Abstract
In vivo stable isotope labeling and computer-assisted metabolic flux analysis were used to investigate the metabolic pathways in petunia (Petunia hybrida) cv Mitchell leading from Phe to benzenoid compounds, a process that requires the shortening of the side chain by a C2 unit. Deuterium-labeled Phe (2H5-Phe) was supplied to excised petunia petals. The intracellular pools of benzenoid/phenylpropanoid-related compounds (intermediates and end products) as well as volatile end products within the floral bouquet were analyzed for pool sizes and labeling kinetics by gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry. Modeling of the benzenoid network revealed that both the CoA-dependent, β-oxidative and CoA-independent, non-β-oxidative pathways contribute to the formation of benzenoid compounds in petunia flowers. The flux through the CoA-independent, non-β-oxidative pathway with benzaldehyde as a key intermediate was estimated to be about 2 times higher than the flux through the CoA-dependent, β-oxidative pathway. Modeling of 2H5-Phe labeling data predicted that in addition to benzaldehyde, benzylbenzoate is an intermediate between l-Phe and benzoic acid. Benzylbenzoate is the result of benzoylation of benzyl alcohol, for which activity was detected in petunia petals. A cDNA encoding a benzoyl-CoA:benzyl alcohol/phenylethanol benzoyltransferase was isolated from petunia cv Mitchell using a functional genomic approach. Biochemical characterization of a purified recombinant benzoyl-CoA:benzyl alcohol/phenylethanol benzoyltransferase protein showed that it can produce benzylbenzoate and phenylethyl benzoate, both present in petunia corollas, with similar catalytic efficiencies.
Phenylpropanoid metabolism comprises a complex series of branching biochemical pathways that provide plants with thousands of compounds, which are often specific to particular plant species. Many are intermediates in the synthesis of structural cell components (e.g. lignin, suberin, and other cell wall-associated phenolics), while others comprise a diverse array of pigments (e.g. flavonoids and anthocyanins), both of which are usually nonvolatile. However, phenylpropanoids that are reduced at the C9 position (to aldehyde, alcohol, or alkane/alkene) and/or that contain alkyl additions to the hydroxyl groups of the benzyl ring or to the carboxyl group (i.e. ethers and esters) are volatile. In addition, many benzenoid compounds, which lack the three-carbon chain and originate from trans-cinnamic acid (CA) as a side branch of the general phenylpropanoid pathway, are also volatile. These volatile phenylpropanoids/benzenoids are common constituents of floral scent (Knudsen et al., 1993). Recently, several enzymes that catalyze the formation of floral benzenoid volatiles have been identified and characterized. They include acetyl-CoA:benzyl alcohol acetyltransferase (BEAT; Dudareva et al., 1998a, 1998b), S-adenosyl-l-Met:salicylic acid carboxyl methyltransferase (Dudareva et al., 1998b; Ross et al., 1999; Negre et al., 2002), S-adenosyl-l-Met:benzoic acid carboxyl methyltransferase (Dudareva et al., 2000; Murfitt et al., 2000), benzoyl-CoA:benzyl alcohol benzoyl transferase (BEBT; D'Auria et al., 2002), and orcinol O-methyl transferase (Lavid et al., 2002), which are responsible for the formation of benzylacetate, methylsalicylate, methylbenzoate, benzylbenzoate, and dimethoxytoluene, respectively. In addition, S-adenosyl-l-Met:(iso)eugenol O-methyltransferase, which catalyzes the final step in the formation of the volatile phenylpropene, methyleugenol, was also isolated and characterized (Wang et al., 1997; Wang and Pichersky, 1998).
The biochemistry and enzymology of floral scent have mainly concentrated on the isolation and characterization of enzymes and genes involved in the final steps of the biosynthesis of scent volatile compounds. Surprisingly, little is known about the biochemical pathways leading to simple benzenoid compounds. The first committed step in the biosynthesis of benzenoid compounds is catalyzed by the well-known and widely distributed enzyme l-Phe ammonia-lyase (PAL; EC 4.3.1.5), just as in the biosynthesis of other phenylpropanoids. PAL catalyzes the deamination of l-Phe to produce CA. Formation of benzenoid compounds from CA requires the shortening of the side chain by a C2 unit, for which several routes have been proposed. The CoA-dependent, β-oxidative pathway is analogous to that underlying β-oxidation of fatty acids. It involves activation of CA via formation of a cinnamoyl-CoA ester, then hydration of the CoA ester to 3-hydroxy-3-phenylpropionyl-CoA, prior to oxidation of the hydroxyl group to a ketone, 3-oxo-3-phenylpropionyl-CoA, and finally cleavage of the β-keto thioester via a reverse Claisen reaction with formation of benzoyl-CoA (Fig. 1). Support for the existence of a β-oxidative pathway came from early studies of cell-free extracts of Quercus pedunculata, which showed that conversion of CA to benzoic acid was stimulated by the addition of ATP and CoA (Alibert and Ranjeva, 1971). A recent investigation with stable isotope-labeled (2H6,18O) 3-hydroxy-3-phenylpropionic acid also suggested that CA chain shortening is probably achieved by β-oxidation in cucumber (Cucumis sativus) and Nicotiana attenuata (Jarvis et al., 2000). Moreover, the β-oxidation pathway and the preceding PAL reaction were recently detected in a marine streptomycene (Hertweck and Moore, 2000; Hertweck et al., 2001).
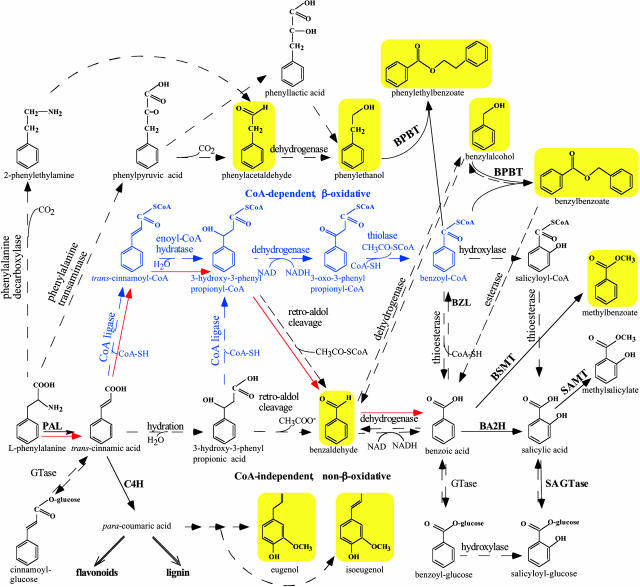
Figure 1.
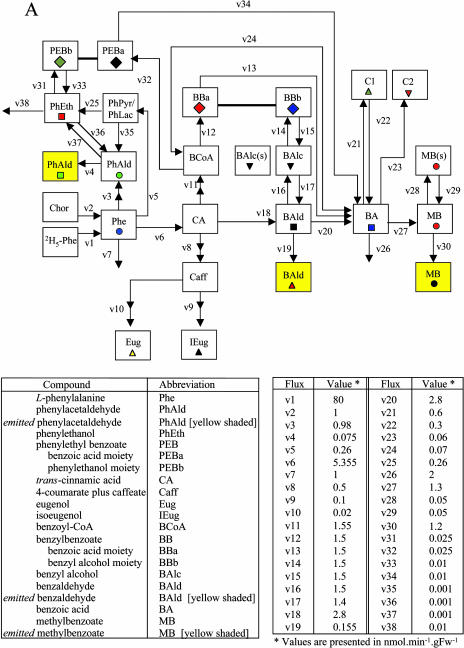
Proposed biosynthetic pathways leading to some benzenoid compounds in petunia. CoA-dependent, β-oxidative pathway of side chain shortening is shown in blue, whereas CoA-independent, non-β-oxidative pathway is shown in black. Red arrows show CoA-dependent and non-β-oxidative pathway. Solid arrows indicate established biochemical reactions, whereas broken arrows indicate possible steps not yet described. The conversion of cinnamoyl-CoA to benzoyl-CoA has already been described in mammals (Mao et al., 1994). BSMT and SAMT, S-adenosyl-l-Met:benzoic acid/salicylic acid and salicylic acid carboxyl methyltransferase, respectively; BA2H, benzoic acid 2-hydroxylase; BZL, benzoate:CoA ligase; C4H, cinnamic acid-4-hydroxylase; SA GTase, UDP-Glc:salicylic acid glucosyltransferase. Volatile benzenoid/phenylpropanoid-related compounds shown with a yellow background were analyzed in petunia floral scent and in petal tissue. Additionally, the pools of l-Phe and benzoic acid were also evaluated.
The alternative CoA-independent, non-β-oxidative pathway involves hydration of the free CA to 3-hydroxy-3-phenylpropionic acid (3-hydroxy-3-phenylpropanoic), side chain degradation via a reverse aldol reaction with formation of benzaldehyde, and its oxidation to benzoic acid (Fig. 1). A non-β-oxidative mechanism was demonstrated in three independent in vitro studies carried out with cell-suspension cultures of carrot (Schnitzler et al., 1992) and cell-free extracts of Lithospermum erythrorizum (Yazaki et al., 1991) and potato (French et al., 1976) for the formation of p-hydroxybenzoic acid, a key intermediate in several biosynthetic pathways, including biosyntheses of ubiquinones and the red naphtoquinone pigment shikonin. This non-β-oxidative pathway is characterized by the presence of p-hydroxybenzaldehyde as an important metabolic intermediate before oxidation to p-hydroxybenzoic acid and requires an aldehyde dehydrogenase for the conversion of an aldehyde to its corresponding carboxylic acid. A NADP+-dependent aldehyde dehydrogenase, which catalyzes the oxidation of coniferaldehyde and sinapaldehyde to ferulic and sinapic acids, respectively, was recently isolated and characterized from Arabidopsis (Nair et al., 2004).
The side chain shortening could also occur via a combination of these two pathways and could be CoA-dependent and non-β-oxidative (Fig. 1), as was recently shown in Hypericum androsaemum cell cultures (Ahmed et al. 2002). To date, contradictory results concerning the metabolic pathways leading to benzenoid compounds suggest that several side chain shortening routes may exist in different plants and even in the same plant depending on physiological conditions. The mechanism of side chain shortening remains an important unresolved question. In addition, the interconnections between benzenoid compounds in this branchway are also unknown.
In this study, we use in vivo stable isotope labeling and metabolic flux analysis in combination with the power of computer-assisted metabolic modeling to investigate the metabolic pathways leading to benzenoid compounds in petunia (Petunia hybrida) cv Mitchell, a member of the Solanaceae family. Petunia cv Mitchell provides an ideal experimental system to investigate the benzenoid network because its floral scent consists almost exclusively of benzenoid/phenylpropanoid-related compounds (dominated by methylbenzoate, benzaldehyde, and phenylacetaldehyde), the levels of which change rhythmically through a daily light/dark cycle with a maximum at midnight (Kolosova et al., 2001; Verdonk et al., 2003). Petal tissue also contains substantial quantifiable endogenous intracellular pools of benzaldehyde, benzyl alcohol, benzylbenzoate, methylbenzoate, phenylacetaldehyde, phenylethanol, phenylethyl benzoate, isoeugenol, and eugenol. By supplying deuterium-labeled Phe to excised petunia cv Mitchell petals, a tissue highly specialized for floral scent biosynthesis, we determined the labeling kinetics of the intracellular pools of benzenoid/phenylpropanoid-related compounds (intermediates and end products) as well as volatile end products within the floral bouquet. Quantitative analysis of isotopic abundances and pool sizes of benzenoid/phenylpropanoid-related compounds after subjection to computer modeling to organize and conceptualize the data revealed that two alternative pathways (CoA-dependent, β-oxidative and CoA-independent, non-β-oxidative) operate in the benzenoid branchway in petunia flowers. This knowledge is essential for the future discovery of enzymes and genes involved in the benzenoid network, which still remain unknown.
RESULTS
Experimental Design
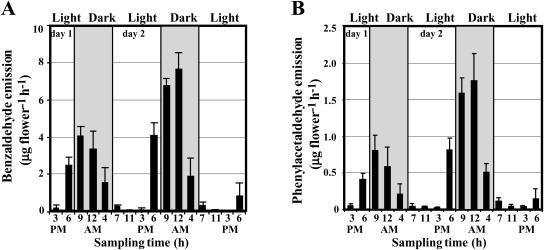
Our previous results show that emission of methylbenzoate, one of the major scent compounds in petunia cv Mitchell flowers, changes rhythmically through a daily light/dark cycle with a maximum level at night (Kolosova et al., 2001). Petunia flowers also emit benzaldehyde and phenylacetaldehyde, which follow the same emission pattern with the highest rate (7.67 ± 0.85 and 1.76 ± 0.37 μg flower−1 h−1, respectively, on day 2 postanthesis) around midnight (Fig. 2). In vivo tracer experiments with radioisotope and/or stable isotope-labeled precursors have long been used to discover unknown biochemical pathways and to aid in discriminating between alternative metabolic pathways (Kocsis et al., 1998; McNeil et al., 2000a); therefore, we used this approach for the investigation of the benzenoid network in petunia. Since benzaldehyde is a key intermediate in the non-β-oxidative pathway of side chain shortening in benzenoid metabolism, its presence in petunia floral scent allowed us to follow its labeling pattern in the volatile fraction after supplying a labeled precursor. Moreover, substantial endogenous intracellular pools of benzaldehyde, benzyl alcohol, benzylbenzoate, methylbenzoate, phenylacetaldehyde, phenylethanol, phenylethyl benzoate, isoeugenol, and eugenol placed us in unique position to identify the interconnections between these compounds within the benzenoid/phenylpropanoid network and to analyze the flux through the benzenoid branchway in situ.
Figure 2.
Emission of benzaldehyde (A) and phenylacetaldehyde (B) in petunia flowers during two normal light/dark cycles. Floral scent was collected from 1- and 2-d-old flowers during 48-h period under normal light/dark conditions. An increase of the maximum amplitude in 2-d-old flowers relative to 1-d-old flowers reflects the developmental changes in emission. Gray and white areas correspond to dark and light, respectively. Each graph represents the average of four independent experiments. sds are indicated by vertical bars.
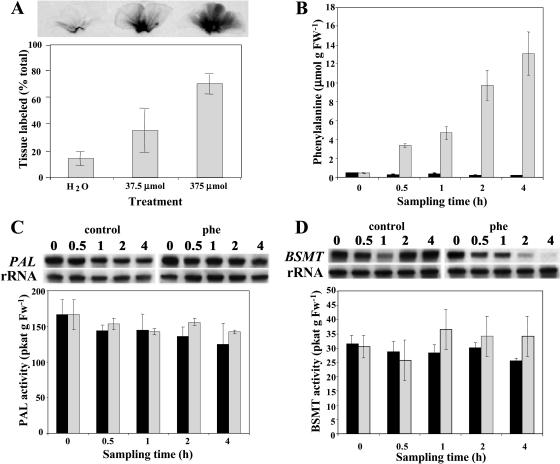
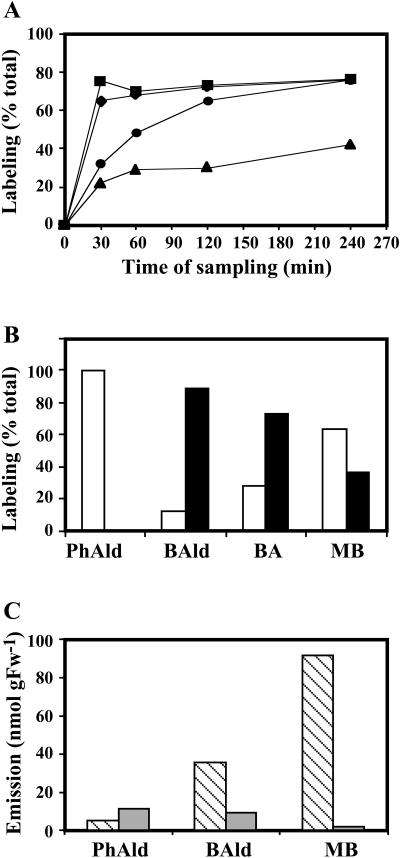
Although multiple mechanisms have been proposed for benzenoid biosynthesis from CA in plants, the first committed step in each of these pathways is the conversion of l-Phe to CA by PAL. Thus, in our labeling experiments we used deuterium ring-labeled Phe (2H5-Phe) supplied to excised petunia corolla limbs, a tissue highly specialized for floral scent biosynthesis that is able to continue producing and emitting volatile compounds after excision. Since petunia cv Mitchell is a nocturnally emitting plant (Kolosova et al., 2001; Verdonk et al., 2003), all feeding experiments were performed under dark conditions. To minimize the contribution of volatile and nonvolatile compounds from unlabeled areas of petal tissue and to limit the effect of natural rhythmic emission fluctuations during the daily light/dark cycle (Kolosova et al., 2001), quick uniform labeling of tissue was essential in these experiments. To find the optimal concentration of Phe for rapid uniform labeling, excised limbs of corolla were fed for 1 h through the cut edge with one of three different amounts of unlabeled Phe (0 μmol as a control, 37.5 μmol, and 375 μmol) combined with a small amount of radiolabeled Phe (2 μL of U-[14C]Phe). Exposure of petal tissues to x-ray film showed that feeding of 375 μmol Phe gave a more uniform distribution of labeled precursor resulting in 70% of the tissue labeled within an hour (Fig. 3A). Also, the amounts of emitted volatiles were slightly lower but comparable to the amounts emitted from flowers attached to the plant. Thus, this amount of Phe was used for further detailed time-course studies, including 30-, 60-, 120-, and 240-min labeling periods to ensure rapid label incorporation into metabolites. This level of exogenous Phe caused a large expansion of the endogenous Phe pool (Fig. 3B), and so it is likely that the flux measurements represent the maximal velocities of the metabolic system in vivo. Interestingly, when excised petunia limbs were not supplied with exogenous Phe, the emission levels of volatile benzenoid compounds were drastically decreased, indicating that petal tissue was not able to emit volatile compounds due to a limiting pool of endogenous Phe. Indeed, when the endogenous pool of Phe was measured in petal tissue at excision and 30 min later, we found that the endogenous pool of Phe rapidly declined from 445 ± 1.47 nmol g fresh weight (FW)−1 to 257 ± 79 nmol g FW−1 (Fig. 3B).
Figure 3.
Effect of Phe feeding on its distribution throughout petal tissue (A), endogenous Phe pool (B), PAL (C), and BSMT (D) mRNA gene expression and enzyme activities. Black and gray bars represent control (nonfed) and Phe-fed petunia flowers, respectively. A, [14C]Phe along with shown concentrations of unlabeled Phe was fed to petunia petals for 1 h. The top autoradiography shows label movement, and quantification of this movement is shown in the graph below. B, Quantification of endogenous Phe pool under experimental conditions. Graph represents the average of three independent experiments. sds are indicated by vertical bars. C and D, RNA blot analysis of PAL and BSMT mRNA levels (top) and PAL and BSMT activities (bottom graphs) in control and Phe-fed petal tissues at time points of sampling used in feeding experiments. Autoradiography of RNA blots was performed overnight. The blots were rehybridized with an 18S rDNA probe (bottom of top) to standardize samples. Each blot represents a typical result of three independent experiments, including the ones shown here. Enzyme assays were run in duplicate for each time point on at least five independent crude extract preparations for both PAL and BSMT activities, and the sds indicated by vertical bars were obtained.
Since exogenously supplied precursors can affect the natural balance of cellular intermediates and increase the flux through the pathways, we investigated the changes in Phe pool levels in petal tissue after 30, 60, 120, and 240 min of feeding and the effect of the resulting endogenous Phe pool on expression and activity of PAL, the first enzyme in the network, and S-adenosyl-l-Met:benzoic acid/salicylic acid carboxyl methyltransferase (BSMT), the enzyme in petunia responsible for the biosynthesis of methylbenzoate (Negre et al., 2003), one of the end products in the network (Fig. 1). Previously, it has been shown that exogenously supplied Phe up-regulates the expression of some genes of the phenylpropanoid pathway, including PAL, in cell suspension culture (Anterola et al., 2002). Analysis of Phe levels in petal tissue after feeding revealed that the endogenous pool of Phe increased linearly almost 30 times by the end of experiment, from about 445 ± 1.47 nmol g FW−1 to about 13,000 ± 2,287 nmol g FW−1 within 4 h (Fig. 3B). PAL expression increased slightly in tissue fed with Phe when compared with that in petal limbs on intact plants, while PAL activity remained unchanged (Fig. 3C). In the case of the BSMT gene, the situation was slightly different. BSMT expression decreased after 2 h of feeding, while similar to PAL activity, BSMT activity was not altered, presumably due to protein stability (Fig. 3D). These results show that PAL and BSMT activities did not change in the time frame of the experiment in response to Phe feeding. For this analysis, samples (total RNA and crude extracts) were prepared from petal tissues fed with Phe for 30, 60, 120, and 240 min under dark conditions and compared with samples isolated from petal tissue harvested at the same time points directly from intact petunia plants grown under a reverse photoperiod resulting in dark conditions at these time points. Feeding experiments were performed when volatile emission was the highest and most constant to eliminate the effect of rhythmicity.
Position of Benzaldehyde in the Benzenoid Network
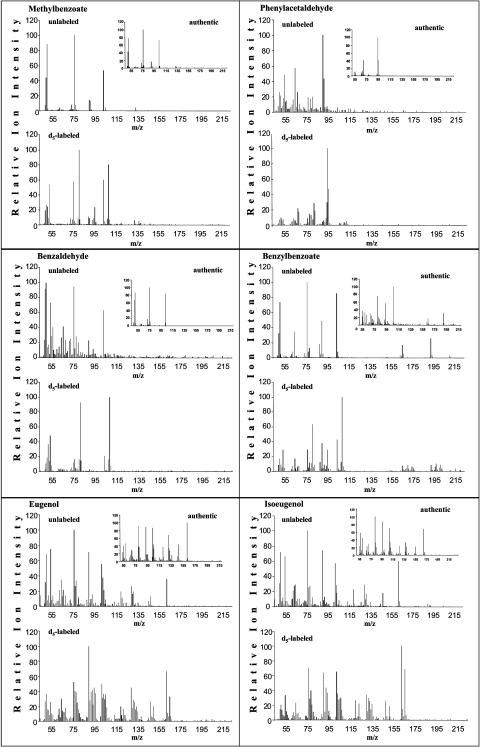
Floral volatiles were collected for 30, 60, 120, and 240 min under dark conditions from excised corolla limbs of 2-d-old petunia flowers continuously fed with 2H5-Phe and analyzed by gas chromatography-mass spectrometry (GC-MS) for pool sizes and isotope abundances of benzaldehyde, methylbenzoate, and phenylacetaldehyde emitted to the gas phase. Because of the nocturnal nature of emission of petunia scent components, plants were grown in growth chambers with a reversed photoperiod, and all experiments began at the same time of the day. After scent collections, petal tissues were extracted with either methanol and analyzed by liquid chromatography-mass spectrometry (LC-MS) to determine the labeling and pool sizes of endogenous nonvolatile metabolites, including benzoic acid and its conjugates, or with hexane, to determine pool sizes and isotope abundances of endogenous intermediates and end products, including phenylacetaldehyde, phenylethanol, phenylethyl benzoate, eugenol, isoeugenol, benzaldehyde, benzyl alcohol, benzylbenzoate, and methylbenzoate. Figure 4 shows that isotopomers synthesized from supplied 2H5-Phe can be easily distinguished from unlabeled compounds based on mass spectra by mass spectrometry (MS). For all compounds except eugenol and isoeugenol, isotope abundance was determined as (2H5 × 100)/(2H5 + 2H0) (atom%). However, newly synthesized labeled eugenol and isoeugenol exhibit a shift of only +3 atomic mass units (amu) because the two hydroxylations of the caffeate moiety remove two deuterium atoms from the ring of the supplied 2H5-Phe. Thus, for eugenol and isoeugenol, isotope abundance was determined as (2H3 × 100)/(2 H3 + 2H0) (atom%; Fig. 4).
Figure 4.
GC-MS analysis of benzenoid and phenylpropanoid compounds produced by petunia petal tissue fed with Phe and 2H5-Phe for 4 h. The panel for each presented compound contains mass spectrum of authentic compound standard, mass spectrum of compound extracted from Phe-fed tissue (unlabeled), and mass spectrum representing a combination of unlabeled and labeled compound extracted from 2H5-Phe fed petal tissue (d5-labeled). All newly synthesized labeled benzenoid compounds exhibit a mass shift by 5 amu, except eugenol and isoeugenol, which exhibit a shift of only by 3 amu.
2H5-Phe was rapidly incorporated into benzaldehyde and phenylacetaldehyde, resulting in approximately 70% labeling for both compounds within 30 min after feeding that increased only slightly (by an additional 5%) during the next 3.5 h of feeding (Fig. 5A). Over a time course the pool of benzaldehyde was more extensively labeled than the benzoic acid pool, which was labeled more heavily than the methylbenzoate pool, suggesting that benzaldehyde is an intermediate in benzoic acid biosynthesis from Phe in petunia petals (Fig. 5A). To confirm the role of benzaldehyde as an intermediate in the pathway from Phe to benzoic acid, 2H5-benzaldehyde (37.5 μmol) was fed to petal tissue for 4 h with a simultaneous collection of volatiles. Supplied 2H5-benzaldehyde was converted to benzoic acid and methylbenzoate with the endogenous benzoic acid pool labeled to a greater extent than emitted methylbenzoate (72.4% and 36.3%, respectively; Fig. 5B). Phenylacetaldehyde was 100% unlabeled, indicating that it cannot be synthesized from benzaldehyde.
Figure 5.
Identification of relationships between compounds within benzenoid network in petunia. A, In vivo labeling kinetics of benzenoid/phenylpropanoid compounds after feeding petal tissue with 2H5-Phe. Squares represent percent of labeled phenylacetaldehyde; diamonds, benzaldehyde; circles, benzoic acid; and triangles, methylbenzoate. Each point represents an average of results from at least three independent experiments. B, In vivo labeling kinetics of benzenoid/phenylpropanoid compounds after feeding petal tissue with 2H5-benzaldehyde. Black and white bars show percent of labeled and unlabeled compounds, respectively, together representing the entire (100%) pool. C, Effect of AIP, an inhibitor of PAL activity, on emission of benzenoid/phenylpropanoid compounds. Hatched and gray bars represent emission from petal tissue pretreated for 40 min with water and AIP, respectively, prior to supplying Phe for 4 h with simultaneous scent collection.
To determine whether formation of benzaldehyde from Phe occurs via CA, we conducted experiments with a specific inhibitor of PAL, 2-aminoindanephosphonate (AIP; Zon and Amrhein, 1992; Appert et al., 2003). In these experiments, excised corolla limbs were pretreated with AIP (25 μmol) for 40 min and then floral scent was collected for 4 h from limbs continuously fed with 2H5-Phe. Treatment with AIP almost completely eliminated emission of methylbenzoate, whereas emission of benzaldehyde decreased by 74% when compared with excised petals pretreated with water instead of AIP (Fig. 5C). AIP did not inhibit the emission of phenylacetaldehyde, and, in fact, it increased it by approximately 2-fold, suggesting that the formation of phenylacetaldehyde from Phe does not occur via CA and that its synthesis may be in competition with CA synthesis for Phe utilization.
Modeling the Benzenoid Network
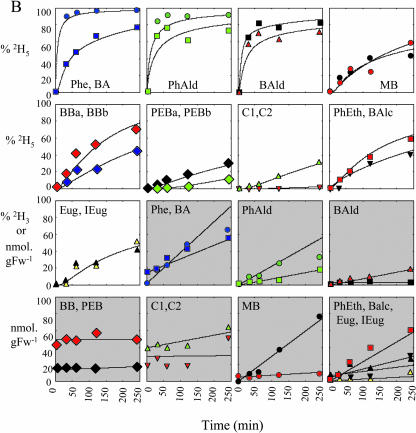
Endogenous intermediates and end products in the benzenoid network were analyzed for pool sizes and isotope abundances after feeding corolla limbs with 2H5-Phe for 30, 60, 120, and 240 min under dark conditions. After hexane or methanol extraction, samples were analyzed by GC-MS or LC-MS, and experimentally obtained results are summarized in Figure 6B. These data were combined with pool sizes and isotope abundances of the volatile fraction and inhibitor studies and subjected to computer-assisted isotopic flux analysis (Stephanopoulos et al., 1998), in which fluxes were assumed to be constant and not dependent on pool sizes. The model was developed to interpret the labeling patterns from supplied 2H5-Phe to establish the route(s) by which petunia petals synthesize benzenoid compounds. Figure 6A shows a schematic representation of precursor-product relationships in this model developed based on 2H5-Phe labeling experiments conducted in this study. Two sets of 2H5-Phe labeling experiments were performed showing similar results, while the model was generated based on one set. Charts with white background on Figure 6B show the fit between the model-generated isotopic labeling curves and experimentally obtained data points, and charts with gray background show the fit between experimentally determined and model simulated pool size changes. The metabolic network presented in Figure 6A is composed of 38 fluxes, designated v1 to v38, which were adjusted until satisfactory fits to the time courses were obtained (McNeil et al., 2000b). Since isoeugenol and eugenol were also detected in endogenous extracts, these two phenylpropenes were also included in the benzenoid network model, as well as phenylpropanoid-related compounds, phenylacetaldehyde, phenylethanol, and phenylethyl benzoate.
Figure 6.
Metabolic modeling of in vivo labeling kinetics and pool sizes of benzenoid and phenylpropanoid compounds in petunia petal tissue supplied with 2H5-l-Phe for up to 4 h in the dark. A shows the metabolic scheme simulated. B, charts with white background show experimentally determined and simulated isotopic labeling of key intermediates and end products; charts with gray background show experimentally determined and simulated pool sizes of intermediates and end products. Symbols of different colors shown in B represent experimentally observed isotope abundances (% 2H5 [or 2H3 in the case of eugenol and isoeugenol]) and pool sizes of various metabolites (as measured by LC-MS or GC-MS) after supplying excised petunia petals with 2H5-Phe in the dark. Metabolites emitted to the gas phase are shaded in yellow in A. Black lines and curves shown in B represent computer-simulated values assuming the model of precursor-product relationships shown in A, and assuming the fluxes defined in the table on A and initial pool sizes defined below. Initial pool sizes at time T = 0 (min) (nmol g FW−1); endogenous l-Phe = 450; endogenous phenylacetaldehyde (PhAld) = 0.1; exogenous phenylacetaldehyde (PhAld; yellow shaded) = 0; endogenous phenylethanol (PhEth) = 0.1; endogenous CA = 10; endogenous 4-coumarate plus caffeate (Caff) = 25; endogenous isoeugenol (IEug) = 5; endogenous eugenol (Eug) = 1; endogenous benzoyl-CoA (BCoA) = 2; endogenous benzylbenzoate (BB) = 200 (benzoic acid moiety = BBa; benzyl alcohol moiety = BBb); endogenous benzyl alcohol (BAlc) = 14 [sum of a metabolic (2 nmol g FW−1)] and storage (s) (12 nmol g FW−1) pools]; endogenous benzaldehyde (BAld) = 6; exogenous benzaldehyde (BAld; yellow shaded) = 0; endogenous benzoic acid (BA) = 60; unidentified nonvolatile benzoic acid conjugate observed in LC-MS analyses of the methanolic extracts of petal tissue (C1) = 160; unidentified nonvolatile benzoic acid conjugate observed in LC-MS analyses of the methanolic extracts of petal tissue (C2) = 294; phenylethyl benzoate = 14; endogenous methylbenzoate (MB) [sum of a metabolic (5 nmol g FW−1)] and storage (s) (15 nmol g FW−1) pools] = 20; exogenous methylbenzoate (MB; yellow shaded) = 0. To display pool sizes on a single scale in charts with gray background, both observed and simulated pool sizes were multiplied by the following scaling factors: endogenous Phe = 1/200; endogenous PhAld = 1/4; exogenous PhAld = 1/4; endogenous PhEth = 1; endogenous benzylbenzoate (BBa and BBb) = 1/2; endogenous MB = 1/4; exogenous MB = 1/4; endogenous Eug and IEug = 1; endogenous C1 = 1/4; endogenous C2 = 1/10; endogenous and exogenous BAld = 1/2; endogenous BA = 1/4; endogenous phenylethyl benzoate (PEBa and PEBb) = 1/2. Pool size and labeling data were not acquired for the following endogenous metabolites in this labeling experiment: BCoA, CA, PhPyr/PhLac, and Caff. Phenylpyruvate and phenyllactic acid have been proposed as intermediates in PhEth synthesis in rose petals (Watanabe et al., 2002). De novo synthesis of unlabeled Phe from chorismate (Chor; v2) (and/or release of unlabeled Phe from protein via protein turnover) is assumed to contribute to a small isotope dilution of the endogenous Phe pool.
In order to simultaneously account for the observed labeling patterns of benzoic acid (BA), endogenous and exogenous benzaldehyde (BAld), endogenous benzylbenzoate, and both the endogenous and exogenous pools of methylbenzoate (MB), both benzylbenzoate and benzaldehyde must be intermediates between Phe and benzoic acid. This indicates that both the CoA-dependent, β-oxidative and CoA-independent, non-β-oxidative pathways are responsible for the biosynthesis of benzenoid compounds in petunia flowers. It was impossible to account for the labeling patterns of these metabolites and observed emission rates when only a single pathway was assumed to be metabolically active. The involvement of benzaldehyde as an intermediate in the biosynthesis of methylbenzoate from Phe was confirmed by the fact that deuterium-labeled benzaldehyde supplied to the petals was converted to benzoic acid and methylbenzoate (Fig. 5B).
The flux from CA → benzaldehyde → benzoic acid is estimated to be about twice that of the flux from CA → benzoyl-CoA → benzylbenzoate → benzoic acid (v18 = 2.8 and v11 = 1.55 nmol min−1 g FW−1, respectively; Fig. 6A). Interestingly, the two moieties of benzylbenzoate (designated BBa and BBb, corresponding to the benzoic acid and benzyl alcohol moieties of the molecule, respectively) become labeled to different extents, with the benzoic acid moiety more heavily labeled than the benzyl alcohol moiety (Fig. 6B). To account for this differential labeling of these two moieties, a small pool of benzyl alcohol derived from both benzaldehyde and benzylbenzoate was required and confirmed experimentally.
The large pool of benzylbenzoate (200 nmol g FW−1) accounted for the reduced labeling of benzoic acid, with both moieties of benzylbenzoate contributing to this dilution via different routes. The benzyl alcohol moiety reduced the labeling of benzoic acid indirectly via the dilution of benzaldehyde labeling. The experimentally obtained benzaldehyde labeling was consistent with the flux from benzyl alcohol to benzaldehyde acting as a reversible step (Fig. 6A). The benzoic acid moiety of benzylbenzoate directly contributed to the dilution of benzoic acid labeling. This moiety of benzylbenzoate is derived from benzoyl-CoA, which is synthesized primarily from the CoA-dependent, β-oxidative pathway, although the CoA-independent, non-β-oxidative pathway also makes a small contribution to its formation via the reverse flux from benzoic acid to benzoyl-CoA (Fig. 6A). The flux to benzoyl-CoA through the non-β-oxidative pathway is predicted to be approximately 20 times lower than that from the CoA-dependent, β-oxidative pathway (v24 = 0.07 versus v11 = 1.55 nmol min−1 g FW−1) because higher fluxes through this route led to an underestimation of the level of labeling of the benzoic acid moiety of benzylbenzoate.
To test the model-derived precursor-product relationships, 2H7-benzyl alcohol (37.5 μmol) was supplied to excised petunia petals for 4 h, and endogenous intermediates and end products in the benzenoid network were analyzed for pool sizes and isotope abundances. Supplied 2H7-benzyl alcohol led to the labeling of benzaldehyde (approximately 90%), methylbenzoate (approximately 42%), and both moieties of benzylbenzoate with the benzyl alcohol moiety more heavily labeled (approximately 56%) than the benzoic acid moiety (approximately 25%). These results confirmed that conversion of benzaldehyde to benzyl alcohol is reversible. The lower labeling of the benzyl alcohol moiety of benzylbenzoate relative to benzaldehyde labeling, each requiring one step with almost the same flux (v14 = 1.5 nmol min−1 g FW−1 and v17 = 1.4 nmol min−1 g FW−1), as was predicted by the model (Fig. 6A), is due to its dilution by the large pool size of unlabeled benzylbenzoate. The labeling of the benzoic acid moiety of benzylbenzoate is possible only if a flux from benzoic acid to benzoyl-CoA exists, confirming the model prediction of the reversibility of benzoyl-CoA → benzoic acid flux. The benzoate:CoA ligase enzyme that catalyzes the thioesterification of benzoic acid to its corresponding CoA derivative was recently partially purified from Clarkia breweri flowers (Beuerle and Pichersky, 2002b).
Over a time course, LC-MS analyses not only revealed changes in endogenous pool sizes and isotope abundances of benzoic acid but also identified two unknown endogenous benzoic acid derivatives (C1 and C2), one of which is likely a glucoside of benzoic acid (Fig. 6A). At least one of these conjugates (C1) is metabolized back to benzoic acid, contributing to isotope dilution of the benzoic acid and methylbenzoate pools. We did not find any other labeled intermediates in LC-MS analyses, which could be due to our extraction procedure, pool sizes below our detection level, or channeling of these compounds. However, the model predicted the existence of other fates of benzoic acid, which were not yet identified.
At the earliest sampling time, the emitted methylbenzoate was more highly labeled (18% at the 30-min time point) than would be expected from the experimentally obtained total internal methylbenzoate labeling (11%), eliminating the possibility of the existence of a single pool. It was necessary to invoke a large (75% of total), metabolically inactive (storage) pool of methylbenzoate, likely localized in the vacuole, in slow exchange (v29 = 0.05 nmol min−1 g FW−1) with a metabolically active pool, resulting in reduced labeling of the total extractable endogenous methylbenzoate (Fig. 6A).
Formation of phenylacetaldehyde from Phe probably occurs via a small pool of the intermediate phenylethylamine or phenylpyruvic acid (Fig. 1). The assumption that phenylacetaldehyde is the only precursor of phenylethanol led to the overestimation of the isotopic abundance of the latter, indicating that phenylacetaldehyde could only make a small contribution to phenylethanol labeling (Fig. 6A). A quantitative explanation of the labeling kinetics of phenylethanol suggests that the major flux to phenylethanol goes through a different route, possibly through phenylpyruvate and phenyllactic acid, as has been reported recently in rose flowers (Watanabe et al., 2002). Phenylethanol together with benzoyl-CoA are precursors of phenylethyl benzoate. However, similar to benzylbenzoate, these two moieties, phenylethanol and benzoyl-CoA, were differentially labeled (Fig. 6B) with the benzoyl-CoA moiety labeled more heavily due to a higher flux from Phe to benzoyl-CoA through the CoA-dependent, β-oxidative pathway than from Phe to phenylethanol (v11 = 1.55 nmol min−1 g FW−1 and v5 = 0.26 nmol min−1 g FW−1, respectively; Fig. 6A). While the label in the benzoyl-CoA moiety was found after 30 min of feeding (approximately 2% 2H5-benzoyl-CoA), which increased up to 10% and 30% after 1 and 2 h, respectively, the appearance of labeling in the phenylethanol moiety was detected only after 2 h of 2H5-Phe feeding (approximately 10%).
The labeling kinetics of isoeugenol and eugenol (Fig. 6B) were consistent with synthesis from trans-cinnamate (via 4-coumarate and caffeic acid derivatives) in a 5:1 flux ratio, respectively. Because methyleugenol and methylisoeugenol were not found in the hexane extracts and were not emitted to the gas phase, and because eugenol and isoeugenol both behaved as metabolically inert end products, it can be assumed that flowers of petunia cv Mitchell lack the (iso)eugenol O-methyltransferase activity that was previously found in C. breweri (Wang et al., 1997).
Isolation and Characterization of Acyltransferase cDNA That Encodes an Enzyme Capable of Synthesizing Benzylbenzoate in Petunia Flowers
To isolate the gene(s) responsible for benzylbenzoate formation in petunia, we searched a recently generated expressed sequence tag (EST) collection (1,584 sequences) from a cDNA library constructed from mRNA isolated from petunia corolla limbs of buds to 2-d-old flowers for potential acyltransferases. This search revealed one EST clone of 1,637 nucleotides (1-1-F06) with 90% amino acid identity to a recently isolated BEBT from Nicotiana tabacum (D'Auria et al., 2002; Fig. 7). This full-length cDNA encodes an open reading frame of 1,380 nucleotides, corresponding to a protein of 460 amino acids with a calculated molecular mass of 51,039 D and pI of 6.12 (GenBank accession no. AY611496). The protein encoded by this cDNA also has 72% and 57% amino acid identity to BEBT from C. breweri and acetyl-CoA:cis-3-hexen-1-ol acetyl transferase (CHAT) from Arabidopsis, respectively (D'Auria et al., 2002; Fig. 7). In addition, it exhibits from 55% to 66% amino acid identity to a putative hypersensitivity-related protein from rice (Oryza sativa; NP_922209) and to putative acyltransferases from pear (Pyrus communis; AAS48090) and muskmelon (Cucumis melo; AAL77060). The tentative petunia BEBT contains the key features of the BAHD family of acyltransferases, the HxxxD motif, which is believed to be involved in catalysis, and the DFGWG motif of unknown function (St-Pierre and De Luca, 2000; Fig. 7).
Figure 7.
Comparison of the predicted amino acid sequence of petunia BPBT with related proteins. Petunia BPBT sequence (PhBPBT) was aligned with BEBT from N. tabacum (NtBEBT, AAN09798), BEBT from C. breweri (CbBEBT, AAN09796), CHAT from Arabidopsis (AtCHAT, AAN09797), and BEAT from C. breweri (CbBEAT, AAC18062) using ClustalW. This alignment was shaded using Boxshade 3.21 software program (Human Genome Sequencing Center, Houston). Residues shaded in black indicate conserved amino acids identical in least two sequences shown, unless two of the others are identical to each other, and residues shaded in gray represent similar matches. Dashes indicate gaps that have been inserted for optimal alignment. Stars and diamonds indicate the HxxxD and DFGWG motifs, respectively.
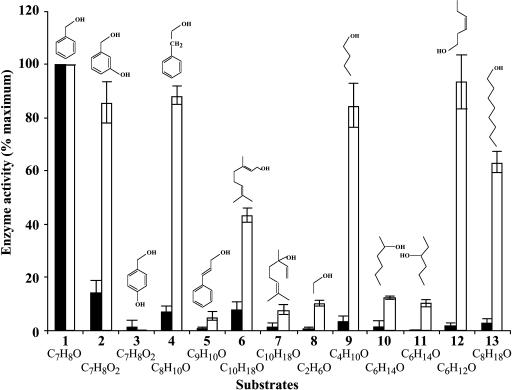
To determine the enzymatic activity of the putative BEBT protein, the coding region of the gene was subcloned into the expression vector pET-11d, expressed in Escherichia coli, and the recombinant protein was purified to near homogeneity using a weak anion-exchange DEAE-cellulose column followed by Mono-Q anion-exchange chromatography and by size-exclusion chromatography. The purified recombinant protein was used to evaluate its ability to catalyze the transfer of benzoyl and acetyl moieties to a large variety of potential substrate alcohols (Fig. 8) and to determine its general catalytic properties and kinetic parameters with the preferred substrates (Table I).
Figure 8.
Comparisons of relative specific activities of petunia BPBT with a variety of substrates. Black and white bars represent activities with acetyl-CoA and benzoyl-CoA, respectively, and indicated alcohol cosubstrates. In both cases activity with benzyl alcohol was set to 100% and was 10.2 ± 1.2 nkat/mg protein and 19.7 ± 3.4 nkat/mg protein in presence of acetyl-CoA and benzoyl-CoA, respectively. Alcohol substrates were as follows: 1, benzyl alcohol; 2, 3-hydroxy-benzyl alcohol; 3, 4-hydroxy-benzyl alcohol; 4, 2-phenylethanol, 5, cinnamyl alcohol; 6, geraniol; 7, linalool; 8, ethanol; 9, butanol; 10, 2-hexanol; 11, 3-hexanol; 12, cis-3-hexen-1-ol; and 13, 1-octanol. Each point is the average of three independent experiments. se values are indicated by vertical bars.
Table I.
Kinetic parameters of petunia BPBT
| Substrate | Km | Vmax | Kcat | Kcat/Km |
|---|---|---|---|---|
| μm | nkat/mg | s−1 | mm−1 s−1 | |
| Acetyl-CoA | 682 ± 68 | 15.0 ± 3.1 | 4.4 ± 0.9 | 6.68 ± 1.6 |
| Benzoyl-CoAa | 1555 ± 108 | 194 ± 10 | 57.1 ± 3.1 | 37.2 ± 2.7 |
| Benzoyl-CoAb | 875 ± 134 | 279 ± 48 | 81.3 ± 14 | 92.2 ± 2.5 |
| Benzyl alcoholc | 28.6 ± 3.5 | 5.52 ± 0.7 | 1.61 ± 0.2 | 56.8 ± 5.5 |
| Benzyl alcohold | 444 ± 44 | 125.5 ± 6 | 36.9 ± 1.8 | 83.1 ± 3.9 |
| Phenyl ethanole | 686 ± 60 | 122.9 ± 3 | 35.8 ± 1 | 52.8 ± 3.6 |
Benzoyl-CoA with benzyl alcohol.
Benzoyl-CoA with phenyl ethanol.
Benzyl alcohol with acetyl-CoA.
Benzyl alcohol with benzoyl-CoA.
Phenyl ethanol with benzoyl-CoA.
With acetyl-CoA, the highest activity of recombinant protein was found with benzyl alcohol as a substrate, and approximately 7- to 14-fold lower activities were detected with 3-hydroxy-benzyl alcohol, geraniol, and 2-phenylethanol (Fig. 8). Activities with the other tested substrates, including butanol, 1-octanol, 4-hydroxy-benzyl alcohol, 2-hexanol, cis-3-hexen-1-ol, and linalool, did not exceed 3% of the highest activity (Fig. 8). With benzoyl-CoA, the protein displayed a broad substrate preference, using efficiently a number of substrates, including benzyl alcohol, 3-hydroxy-benzyl alcohol, 2-phenylethanol, butanol, cis-3-hexen-1-ol, and 1-octanol (Fig. 8).
Kinetic characterization of the purified recombinant protein revealed that the apparent Km for acetyl-CoA is very similar to the value recently reported for C. breweri BEBT (682 μm and 818 μm for petunia and Clarkia proteins, respectively [D'Auria et al., 2002]), whereas the apparent Km for benzoyl-CoA is 75 times higher for the petunia enzyme (1.5 mm) than that for BEBT from Clarkia (20.5 μm; Table I). The apparent catalytic efficiency (Kcat:Km ratio) of the petunia putative BEBT with benzoyl-CoA was almost 6-fold higher than with acetyl-CoA (Table I) as was found for C. breweri BEBT (D'Auria et al., 2002), indicating that benzoyl-CoA is the preferred substrate. In the presence of benzoyl-CoA, the apparent Km value for benzyl alcohol was 444 μm, which is 15 times higher than the apparent Km value for benzyl alcohol with acetyl-CoA (28.6 μm); however, the protein can use benzyl alcohol 1.5 times more efficiently with benzoyl-CoA as a cosubstrate than with acetyl-CoA. These results show that benzoyl-CoA and benzyl alcohol are likely in vivo substrates of BEBT. Moreover, a substantial internal pool of benzylbenzoate (76 μg g FW−1 on day 2) was found in petunia flowers, whereas benzylacetate (0.22 μg g FW−1 on day 2) was barely detectable. Petunia flowers also produce phenylethyl benzoate (4.6 μg g FW−1 on day 2), and the putative petunia BEBT protein can be responsible for its formation since it exhibits strong activity with phenylethanol and benzoyl-CoA as substrates (Fig. 8). The apparent Km value of putative BEBT for phenylethanol in the presence of benzoyl-CoA was 686 μm, which is 1.5 times higher than the Km value for benzyl alcohol (Table I). The catalytic efficiency of the enzyme with benzyl alcohol and phenylethanol in the presence of benzoyl-CoA differed by only 1.6 times, slightly favoring benzylbenzoate as a product, suggesting that both alcohols could be used as substrates in vivo. Since the isolated putative BEBT gene is most likely responsible for the biosynthesis of both benzylbenzoate and phenylethyl benzoate in petunia, we designated it as benzoyl-CoA:benzyl alcohol/phenylethanol benzoyltransferase (BPBT). To evaluate the specificity of the transferase for the acyl donor, we also checked larger acyl CoA substrates, butyryl-, malonyl-, and hexanoyl-CoA, in the presence of benzyl alcohol and found that petunia BPBT can use butyryl- and hexanoyl-CoA at lower rates (40% and 31% of BPBT activity with benzoyl-CoA, respectively).
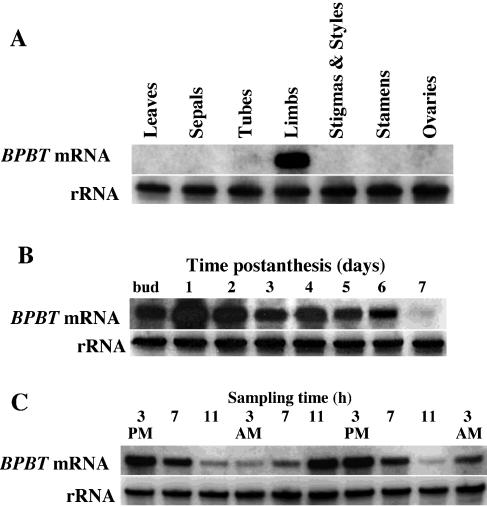
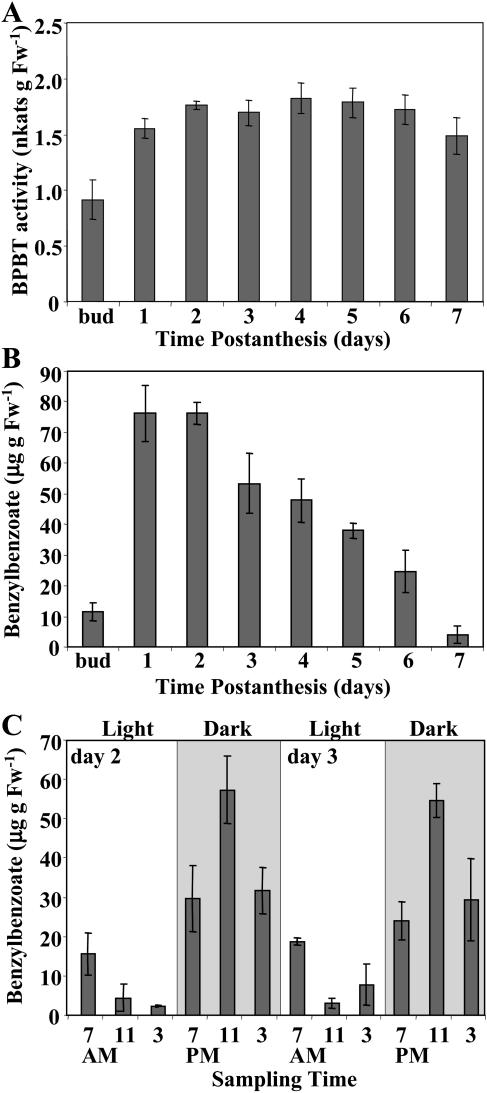
Analysis of BPBT gene expression in leaves and different flower organs of 2-d-old petunia flowers by RNA gel-blot analysis using the BPBT coding region as a probe revealed BPBT mRNA transcripts predominantly in the limb of petunia corollas (Fig. 9A), the parts of the flower that were previously shown to be primarily responsible for scent production and emission in petunia (Kolosova et al., 2001; Negre et al., 2003; Verdonk et al., 2003). The steady-state BPBT mRNA level in corolla limbs was developmentally regulated, peaking 1 to 2 d after anthesis (Fig. 9B) and changing rhythmically during a daily light/dark cycle (Fig. 9C) closely correlating with the pattern of BPBT activity (Fig. 10A) and benzylbenzoate accumulation (Fig. 10, B and C).
Figure 9.
Characterization of petunia BPBT gene expression. A, Tissue specificity of BPBT mRNA. RNA gel blot of total RNA (5 μg per lane) isolated from young leaves, sepals, tubes, and limbs of corollas, pistil, stamens, and ovaries of 2-d-old petunia flowers. The top gel represents the results of hybridization with a coding region of the BPBT genes as a probe. Autoradiography was performed overnight. The blot shown here as well as in B and C were rehybridized with an 18S rDNA probe (bottom) to standardize samples. B, Developmental changes in steady-state BPBT mRNA level in limbs of petunia corollas. Total RNA was isolated at different stages of flower development, from mature buds to day 7 postanthesis. Each lane contained 5 μg of total RNA. Autoradiography was performed overnight. C, RNA gel-blot analysis of steady-state BPBT mRNA levels in petunia flowers during a normal light/dark cycle. Total RNA was isolated from limbs of 2- and 3-d-old flowers at time points indicated at top of figure, and 5 μg of total RNA was loaded in each lane. Autoradiography was performed overnight.
Figure 10.
Characterization of the BPBT activity (A) and internal pools of benzylbenzoate during flower development (B) and two daily light/dark cycles (C) in petunia petals. Each graph represents the average of three independent experiments. sds are indicated by vertical bars. Gray and white areas in C correspond to dark and light, respectively.
DISCUSSION
Benzenoid Network in Petunia Flowers
Benzenoid compounds are among the widespread fragrant components in the plant kingdom, where they contribute significantly to total floral scent output (Knudsen et al., 1993). They also determine the aroma of numerous fruits, thereby enriching their overall flavor quality (Croteau, 1977; Herrmann, 1990). Several enzymes responsible for the final steps in the formation of benzenoid compounds were isolated and characterized (Dudareva, 2002; D'Auria et al., 2002; Lavid et al., 2002); however, the biochemical pathways leading to these simple benzenoid compounds remain unknown. Thus, to establish the routes by which plants make benzenoid compounds in vivo, we used petunia flowers as a model system, which produce a diverse blend of benzenoid/phenylpropanoid-related compounds, some of which are emitted from petunia petals (benzaldehyde, methylbenzoate, and phenylacetaldehyde; Fig. 2; Kolosova et al., 2001; Verdonk et al., 2003) and some of which accumulate within the tissue (benzaldehyde, benzyl alcohol, benzylbenzoate, methylbenzoate, phenylacetaldehyde, phenylethanol, phenylethyl benzoate, isoeugenol, and eugenol). In this study, we applied metabolic flux analysis with stable isotope labeling not only to determine the biochemical pathways leading to the formation of benzenoid compounds in petunia but also to determine the interconnections between intermediates within the benzenoid network. Recent examples of the application of these combined techniques in plant biochemistry include the use of radiolabeling and mathematical modeling for the elucidation of the pathway of synthesis of 3-dimethylsulfoniopropionate in Spartina alterniflora (Kocsis et al., 1998) and for the quantification of the in vivo metabolic fluxes via the network of choline, phosphocholine, and phosphatidylcholine biosynthesis and metabolism in tobacco (McNeil et al., 2000a).
Mathematical models can handle the large number of interacting variables of metabolic networks, assist in explaining the sometimes counter-intuitive behavior of metabolic networks in response to genetic or other perturbations, and provide information on the relative flux of carbon through a number of key reactions and intermediates (Bailey, 1998, 1999). Consequently, computer-assisted metabolic flux and control analysis have become essential tools for microbial metabolic engineers (Stephanopoulos et al., 1998; Gombert and Nielsen, 2000). The utility of these tools in plant metabolic engineering is receiving increased attention (Roscher et al., 2000; Morgan and Rhodes, 2002; Rontein et al., 2002; Schwender and Ohlrogge, 2002; Schwender et al., 2003).
Formation of benzenoid compounds from Phe requires the shortening of the side chain by a C2 unit that could occur via the CoA-dependent, β-oxidative pathway, CoA-independent, non-β-oxidative pathway, or via the combination of these pathways (Fig. 1). In our experiments, 2H5-Phe was supplied to excised petunia petals, and the labeling kinetics of the endogenous pools of phenylpropanoid/benzenoid metabolites and the volatiles emitted to the gas phase were monitored using GC-MS and LC-MS. It should be noted that not all intermediates were detected in our labeling experiments, since no currently available LC-based methods allow determination of all these compounds due to differences in solubility and stability, as well as a wide concentration range of these molecules. Metabolic pool sizes and labeling patterns of detected intermediates and end products (Fig. 6B) were incorporated in the flux model (Fig. 6A), which was developed in Visual Basic (described at http://www.hort.purdue.edu/cfpesp/models/models.htm) and used to interpret the results of the labeling experiments and to identify which of the pathways in Figure 1 carries most of the biosynthetic flux. While the fluxes in the model were adjusted to fit experimentally determined labeling and pool sizes of benzenoid compounds, PAL and BSMT activities in fed petunia tissue allowed us to test predetermined computer-simulated fluxes. PAL activity of approximately 152 pkat g FW−1 (average over 4 h; Fig. 3C) provided a flux of approximately 9 nmol min−1 g FW−1, which is less than a 2-fold difference from the flux proposed by the model (v6 = 5.355 nmol min−1 g FW−1; Fig. 6A). Similarly, BSMT activity of approximately 32 pkat g FW−1 (average over 4 h; Fig. 3D) provided a flux of approximately 1.92 nmol min−1 g FW−1, which fits the proposed flux of 1.3 nmol min−1 g FW−1 (v27; Fig. 6A). The agreement between measured enzyme activities and model-determined fluxes provides additional support of the model assumptions in both an early and late stage of the network.
The metabolic flux model developed in this study, combined with the PAL inhibition data, revealed that the formation of phenylpropanoid-related compounds (phenylacetaldehyde, phenylethanol, and the phenylethanol moiety of phenylethyl benzoate) from Phe does not occur via CA (Figs. 5C and 6A). Modeling also revealed that both the CoA-dependent, β-oxidative and CoA-independent, non-β-oxidative pathways contribute to the formation of benzenoid compounds in petunia flowers. According to the model, the flux through the CoA-independent, non-β-oxidative pathway with benzaldehyde as a key intermediate was estimated to be about 2 times higher than the flux through the CoA-dependent, β-oxidative pathway, which relies on the formation of CoA esters. Since the emission of benzenoid compounds occurs predominantly at night, the possibility exists that the degree of contribution of each biosynthetic pathway may change during the light/dark cycle. At present it remains unknown whether the formation of benzaldehyde occurs only via the β-oxidative (CA → cinnamoyl-CoA → 3-hydroxy-3-phenylpropionyl-CoA → benzaldehyde) or only via the non-β-oxidative (CA → 3-hydroxy-3-phenylpropionic → benzaldehyde) pathway, or by the combination of both (Fig. 1). Further identification of the intermediates in benzaldehyde biosynthesis in petunia flowers should enable us to answer this question.
Modeling of 2H5-Phe labeling data indicated that benzylbenzoate is one of the intermediates between l-Phe and benzoic acid. Benzylbenzoate is formed from benzyl alcohol and benzoyl-CoA in a reaction catalyzed by a benzoyl transferase, activity for which was found in petunia petal tissue (Fig. 10A). The activity of this enzyme over flower development showed a typical profile of scent biosynthetic enzymes, where high enzymatic activities were found in old flowers without concomitant emission of corresponding products (Dudareva and Pichersky, 2000). While a small amount of benzylbenzoate was found in the petunia floral scent bouquet, there was a large extractable internal pool in the petal tissue. This internal benzylbenzoate pool (Fig. 10, B and C) positively correlated with developmental (data not shown) and rhythmic methylbenzoate emission (Kolosova et al., 2001). The developmental activity of the enzyme responsible for benzylbenzoate formation (Fig. 10A) did not closely correlate with developmental changes in the internal pool sizes (Fig. 10B). BPBT activity toward benzyl alcohol remained high during flower development, while the internal pool of benzylbenzoate gradually decreased starting from the third DPA, which is likely due to regulation at the level of substrates, as was shown previously for methylbenzoate formation (Dudareva et al., 2000).
Using a functional genomic approach, we have isolated a BPBT gene from a petunia petal-specific EST collection that encodes a protein capable of synthesizing benzylbenzoate (Fig. 7). This gene was expressed almost exclusively in the limbs of petunia corollas (Fig. 9A), and its expression was developmentally and rhythmically regulated (Fig. 9, B and C). During a daily light/dark cycle, its expression peaked around 3 pm, several hours before maximum benzylbenzoate accumulation (Fig. 10C). This earlier expression of the BPBT gene is probably required for building a large pool of benzylbenzoate (200 nmol g FW−1), which contributes to methylbenzoate emission at night via benzoic acid.
Biochemical characterization of the BPBT protein showed that it can use a broad range of alcohols in the presence of benzoyl-CoA, one of which is 2-phenylethanol (Fig. 8). Benzoylation of phenylethanol results in the formation of phenylethyl benzoate, a phenylpropanoid-related compound found in petunia petal tissue. The BPBT catalytic efficiency with phenylethanol was close to that with benzyl alcohol (Table I), suggesting that this enzyme is most likely responsible for the formation of both phenylethyl benzoate and benzylbenzoate in vivo. However, the internal pool of benzylbenzoate (200 nmol g FW−1) was 14 times larger than that of phenylethyl benzoate (14 nmol g FW−1) due to the larger amount of metabolically active benzyl alcohol (2 nmol g FW−1) relative to phenylethanol (0.1 nmol g FW−1) available to the enzyme.
The model developed in this study indicated that benzoyl-CoA, one of the BPBT substrates for formation of both phenylethyl benzoate and benzylbenzoate, is synthesized via the CoA-dependent, β-oxidative pathway, although the CoA-independent, non-β-oxidative pathway also makes a small contribution to its formation via the reverse flux from benzoic acid to benzoyl-CoA (Fig. 6A). Benzyl alcohol, the other substrate of BPBT for benzylbenzoate formation, is derived from both benzaldehyde and benzylbenzoate, as was predicted by modeling and tested by feeding with 2H5-benzyl alcohol. Phenylethylalcohol, the other substrate of BPBT for phenylethyl benzoate formation, is synthesized from phenylethanol, for which two potential routes were included in the model via phenylpyruvate and phenyllactic acid or via phenylacetaldehyde (Fig. 6A).
Although our results do not show any existence of channeling of metabolic intermediates in our established steps of the benzenoid branchway, the possibility exists that channeling of metabolic intermediates occurs through multienzyme complexes early in the pathway (before benzaldehyde), which may result in little or no release of the label into the bulk extractable pools (Hrazdina, 1992; Burbulis and Winkel-Shirley, 1999; Rasmussen and Dixon, 1999). Modeling of metabolic fluxes is a powerful tool for revealing evidence of channeling and quantifying the presence of small rapidly turning-over metabolic pools of intermediates in slow exchange with larger metabolically inactive pools (Kocsis et al., 1998; McNeil et al., 2000a, 2000b). Our model suggests the existence of metabolically active and inactive pools of both methylbenzoate and benzyl alcohol (Fig. 6A). It is possible that there is similar partitioning of Phe, but supplied 2H5-Phe may have masked this. Although the computer simulation model generated here was designed to determine the maximal velocities of the network involved in the production of benzenoid/phenylpropanoid-related compounds in petunia by supplying saturating doses of 2H5-Phe, labeling experiments with different doses of precursors in the presence or absence of metabolic inhibitors and unlabeled trapping pools will greatly assist in revealing evidence for channeling.
Our model did not address the regulatory architecture of the benzenoid network (Stephanopoulos and Vallino, 1991). Also, the interaction between the benzenoid branchway and the general phenylpropanoid pathways remains unknown. Recently, it was shown that blocking the anthocyanin biosynthetic pathway in carnation led to increased methylbenzoate production and emission (Zuker et al., 2002). Metabolic modeling in combination with future genetic perturbations via a reverse genetic approach will help us to understand the reversible, branching, and parallel pathways in the benzenoid network, the results of which will be used to refine the model.
MATERIALS AND METHODS
Plant Material, Chemicals, and Radiochemicals
Petunia (Petunia hybrida) cv Mitchell (Ball Seed, West Chicago, IL) were grown under standard greenhouse conditions as described previously (Kolosova et al., 2001). Headspace collections of floral volatiles were performed in the greenhouse under previously described conditions (Dudareva et al., 2003). For labeling experiments, plants were grown in growth chambers with a reversed photoperiod. Two-day-old flowers were harvested, the tubes were removed, and the corolla limbs were used for all labeling experiments.
Deuterium-labeled Phe (l-Phe-ring-2H5) and benzyl alcohol (2H7-benzyl alcohol) were purchased from Cambridge Isotope Laboratories (Andover, MA), deuterium-labeled benzaldehyde (benzaldehyde-2H6) was from Isotec (Miamisburg, OH), l-Phe was from Sigma (St. Louis), and radiolabeled Phe, U-[14C]Phe (425 mCi/mmol), was from American Radiolabeled Chemicals (St. Louis). Radiolabeled [acetyl-1-14C]CoA (55 mCi/mmol) was purchased from MP Biomedicals (Irvine, CA) and S-[methyl-14C]adenosyl-l-Met (52.7 mCi/mmol) was from Perkin-Elmer Life Sciences (Boston). [7-14C]benzoyl-CoA (53 mCi/mmol) was enzymatically synthesized from [7-14C]benzoic acid (Beuerle and Pichersky, 2002a) and kindly provided to us by Drs. Y. Iijima and E. Pichersky (University of Michigan, Ann Arbor). The PAL specific inhibitor AIP (Zon and Amrhein, 1992) was also a gift from Dr. E. Pichersky. Other chemicals and reagents were purchased from Sigma or Aldrich (Milwaukee, WI), unless otherwise noted.
Labeling Experiments
Stable isotope labeling was conducted in a glass container by placing the cut surface of excised corollas of 2-d-old petunia flowers on moist filter paper supplied with 2H5-Phe (total 10 corollas of 0.2 g each per experiment). A constant stream of air flowing at 450 mL min−1 was drawn through the container with a vacuum pump. The air exiting the sample-filled container was passed through an adsorbent trap consisting of a 120- × 7-mm glass tube containing 100 mg of polymer Porapak Type Q (80/100 mesh; Alltech, Deerfield, IL) held in place with plugs of silanized glass wool. An additional adsorbent trap of identical construction was placed at the inlet of the container to purify incoming air. Trials with two such traps connected in series at the container exit indicated that there was no detectable breakthrough (i.e. no loss of floral volatiles from the first trap due to overloading, even when collections were carried out for periods of up to 24 h). Since petunia cv Mitchell is a nocturnally emitting plant (Kolosova et al., 2001), all labeling experiments were conducted under dark conditions.
Identification and Quantification of Labeled and Unlabeled Compounds
Trapped volatiles were eluted from the trap with hexane, and the amount and isotope abundance of benzenoids emitted to the gas phase were analyzed by GC-MS with a FinniganMAT GCQ instrument (Thermoquest, San Jose, CA; injector temperature 230°C, injector volume 1 μL, and split ratio 50:1) using a DB-1 nonpolar capillary column (30 m × 0.25 mm [i.d.]; film thickness 0.25 μm). Ionization energy was set at 70 eV. Column temperature was held at 50°C for 1 min and then heated to 240°C at 10°C min−1. Mass spectra were obtained in scan mode scanning across 41 to 400 amu. Components were first identified from a computer database containing several thousand mass spectra and were then confirmed by comparing retention times and mass spectra with those of authentic standards. Quantification was based on flame ionization detector peak areas and the internal standards, toluene and napthalene. Single ion monitoring was generally used for the measurements of mass isotopomers. All newly synthesized labeled benzenoid compounds exhibit a mass shift by 5 amu, except eugenol and isoeugenol, which exhibit a shift of only by 3 amu. Two moieties of benzylbenzoate corresponding to the benzoic acid and benzyl alcohol moieties of the molecule were labeled to different extents, and their labeling was determined separately. The percentage of labeling was determined as the intensity of the shifted representative molecular ion divided by the sum of intensities for unshifted and shifted representative molecular ions.
To determine pool sizes and isotope abundance of endogenous pools of intermediates and end products, including phenylacetaldehyde, phenylethanol, eugenol, isoeugenol, benzaldehyde, benzylbenzoate, and methylbenzoate, corolla tissues after feeding were extracted with hexane. Tissue was ground in liquid nitrogen and hexane was added to 4 mL/g of tissue. Tissue was extracted on an Orbit shaker at 170 rpm for 1 h and extract was centrifuged at 10,000 rpm for 10 min, followed by filtration through a 25-mL syringe with a 0.2 μm sterile nylon filter. Filtered extract was evaporated down to 160 μL, and samples were then analyzed by GC-MS as mentioned above. All samples were corrected for recovery as determined using internally spiked samples.
To determine the labeling of endogenous nonvolatile metabolites, including benzoic acid and its conjugates, labeled corolla tissue was extracted with methanol and analyzed by LC-MS.
To determine the internal pool sizes of Phe, corolla tissues were extracted with methanol followed by phase separation with chloroform and water (10 mL of methanol:5 mL of chloroform:6 mL of water; Rhodes et al., 1987). Amino acids in the aqueous phase were purified by Dowex-50-H+ ion-exchange chromatography. The pool size and the isotope abundance of Phe were quantified by GC-MS of N(O,S)-heptafluorobutyryl isobutyl amino acid derivatives, using α-amino-n-butyrate as an internal standard, as previously described (Rhodes et al., 1987).
LC-MS Analysis
After scent collection, petal tissue was extracted with methanol and the isotope abundance of endogenous nonvolatile intermediates was analyzed by LC-MS. Prior to entrance into the mass spectrometer, extracted compounds were separated using a Supelco Discovery HS C18 column (15 cm × 2.1 mm i.d.) attached to a Waters 2690 separations module (Milford, MA) with attached column oven. Compound elution was monitored at 210 and 280 nm with a Waters 996 UV/Visible photodiode array detector. Complete baseline separation of all phenylpropanoid compounds was achieved at a flow rate of 0.25 mL min−1 with the column incubated at constant temperature of 40°C. Solvent A was 0.05% formic acid in water; solvent B was 100% acetonitrile. The column was pre-equilibrated with 5% solvent B in solvent A. After injection of up to 25 μL of aqueous sample, the column was washed with 0.5 mL of pre-equilibration solvent. Compounds were eluted from the column with a linear gradient from 5% to 66% solvent B over 13.75 mL. The column was then washed by increasing solvent B to 100% (linear gradient in 0.75 mL) and holding at 100% solvent B for 0.75 mL. The column was then re-equilibrated by returning the column to 5% solvent B (over 0.75 mL) followed by a 2.5 mL wash with this solvent. Total run time was 70 min.
In-line mass spectrometry (LC-MS) was performed on HPLC eluents using a Micromass Quattro LCZ triple quadruple mass spectrometer (Micromass, Beverly, MA). Flow splitting (10:1) after the Waters 996 photodiode array detector resulted in an inlet flow rate into the mass spectrometer electrospray source (ESI Z-Spray; Micromass) of approximately 20 to 25 μL min−1. This dramatically enhanced the signal when compared to nonsplit samples. Ionization of target molecules in negative ion mode was achieved with a capillary voltage of 3.0 kV and a cone voltage of 30 V. The desolvation and cone gases were set at 430 and 60 L h−1, respectively, and the desolvation and source temperatures were 250°C and 120°C, respectively. Mass detection was performed in negative ion scanning mode, at 450 amu s−1, with 0.1-s interscan delay. All other electrospray source and instrument parameters were set as recommended by the instrument manufacturer. Data analysis was performed using MassLynx computer software (Micromass).
Computer Modeling of Labeling Data
The computer model used was similar to the metabolic flux analysis models described by Kocsis et al. (1998) and McNeil et al. (2000a). Programs were written in Microsoft Visual Basic (Redmond, WA). Key model parameters are the initial pool sizes and their isotopic abundance, and the flux rates connecting the various pools.
For all metabolites, the rate of change of the concentration of metabolite M is taken as
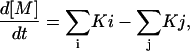 |
where the Kis are the fixed rates of production of M from its various precursor pools, and the Kjs are the fixed rates of conversion of M to its various fates. For the compounds emitted to the gas phase, the Kjs are zero.
During short time intervals (0.125-min iterations), material of the current isotopic abundance was drawn from one pool to another at specified rates, new isotopic abundances and pool sizes were computed, and isotopic abundance in each pool was plotted (superimposed on observed data) as a function of time. The model was primarily constrained by observed rates of emission of methylbenzoate, benzaldehyde, and phenylacetaldehyde to the gas phase (as determined by GC and GC-MS), and observed pool sizes of benzoic acid and its conjugates (C1 and C2; as determined by LC-MS). Other flux rates and pool sizes were progressively adjusted until a close match between observed and simulated isotopic abundance was obtained, as judged graphically or by computing absolute deviations between observed and simulated values. Further details on the development of the type of metabolic models used here are given at http://www.hort.purdue.edu/cfpesp/models/models.htm.
cDNA Library Construction and DNA Sequencing
A directional cDNA library was constructed from mRNA isolated from petunia petal tissues of buds 1 d before opening to 2-d-old flowers using ZAP express cDNA synthesis kit (Stratagene, La Jolla, CA). The titer of the unamplified library was 2.8 × 106 plaque forming units. This primary library was amplified, and the amplified library had a titer of 7 × 109 plaque forming units. Mass excision of pBluescript phagemids (Stratagene) from the amplified library resulted in a stock, which was used for plating and random colony picking for DNA sequencing. A total of 1,584 cDNAs were automatically isolated and sequenced from their 5′ end using T3 primer. After vector removal, the resulting petunia ESTs were compared with GenBank and dbEST using the BLASTX and TBLASTX search algorithms.
Functional Expression of BPBT in Escherichia coli and Purification of Recombinant Protein
The coding region of petunia BPBT was amplified by PCR using the forward primer 5′-CTTCCATGGATTCAAAGCAATCATCAGA-3′, which introduced an NcoI site at the initiating ATG codon in combination with the reverse primer 5′-CCTAGCTCATAGGGCAGGTGTGATAAAGGC-3′, which introduced a BamHI site downstream of the stop codon and subcloned into the expression vector pET 11d (Novagen, Madison, WI). Sequencing revealed that no errors had been introduced during PCR amplifications.
For functional expression, E. coli BL834 (DE3) cells were transformed with the resulting recombinant plasmid and pET vector without an insert as control, and grown in Luria-Bertani medium with 100 μg/mL ampicilin at 37°C. When the culture density reached OD600 of 0.5, the expression of BEBT cDNA was induced by addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 0.4 mm. After 20-h incubation with shaking (200 rpm) at 20°C, E. coli cells were harvested by centrifugation and resuspended in buffer A [3:1 (v/w) buffer:cells] containing 50 mm Bis-Tris-HCl, pH 6.9, 10% glycerol, and 10 mm β-mercaptoethanol. Resuspended cells were broken by French press (three passes at 1,200 psi), and the cell debris was removed by centrifugation (30 min at 24,000g). The supernatant was incubated with protamine sulfate [1% (w/v)] for 30 min at 4°C. After centrifugation (30 min at 24,000g), supernatant was dialyzed against 2 L of buffer A overnight at 4°C. BPBT protein was purified using a weak anion-exchange DEAE-cellulose column followed by Mono-Q anion-exchange and size-exclusion chromatographies.
First, BPBT protein was loaded onto a DEAE-cellulose column (18 mL of DE53; Whatman, Clifton, NJ) preequilibrated with buffer A at a flow rate of about 1 mL/min. After washing off unabsorbed material from the column with 60 mL of buffer A, BPBT was eluted with a linear gradient (120 mL) from 0 to 500 mm KCl in buffer A. Fractions (3 mL) were collected and assayed for BPBT activity with benzyl alcohol as a substrate. Fractions with the highest BPBT activity in the 140 to 225 mm KCl range were pooled (total of 18 mL), dialyzed against buffer A, and subjected to ion-exchange chromatography on a Mono-Q column (gel bed volume 1 mL) (Amersham Biosciences, Piscataway, NJ) using the FPLC system. BPBT protein was eluted from the column at 80 to 160 mm KCl using a 15-mL linear (0–400 mm) gradient of KCl in buffer A at flow rate of 0.5 mL/min. Fractions with highest BPBT activity in the 75 to 165 mm KCl range (total of 4 mL) were pooled and subjected to size-exclusion chromatography (Superdex 200; 16 mm × 60 cm). Fractions of 2 mL were collected at a flow rate of 1 mL/min and analyzed for BPBT activity. Fractions with highest BPBT activity were examined by SDS-PAGE gel electrophoresis followed by Coomassie Brilliant Blue staining of the gel.
For kinetic analysis, an appropriate enzyme concentration was chosen so that the reaction velocity was proportional to the enzyme concentration and was linear with respect to incubation time for at least 30 min. Protein concentrations were determined by the Bradford method (Bradford, 1976) using the Bio-Rad (Hercules, CA) protein reagent and bovine serum albumin as a standard. Kinetic data were evaluated using Lineweaver-Burk, Eadie-Hofstee, and Hanes plots (Segel, 1993).
Enzyme Assays
Crude protein extracts were made from corolla limbs of petunia flowers as described previously (Dudareva et al., 2000) and used immediately or stored at −80°C until needed. Crude extracts for PAL activity were passed through small Sephadex G-25 spin columns to remove the excess of Phe from fed tissue. PAL activity was determined by measuring the formation of U-14C-CA from U-14C-l-Phe (Kolosova et al., 2001), and BSMT was determined by measuring transfer of the 14C-labeled methyl group of S-[methyl-14C]adenosyl-l-Met to the carboxyl group of benzoic acid (Dudareva et al., 2000). BPBT activity was determined as described previously (D'Auria et al., 2002). Acetyltransferase assays for BPBT were performed according to Dudareva et al. (1998a). All assays were carried out at room temperature for 30 min. The radioactively labeled products were extracted with hexane and counted in a liquid scintillation counter (model LS 3801; Beckman, Fullerton, CA). The raw data (cpm) were converted to picokatals (picomoles of product produced per second) based on the specific activity of the substrate and efficiency of counting. Controls included assays with boiled extracts and without substrate, and background radioactivity produced in such assays was subtracted from all of the results.
Verification of products produced by BPBT in benzoylation and acylation reactions was achieved by performing enzyme assays as described above only using nonradioactive benzoyl-CoA and acetyl-CoA instead of radioactive ones and analyzing the products by GC-MS.
RNA Isolation and RNA Gel-Blot Analysis
Total RNA was isolated and analyzed as previously described (Dudareva et al., 1996, 1998a, 2000; Kolosova et al., 2001) from petal tissues fed with Phe and petal tissue harvested at the same time points directly from intact petunia plants, from floral tissues and from petals at different stages of flower development, and 10 time points during a daily light/dark cycle. A 1-kb EcoRI fragment containing the coding region of the BSMT genes, a 2.2-kb EcoRI/XhoI fragment containing the PAL gene coding region, and a 1.9-kb EcoRI fragment containing the coding region of the BPBT gene were used as probes in RNA gel-blot analyses. Five micrograms of total RNA was loaded in each lane. The blots were rehybridized with 18S rDNA for loading control.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY611496.
Acknowledgments
We thank Dr. Yoko Iijima and Dr. Eran Pichersky for their generous gift of [7-14C]benzoyl-CoA, and Dr. Eran Pichersky for sharing with us 2-aminoindane-phosphonate. We also thank Dr. John C. D'Auria for his helpful discussions and Dr. Stanislav Zakharov for his help with the gel filtration chromatography.
This work was supported by the U.S. Department of Agriculture National Research Initiative (grant no. 2003–35318–13619) and by the Fred Gloeckner Foundation (grant to N.D.). This paper is contribution Number 17397 from the Purdue University Agricultural Experimental Station.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.045468.
References
- Ahmed MA, El-Mawla A, Beerhues L (2002) Benzoic acid biosynthesis in cell cultures of Hypericum androsaemum. Planta 214: 727–733 [DOI] [PubMed] [Google Scholar]
- Alibert G, Ranjeva R (1971) Research on the enzymes catalyzing the biosynthesis of phenolic acids in Quercus pedunculata (Ehrh.): I – Formation of the first members of the cinnamic series and benzoic series. FEBS Lett 19: 11–14 [DOI] [PubMed] [Google Scholar]
- Anterola AM, Jeon JH, Davin LB, Lewis NG (2002) Transcriptional control of monolignol biosynthesis in Pinus taeda: factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J Biol Chem 277: 18272–18280 [DOI] [PubMed] [Google Scholar]
- Appert C, Zon J, Amrhein N (2003) Kinetic analysis of the inhibition of phenylalanine ammonia-lyase by 2-aminoindan-2-phosphonic acid and other phenylalanine analogues. Phytochemistry 62: 415–422 [DOI] [PubMed] [Google Scholar]
- Bailey JE (1998) Mathematical modeling and analysis in biochemical engineering: past accomplishments and future opportunities. Biotechnol Prog 14: 8–20 [DOI] [PubMed] [Google Scholar]
- Bailey JE (1999) Lessons from metabolic engineering for functional genomics and drug discovery. Nat Biotechnol 17: 616–618 [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E (2002. a) Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem 302: 305–312 [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E (2002. b) Purification and characterization of benzoate: coenzyme A ligase from Clarkia breweri. Arch Biochem Biophys 400: 258–264 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burbulis IE, Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96: 12929–12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau R (1977) Biosynthesis of benzaldehyde, benzyl alcohol and benzyl benzoate from benzoic acid in cranberry (Vaccinium macrocarpon). J Food Biochem 1: 317–326 [Google Scholar]
- D'Auria JC, Chen F, Pichersky E (2002) Characterization of an acyltransferase capable of synthesizing benzylbenzoate and other volatile esters in flowers and damaged leaves of Clarkia breweri. Plant Physiol 130: 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N (2002) Molecular control of floral fragrance. In A Vainstein, ed, Breeding for Ornamentals: Classical and MolecularApproaches. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 295–309
- Dudareva N, Cseke L, Blanc VM, Pichersky E (1996) Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, D'Auria JC, Nam KH, Raguso RA, Pichersky E (1998. a) Acetyl-CoA:benzylalcohol acetyltransferase – an enzyme involved in floral scent production in Clarkia breweri. Plant J 14: 297–304 [DOI] [PubMed] [Google Scholar]
- Dudareva N, Martin D, Kish CM, Kolosova N, Gorenstein N, Faldt J, Miller B, Bohlman J (2003) (E)-b-Ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new TPS-subfamily. Plant Cell 15: 1227–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Murfitt LM, Mann CJ, Gorenstein N, Kolosova N, Kish CM, Bonham C, Wood K (2000) Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell 12: 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E (2000) Biochemical and molecular genetic aspects of floral scents. Plant Physiol 122: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Raguso RA, Wang J, Ross JR, Pichersky E (1998. b) Floral scent production in Clarkia breweri. III. Enzymatic synthesis and emission of benzenoid esters. Plant Physiol 116: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CJ, Vance CP, Towers GHN (1976) Conversion of p-coumaric acid to p-hydroxybenzoic acid by cell free extracts of potato tubers and Polyporus hispidus. Phytochemistry 15: 564–566 [Google Scholar]
- Gombert AK, Nielsen J (2000) Mathematical modeling of metabolism. Curr Opin Biotechnol 11: 180–186 [DOI] [PubMed] [Google Scholar]
- Herrmann K (1990) Salicyclic-acid and other hydroxybenzoic acids and their naturally-occurring compounds in food. Ernahrungs-Umschau 37: 108–112 [Google Scholar]
- Hertweck C, Jarvis AP, Xiang LK, Moore BS, Oldham NJ (2001) A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid beta-oxidation. Chembiochem 2: 784–786 [DOI] [PubMed] [Google Scholar]
- Hertweck C, Moore BS (2000) A plant-like biosynthesis of benzoyl-CoA in the marine bacterium ‘Streptomyces maritimus’. Tetrahedron 56: 9115–9120 [Google Scholar]
- Hrazdina G (1992) Compartmentation in aromatic metabolism. Recent Adv Phytochem 26: 1–23 [Google Scholar]
- Jarvis AP, Schaaf O, Oldham NJ (2000) 3-Hydroxy-3-phenylpropanoic acid is an intermediate in the biosynthesis of benzoic acid and salicylic acid but benzaldehyde is not. Planta 212: 119–126 [DOI] [PubMed] [Google Scholar]
- Knudsen JT, Tollsten L, Bergstrom G (1993) Floral scents – a checklist of volatile compounds isolated by headspace techniques. Phytochemistry 33: 253–280 [Google Scholar]
- Kocsis MG, Nolte KD, Rhodes D, Shen TL, Gage DA, Hanson AD (1998) Dimethylsulfoniopropionate biosynthesis in Spartina alterniflora. Evidence that S-methylmethionine and dimethylsulfoniopropylamine are intermediates. Plant Physiol 117: 273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova N, Gorenstein N, Kish CM, Dudareva N (2001) Regulation of circadian methylbenzoate emission in diurnally and nocturnally emitting plants. Plant Cell 13: 2333–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavid N, Wang JH, Shalit M, Guterman I, Bar E, Beuerle T, Menda N, Shafir S, Zamir D, Adam Z, et al (2002) O-methyltransferases involved in the biosynthesis of volatile phenolic derivatives in rose petals. Plant Physiol 129: 1899–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao LF, Chu CH, Schulz H (1994) Hepatic beta-oxidation of 3-phenylpropionic acid and the stereospecific dehydration of (R)-3-hydroxy-3-phenylpropionyl-CoA and (S)-3-hydroxy-3-phenylpropionyl-CoA by different enoyl–CoA hydratases. Biochemistry 33: 3320–3326 [DOI] [PubMed] [Google Scholar]
- McNeil SD, Nuccio ML, Rhodes D, Shachar-Hill Y, Hanson AD (2000. a) Radiotracer and computer modeling evidence that phospho-base methylation is the main route of choline synthesis in tobacco. Plant Physiol 123: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil SD, Rhodes D, Russell BL, Nuccio ML, Shachar-Hill Y, Hanson AD (2000. b) Metabolic modeling identifies key constraints on an engineered glycine betaine synthesis pathway in tobacco. Plant Physiol 124: 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JA, Rhodes D (2002) Mathematical modeling of plant metabolic pathways. Metab Eng 4: 80–89 [DOI] [PubMed] [Google Scholar]
- Murfitt LM, Kolosova N, Mann CJ, Dudareva N (2000) Purification and characterization of S-adenosyl-L-methionine:benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methyl benzoate in flowers of Antirrhinum majus. Arch Biochem Biophys 382: 145–151 [DOI] [PubMed] [Google Scholar]
- Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C (2004) The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre F, Kish CM, Boatright J, Underwood B, Shibuya K, Wagner C, Clark DG, Dudareva N (2003) Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. Plant Cell 15: 2992–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre F, Kolosova N, Knoll J, Kish CM, Dudareva N (2002) Novel S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme responsible for biosynthesis of methyl salicylate and methyl benzoate, is not involved in floral scent production in snapdragon flowers. Arch Biochem Biophys 406: 261–270 [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Dixon RA (1999) Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11: 1537–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Hogan AL, Deal L, Jamieson GC, Haworth P (1987) Amino acid metabolism of Lemna minor L. II. Responses to chlorsulfuron. Plant Physiol 84: 775–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscher A, Kruger NJ, Ratcliffe RG (2000) Strategies for metabolic flux analysis in plants using isotopic labeling. J Biotechnol 77: 81–102 [DOI] [PubMed] [Google Scholar]
- Rontein D, Dieuaide-Noubhani M, Dufourc EJ, Raymond P, Rolin D (2002) The metabolic architecture of plant cells – stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J Biol Chem 277: 43948–43960 [DOI] [PubMed] [Google Scholar]
- Ross JR, Nam KH, D'Auria JC, Pichersky E (1999) S-adenosyl-L-methionine: salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch Biochem Biophys 367: 9–16 [DOI] [PubMed] [Google Scholar]
- Schnitzler J-P, Madlung J, Rose A, Seitz HU (1992) Biosynthesis of p-hydroxybenzoic acid in elicitor-treated carrot cell cultures. Planta 188: 594–600 [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB (2002) Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing Brassica napus embryos. Plant Physiol 130: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y (2003) A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J Biol Chem 278: 29442–29453 [DOI] [PubMed] [Google Scholar]
- Segel IH (1993) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. John Wiley and Sons, New York
- Stephanopoulos GN, Aristidou AA, Nielsen J (1998) Metabolic Engineering Principles and Methodologies. Academic Press, San Diego
- Stephanopoulos GN, Vallino JJ (1991) Network rigidity and metabolic engineering in metabolite overproduction. Science 252: 1675–1681 [DOI] [PubMed] [Google Scholar]
- St-Pierre B, De Luca V (2000) Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. In JT Romeo, R Ibrahim, L Varin, V De Luca, eds, Evolution of Metablic Pathways, Ed 1, Vol 34. Elsevier Science, Oxford, pp 285–315
- Verdonk JC, Ric de Vos CH, Verhoeven HA, Haring MA, van Tunen AJ, Schuurink RC (2003) Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry 62: 997–1008 [DOI] [PubMed] [Google Scholar]
- Wang J, Dudareva N, Bhakta S, Raguso RA, Pichersky E (1997) Floral scent production in Clarkia breweri (Onagraceae). II. Localization and developmental modulation of the enzyme S-adenosyl-L-methionine: (iso)eugenol O-methyltransferase and phenylpropanoid emission. Plant Physiol 114: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pichersky E (1998) Characterization of S-adenosyl-L-methionine: (iso)eugenol O-methyltransferase involved in floral scent production in Clarkia breweri. Arch Biochem Biophys 349: 153–160 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Hayashi K, Yagi K, Asai T, Mactavish H, Picone J, Turnball C, Watanabe N (2002) Biogenesis of 2-phenylethanol in rose flowers: incorporation of [2H8]L-phenylalanine into 2-phenylethanol and its beta-D-glucopyranoside during the flower opening of Rosa ‘Hoh-Jun’ and Rosa damascena Mill. Biosci Biotechnol Biochem 66: 943–947 [DOI] [PubMed] [Google Scholar]
- Yazaki K, Heide L, Tabata M (1991) Formation of p-hydroxybenzoic acid from p-coumaric acid by cell free extract of Lithospermum erythrorhizon cell cultures. Phytochemistry 30: 2233–2236 [Google Scholar]
- Zon J, Amrhein N (1992) Inhibitors of phenylalanine ammonia-lyase: 2-aminoindan-2-phosphonic acid and related compounds. Liebigs Ann Chem 6: 625–628 [Google Scholar]
- Zuker A, Tzfira T, Ben-Meir H, Ovadis M, Shklarman E, Itzhaki H, Forkmann G, Martens S, Neta-Sharir I, Weiss D, et al (2002) Suppression of anthocyanin synthesis by antisense fht enhances flower fragrance. Mol Breed 9: 33–41 [Google Scholar]