spp. (Bouwmeester et al., 2003). The bulk of exuded root metabolites reported have a high solubility in water. By contrast, terpenes are generally lipophilic compounds with low water solubility. For example, the solubility of monoterpene olefins, such as the AtTPS-Cin products, limonene, myrcene, and (E)-β-ocimene, is very low in water (<20 ppm), though that of oxygenated monoterpenes, such as 1,8-cineole, is significantly higher (332 ppm; Weidenhamer et al., 1993). Yet most monoterpenes have high vapor pressures, and this may allow them to move rapidly through the soil in the gaseous phase and might explain their lack of detection in root exudates. In many soil types, gas phase movement may be more rapid and predictable than transport by water under typical conditions.
At3g25820 and At3g25830 Are Closely Related to the Flower-Specific Monoterpene Synthase Gene At3g25810
The close chromosomal proximity of At3g25810 to At3g25820 and At3g25830 and their high levels of coding sequence similarity (78% identity on the protein level), as well as intron number, position, and sequence, suggest that a relatively recent tandem duplication gave rise to a pair of genes composed of At3g25810 and the At3g25820/At3g25830 progenitor, followed by an even more recent tandem duplication that gave rise to the two genes At3g25820 and At3g25830, which are presently still identical (Aubourg et al., 2002). However, the promoter region of At3g25810 upstream of the presumed TATA box (present at approximately 65 nucleotides upstream of the initiating ATG codon) shows practically no sequence similarity to the corresponding promoter region of genes At3g25820 and At3g25830. This observation could explain their divergent modes of expression. It is not clear, however, whether the original tandem duplication did not include promoter regions, or if the promoter was part of the initial duplicated region and subsequent mutations and selection resulted in the differences in the promoter regions observed today.
In addition to divergence in the mode of expression, genes At3g25810 and At3g25820/At3g25830 have also diverged with respect to the enzymatic reaction catalyzed by the proteins they encode. The major product of the reaction catalyzed by AtTPS-Cin is 1,8-cineole, which is not produced by the At3g25810-encoded protein. However, the nine other enzyme products of AtTPS-Cin (Fig. 3) are also produced by the At3g25810 protein (Fig. 7). The TPS encoded by gene At3g25810 was originally reported to catalyze the formation of myrcene and (E)-β-ocimene principally, plus six other monoterpenes (Chen et al., 2003b). When the products were reanalyzed for the present study, the enzyme was also found to produce small amounts of (+)-α-thujene and α-terpineol (data not shown; Fig. 7). Although AtTPS-Cin and the At3g25810-encoded TPS are 22% divergent on the protein level, they appear to have a very similar mechanism of action (Fig. 7). The differences between the two enzymes lie in the apparent ability of AtTPS-Cin to promote the capture of a water molecule by the α-terpinyl carbocation at a greater rate than the At3g25810-encoded enzyme. This reaction step leads via deprotonation to the formation of α-terpineol, a minor product of both enzymes. In AtTPS-Cin, most of the α-terpineol undergoes internal addition, possibly facilitated by protonation of the double bond, and a further deprotonation to generate the cyclic ether function of 1,8-cineole.
Peters and Croteau (2003) have recently identified a region of approximately 70 amino acid residues in the S. officinalis cineole synthase which, when swapped into the corresponding region of S. officinalis sabinene synthase, imparted the ability to form 1,8-cineole. Sequence alignments of AtTPS-Cin and S. officinalis cineole synthase showed only 58% identity of both proteins in this 74-amino acid region, whereas in the same sequence domain, AtTPS-Cin and the At3g25810-encoded protein share 85% amino acid sequence identity and differ in only 11 amino acids (Fig. 2). Of these 11 amino acids, 5 are identical between AtTPS-Cin and the S. officinalis cineole synthase, but are also partly conserved in other monoterpene synthases (Fig. 2). Whether the formation of 1,8-cineole in AtTPS-Cin is determined in part by this 74-residue region cannot be decided on the basis of sequence similarity alone, but needs to be clarified by site-directed mutagenesis and domain-swapping experiments. It is likely that the ability of AtTPS-Cin to form 1,8-cineole has evolved independently of S. officinalis cineole synthase and might be dependent on other sequence regions in the C-terminal domain that directly or indirectly affect the configuration of the active site of the enzyme.
In conclusion, the observed differences in spatial expression and product specificity of the two closely related enzymes AtTPS-Cin and At3g25810 TPS demonstrate the ongoing evolutionary process of duplication and divergence in gene families involved in plant secondary metabolism.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (L.) Heynh (ecotype Columbia) plants were grown from seed on potting soil in a climate-controlled growth chamber (22°C, 55% relative humidity, 150 μmol m−2 s−1 photosynthetically active radiation, 16-h/8-h photoperiod), if not stated otherwise. Roots were collected from 6-week-old bolted mature plants for RNA extraction.
Reagents
GPP and FPP were purchased from Echelon Biosciences (Salt Lake City). [1-3H]GPP and [1-3H]FPP were products of American Radiolabeled Chemicals (St. Louis).
Isolation and Cloning of At3g25820/At3g25830 cDNA
Total RNA was isolated from roots of mature Arabidopsis plants with an RNeasy plant mini kit (Qiagen, Valencia, CA). DNA contamination was removed with deoxyribonuclease (Qiagen) treatment. Five micrograms of the purified RNA sample were used to synthesize cDNA with an oligo(dT) primer and a First-Strand cDNA Synthesis kit (Amersham Biosciences, Uppsala) in a 15-μL reaction. Two primers, 5′-ATGGCAACTTTACGTATAAG-3′ and 5′-TCAACCAAGACGATTTACATC-3′, which correspond to the start and the end of the coding region of At3g25820 and At3g25830, respectively, were subsequently used for PCR amplification of At3g25820/At3g25830 cDNA. The PCR reaction volume was 25 μL and contained 100 ng of each primer, 0.2 mm of each deoxynucleoside triphosphate, 1 μL cDNA, and 0.75 units of Taq DNA Polymerase (Fisher, Pittsburgh). A programmable thermal controller (PTC-100; MJ Research, Waltham, MA) was used with an initial denaturation step of 96°C for 1 min, followed by 30 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 60 s, and a final elongation step of 72°C for 10 min. The resulting cDNA fragment was cloned into the expression vector pCRT7/CT-TOPO (Invitrogen, Carlsbad, CA) for sequencing.
At3g25820/At3g25830 Protein Expression in Escherichia coli
A truncated version of the At3g25820/At3g25830 cDNA was prepared in which the first 46 codons were removed and an ATG codon was added immediately upstream of the two Arg codons 47 and 48. This was accomplished by PCR using a primer pair with the forward primer 5′-ATGCGACGCTCCGGAAACTATCAAC-3′ and the reverse primer 5′-TCAACCAAGACGATTTACATC-3′ and a second primer pair with the forward primer 5′-CACCATGCGACGCTCCGGAAACTATCAACCT-3′ and the reverse primer 5′-TCAACCAAGACGATTTACATCTAA-3′, respectively. The product of the reaction with the first primer pair was inserted into the pCRT7/CT-TOPO expression vector (Invitrogen), while the product amplified in the reaction with the second primer pair was inserted into the expression vector pET101/D-TOPO (Invitrogen). The resulting plasmids were transformed into the E. coli BL21 Codon Plus strain (Stratagene, La Jolla, CA). Bacterial growth conditions, induction of expression of the introduced gene, and protein extraction procedures were as previously described (Chen et al., 2003b).
TPS Assay Product Identification
TPS assays and volatile terpene product separation and analysis were carried out essentially as previously described (Chen et al., 2003b).
Purification of the AtTPS-Cin Enzyme
Protein purification protocols essentially followed the procedures previously described (Chen et al., 2003a). After overnight expression of the protein in E. coli BL21 Codon Plus cultures, the clarified bacterial lysate (30 mL) was loaded onto a DEAE cellulose column (DE53; Whatman, Clifton, NJ) preequilibrated with buffer A (50 mm Bis-Tris, pH 7.0, 10% glycerol [v/v], and 5 mm DTE). After a washing step with buffer A, proteins were eluted with a 0 to 0.45 m KCL gradient in a total volume of 100 mL buffer A. Fractions with the highest activities were pooled (10 mL), diluted 5-fold with buffer A, and loaded onto a Mono-Q column (0.5-cm diameter×6.0 cm; Amersham Biosciences). The enzyme was eluted with a 0.1- to 0.45-m KCl gradient in a total volume of 100 mL buffer A. Active fractions (2 mL each) were collected and the protein purity was examined by SDS-PAGE, followed by Coomassie Brilliant Blue staining. The fraction with the highest degree of purity was used for enzyme characterization studies. Protein concentrations were determined by the Bradford method (Bradford, 1976), using reagents obtained from Bio-Rad (Richmond, CA) and bovine serum albumin as calibration standard.
Enzyme Characterization
Purified AtTPS-Cin enzyme was used for biochemical characterization. A standard assay for determining biochemical properties was carried out in a final volume of 50 μL with 0.1 μg purified enzyme, and 10 μm of [1-3H]GPP (2 or 9 MBq μmol−1) or [1-3H]FPP (9 MBq μmol−1). Buffer, salt, and incubation conditions were the same as previously described (Chen et al., 2003b). Following incubation for 20 min, the reaction products were extracted with 100 μL hexane. Total radioactivity of the reaction products was determined by scintillation counting.
For pH optimum determination, assays with saturated substrate were carried out in 100 mm Bis-Tris propane buffers with pH 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, and 9.0. For determination of the metal ion requirement, assays were carried out in buffer A in the presence of MgCl2 (concentrations from 0 to 50 mm) or MnCl2 (concentrations from to 0 to 8 mm). For kinetic assays, appropriate enzyme concentrations and incubation times were determined so that the reaction velocity was linear during the reaction time period. To determine the Km value for GPP, different concentrations of GPP were applied in standard assays in three replications. Calculation of the apparent Km value was obtained by Lineweaver-Burk plot analysis.
Determination of Gene Expression by RT-PCR
Procedures for total RNA isolation from mature plants and cDNA synthesis were the same as those described for cDNA cloning. For RT-PCR analysis of At3g25820/At3g25830 expression in different organs, primers designed to amplify a fragment of approximately 0.9 kb were as follows: forward primer 5′-TATTTGATGTGATCATCGACC-3′ and reverse primer 5′-GGAACACTTAAGATATAAAAGGT-3′. The two primers used for PCR amplification of 18S rRNA were: forward 5′-GACGGAGAATTAGGGTTCGATTC-3′ and reverse 5′-CCAACTAAGAACGGCCATGCAC-3′. Initially, PCR reactions were performed with At3g25820/At3g25830-specific primers with 0.1, 0.2, 0.5, and 1.0 μL cDNA for 30 cycles with an annealing temperature of 54°C. Each PCR reaction of 25 μL contained 0.4 μm forward primer, 0.4 μm reverse primer, and 0.2 mm of dATP, dTTP, dGTP, and dCTP, respectively. Amplified products were separated on a 1.0% agarose gel and quantified using the Bio-Rad Quantity One program (Hercules, CA). Analysis showed that the amounts of amplified products increased linearly with increasing amounts of cDNA. 0.2 μL cDNA was chosen as the optimal amount of template for further PCR reactions. For 18S rRNA, PCR reactions were carried out in separate tubes under conditions similar to those described for At3g25820/At3g25830, except that the reactions were performed for 20 cycles.
Construction of the At3g25820/At3g25830 Promoter-GUS Reporter Gene Fusion and Histochemical Localization of GUS Activity
Genomic DNA was isolated from Arabidopsis (ecotype Col) leaves as described by Rogers and Bendich (1985). A 2.5-kb promoter region upstream of the At3g25820/At3g25830 translation start site was isolated via PCR from genomic DNA using the forward primer 5′-ATATTTCTAGAATGTCAATTTTCATGGCACATC-3′, containing a XbaI site, and the reverse primer 5′-ATATTGGATCCATTGATTTAGTAGACTATTCTC-3′, containing the At3g25820/At3g25830 start codon and a BamHI site. The resulting PCR product was inserted into the pCR-BluntII-TOPO vector (Invitrogen) for sequencing. The promoter region was then cut out of the pCR-BluntII-TOPO vector with BamHI and XbaI and inserted into the binary vector pDW137 (Blazquez et al., 1997) upstream of the uidA (GUS) gene. The construct was introduced into the Agrobacterium tumefaciens ASE strain, which was used to transform Arabidopsis (ecotype Col) plants by floral vacuum infiltration (Bechthold et al., 1993). Transgenic lines were selected on kanamycin resistance and transformation was additionally confirmed by PCR analysis. Control lines were established carrying the promoterless insertion cassette only. Assays for the localization of GUS enzyme activity were performed as described previously (Chen et al., 2003b). For the determination of promoter-GUS activity, seedlings of four independent T2 lines were grown for approximately 12 d under short-day conditions (10-h/14-h photoperiod) on 1× Murashige and Skoog plates with or without 1% (w/v) Suc and 0.075 mg mL−1 kanamycin, respectively.
For sectioning of GUS-stained roots, root tissue was embedded in a glycol methacrylate resin as described by Beeckman and Viane (2000) using the Technovit 7100 kit (Heraeus Kulzer, Wehrheim, Germany). Four-micrometer-thick sections were prepared on a Leica (Wetzlar, Germany) microtome, mounted, and photographed under bright-field illumination.
Extraction and Analysis of Arabidopsis Roots
Soil-grown roots were harvested from 4-week-old prebolting Arabidopsis plants (ecotype Col). Soil particles were removed from roots by repeated submersion and rinsing in tap water and the tissue was immediately analyzed. Alternatively, roots were harvested from plants in the rosette stage grown hydroponically under short-day conditions for 4 to 5 weeks in Hoagland solution (Gibeaut et al., 1997), and then frozen in liquid N2 and stored at −80°C prior to extraction. Approximately 2 g of roots from hydroponically grown plants were ground in a mortar and then extracted for 2 h under vigorous shaking with 3 mL diethyl ether-pentane (3:1). Extracts were dried over Na2SO4 and 0.2 μg linalool was added as internal standard. After concentration under nitrogen to approximately 40 μL, the samples were analyzed by GC-MS as previously described (Chen et al., 2003b). Solid-phase microextraction was performed on separately harvested 1-g batches of ground fresh roots from soil-grown plants or hydroponically grown roots enclosed in 7-mL glass Teflon screw-capped reaction vials in the presence of 20 ng linalool. A polydimethylsiloxane fiber (PDMS-100; Supelco, Bellefonte, PA) was inserted into the vial during incubation at 30°C for 1 h, followed by an incubation at 42°C for 15 min. The fiber was then inserted directly into the GC-MS and analyzed as previously described (Chen et al., 2003b).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY691947.
Acknowledgments
We thank Katrin Heisse for technical assistance, Aleksandra Skirycz for help with embedding and sectioning of Arabidopsis roots, and Jorge Vivanco for stimulating discussion and unpublished data.
This work was supported by the National Science Foundation (grant no. IBN–0211697 to E.P.), by the Max Planck Society (to J.P., J.G., and D.T.), and by the Natural Sciences and Engineering Research Council of Canada (to J.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044388.
References
- Arimura G, Huber DPW, Bohlmann J (2004) Forest tent caterpillars (Malacosoma disstria) induce local and systemic diurnal emissions of terpenoid volatiles in hybrid poplar (Populus trichocarpa x deltoides): cDNA cloning, functional characterization, and patterns of gene expression of (−)-germacrene D synthase, PtdTPS1. Plant J 37: 603–616 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267: 730–745 [DOI] [PubMed] [Google Scholar]
- Bais HP, Park SW, Weir TL, Callaway RM, Vivanco JM (2004) How plants communicate using the underground information superhighway. Trends Plant Sci 9: 26–32 [DOI] [PubMed] [Google Scholar]
- Bais HP, Vepachedu R, Gilroy S, Callaway RM, Vivanco JM (2003) Allelopathy and exotic plant invasion: from molecules and genes to species interactions. Science 301: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Life Sci 316: 1194–1199 [Google Scholar]
- Beeckman T, Viane R (2000) Embedding thin plant specimens for oriented sectioning. Biotech Histochem 75: 23–26 [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Soowal LN, Lee I, Weigel D (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Martin D, Oldham NJ, Gershenzon J (2000) Terpenoid secondary metabolism in Arabidopsis thaliana: cDNA cloning, characterization, and functional expression of a myrcene/(E)-β-ocimene synthase. Arch Biochem Biophys 375: 261–269 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann J, Stauber EJ, Krock B, Oldham NJ, Gershenzon J, Baldwin IT (2002) Gene expression of 5-epi-aristolochene synthase and formation of capsidiol in roots of Nicotiana attenuata and N. sylvestris. Phytochemistry 60: 109–116 [DOI] [PubMed] [Google Scholar]
- Bos R, Koulman A, Woerdenbag HJ, Quax WJ, Pras N (2002) Volatile components of Anthriscus sylvestris (L.) Hoffm. J Chromatogr A 966: 233–238 [DOI] [PubMed] [Google Scholar]
- Bouwmeester HJ, Matusova R, Zhongkui S, Beale MH (2003) Secondary metabolite signalling in host-parasitic plant interactions. Curr Opin Plant Biol 6: 358–364 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for quantification of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cane DE (1999) Sesquiterpene biosynthesis: cyclization mechanisms. In DE Cane, ed, Isoprenoids Including Carotenoids and Steroids. Comprehensive Natural Products Chemistry, Vol 2. Elsevier Science, Amsterdam, pp 155–200
- Chen F, D'Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E (2003. a) An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J 36: 577–588 [DOI] [PubMed] [Google Scholar]
- Chen F, Tholl D, D'Auria JC, Farooq A, Pichersky E, Gershenzon J (2003. b) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau R, Alonso WR, Koepp AE, Johnson MA (1994) Biosynthesis of monoterpenes: partial purification, characterization, and mechanism of action of 1,8-cineole synthase. Arch Biochem Biophys 309: 184–192 [DOI] [PubMed] [Google Scholar]
- Davis EM, Croteau R (2000) Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. In FJ Leeper, JC Vederas, eds, Topics in Current Chemistry: Biosynthesis: Aromatic Polyketides, Isoprenoids, Alkaloids, Vol 209. Springer-Verlag, Heidelberg, pp 53–95
- Dicke M, Van Loon JJA (2000) Multitrophic effects of herbivore-induced plant volatiles in an evolutionary context. Entomol Exp Appl 97: 237–249 [Google Scholar]
- Dobson HEM (1994) Floral volatiles in insect biology. In E Bernays, ed, Insect-Plant Interactions, Vol 5. CRC Press, Boca Raton, FL, pp 47–81
- Dudareva N, Martin D, Kish CM, Kolosova N, Gorenstein N, Fäldt J, Miller B, Bohlmann J (2003) (E)-β-Ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 15: 1227–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E (2000) Biochemical and molecular genetic aspects of floral scents. Plant Physiol 122: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberth J, Koch T, Schuler G, Bachmann N, Rechtenbach J, Boland W (2001) Ion channel-forming alamethicin is a potent elicitor of volatile biosynthesis and tendril coiling. Cross talk between jasmonate and salicylate signaling in lima bean. Plant Physiol 125: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fäldt J, Arimura GI, Gershenzon J, Takabayashi J, Bohlmann J (2003) Functional identification of AtTPS03 as (E)-β-ocimene synthase: a new monoterpene synthase catalyzing jasmonate- and wound-induced volatile formation in Arabidopsis thaliana. Planta 216: 745–751 [DOI] [PubMed] [Google Scholar]
- Fischer NH (1991) Plant terpenoids as allelopathic agents. In JB Harborne, FA Tomas-Barberan, eds, Ecological Chemistry and Biochemistry of Terpenoids, Proceedings of the Phytochemical Society of Europe, Vol 31. Oxford University Press, Oxford, pp 377–398
- Gershenzon J, Kreis W (1999) Biochemistry of terpenoids: monoterpenes, sesquiterpenes, diterpenes, sterols, cardiac glycosides and steroid saponins. In M Wink, ed, Biochemistry of Plant Secondary Metabolism, CRC Press, Boca Raton, FL, pp 222–299
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer KA, Carson CF, Riley TV (2003) Antifungal activity of the components of Melaleuca alternifolia (tea tree) oil. J Appl Microbiol 95: 853–860 [DOI] [PubMed] [Google Scholar]
- Hans J, Hause B, Strack D, Walter MH (2004) Cloning, characterization, and immunolocalization of a mycorrhiza-inducible 1-deoxy-D-xylulose 5-phosphate reductoisomerase in arbuscule-containing cells of maize. Plant Physiol 134: 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kovacevic N, Pavlovic M, Menkovic N, Tzakou O, Couladis M (2002) Composition of the essential oil from roots and rhizomes of Valeriana pancicii Halácsy & Bald. Flavour Frag J 17: 355–357 [Google Scholar]
- Langenheim JH (1994) Higher plant terpenoids: a phytocentric overview of their ecological roles. J Chem Ecol 20: 1223–1280 [DOI] [PubMed] [Google Scholar]
- Martin D, Fäldt J, Bohlmann J (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135: 1908–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan K, Basheer C, Bajic VB, Swarup S (2003) Enhancement of plant-microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol 132: 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare PW, Tumlinson JH (1999) Plant volatiles as a defense against insect herbivores. Plant Physiol 121: 325–331 [PMC free article] [PubMed] [Google Scholar]
- Peters RJ, Croteau RB (2003) Alternative termination chemistries utilized by monoterpene cyclases: chimeric analysis of bornyl diphosphate, 1,8-cineole, and sabinene synthases. Arch Biochem Biophys 417: 203–211 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gershenzon J (2002) The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol 5: 237–243 [DOI] [PubMed] [Google Scholar]
- Pina-Vaz C, Rodrigues AG, Pinto E, Costa-de-Oliveira S, Tavares C, Salgueiro L, Cavaleiro C, Goncalves MJ, Martinez-de-Oliveira J (2004) Antifungal activity of thymus oils and their major compounds. J Eur Acad Dermatol Venereol 18: 73–78 [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5: 69–76 [DOI] [PubMed] [Google Scholar]
- Romagni JG, Allen SN, Dayan FE (2000) Allelopathic effects of volatile cineoles on two weedy plant species. J Chem Ecol 26: 303–313 [Google Scholar]
- Schuler G, Gorls H, Boland W (2001) 6-Substituted indanoyl isoleucine conjugates mimic the biological activity of coronatine. Eur J Org Chem 9: 1663–1668 [Google Scholar]
- Singh HP, Batish DR, Kaur S, Ramezani H, Kohli RK (2002) Comparative phytotoxicity of four monoterpenes against Cassia occidentalis. Ann Appl Biol 141: 111–116 [Google Scholar]
- Steeghs M, Bais HP, de Gouw J, Goldan P, Kuster W, Northway M, Fall R, Vivanco JM (2004) Proton-transfer-reaction mass spectrometry as a new tool for real time analysis of root-secreted volatile organic compounds in Arabidopsis. Plant Physiol 135: 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CL, Crock J, Bohlmann J, Croteau R (1998) Sesquiterpene synthases from grand fir (Abies grandis)—comparison of constitutive and wound-induced activities, and cDNA isolation, characterization and bacterial expression of δ-selinene synthase and γ-humulene synthase. J Biol Chem 273: 2078–2089 [DOI] [PubMed] [Google Scholar]
- Tripathi AK, Prajapati V, Aggarwal KK, Kumar S (2001) Toxicity, feeding deterrence, and effect of activity of 1,8-cineole from Artemisia annua on progeny production of Tribolium castanaeum (Coleoptera: Tenebrionidae). J Econ Entomol 94: 979–983 [DOI] [PubMed] [Google Scholar]
- Walker TS, Bais HP, Grotewold E, Vivanco JM (2003. a) Root exudation and rhizosphere biology. Plant Physiol 132: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TS, Bais HP, Halligan KM, Stermitz FR, Vivanco JM (2003. b) Metabolic profiling of root exudates of Arabidopsis thaliana. J Agric Food Chem 51: 2548–2554 [DOI] [PubMed] [Google Scholar]
- Weidenhamer JD, Macias FA, Fischer NH, Williamson GB (1993) Just how insoluble are monoterpenes? J Chem Ecol 19: 1799–1807 [DOI] [PubMed]
- Wichtmann EM, Stahl-Biskup E (1987) Composition of the essential oils from caraway herb and root. Flavour Frag J 2: 83–89 [Google Scholar]
- Wiggins NL, McArthur C, McLean S, Boyle R (2003) Effects of two plant secondary metabolites, cineole and gallic acid, on nightly feeding patterns of the common brushtail possum. J Chem Ecol 29: 1447–1464 [DOI] [PubMed] [Google Scholar]
- Williams DC, McGarvey D, Katahira EJ, Croteau R (1998) Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37: 12213–12220 [DOI] [PubMed] [Google Scholar]
- Wise ML, Croteau R (1999) Monoterpene biosynthesis. In DE Cane, ed, Isoprenoids Including Carotenoids and Steroids. Comprehensive Natural Products Chemistry, Vol 2. Elsevier Science, Amsterdam, pp 97–153
- Wise ML, Savage TJ, Katahira E, Croteau R (1998) Monoterpene synthases from common sage (Salvia officinalis). cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1,8-cineole synthase, and (+)-bornyl diphosphate synthase. J Biol Chem 273: 14891–14899 [DOI] [PubMed] [Google Scholar]