Abstract
Studies have revealed in plant chloroplasts, mitochondria, cell walls, and cytoplasm the existence of transglutaminase (TGase) activities, similar to those known in animals and prokaryotes having mainly structural roles, but no protein has been associated to this type of activity in plants. A recent computational analysis has shown in Arabidopsis the presence of a gene, AtPng1p, which encodes a putative N-glycanase. AtPng1p contains the Cys-His-Asp triad present in the TGase catalytic domain. AtPng1p is a single gene expressed ubiquitously in the plant but at low levels in all light-assayed conditions. The recombinant AtPng1p protein could be immuno-detected using animal TGase antibodies. Furthermore, western-blot analysis using antibodies raised against the recombinant AtPng1p protein have lead to its detection in microsomal fraction. The purified protein links polyamines—spermine (Spm) > spermidine (Spd) > putrescine (Put)—and biotin-cadaverine to dimethylcasein in a calcium-dependent manner. Analyses of the γ-glutamyl-derivatives revealed that the formation of covalent linkages between proteins and polyamines occurs via the transamidation of γ-glutamyl residues of the substrate, confirming that the AtPng1p gene product acts as a TGase. The Ca2+- and GTP-dependent cross-linking activity of the AtPng1p protein can be visualized by the polymerization of bovine serum albumine, obtained, like the commercial TGase, at basic pH and in the presence of dithiotreitol. To our knowledge, this is the first reported plant protein, characterized at molecular level, showing TGase activity, as all its parameters analyzed so far agree with those typically exhibited by the animal TGases.
Transglutaminases (TGases; E.C. 2.3.2.13) catalyze protein cross-linking by the interaction of an acyl acceptor glutamyl residue and an amine donor, a lysyl residue, of the same or another protein, or the formation of a link between a protein and a free primary amine, like a polyamine (PA). The terminal amino-groups of PAs conjugate one or two glutamyl residues giving rise either to mono-(γ-glutamyl)-polyamines (mono-PAs) or bis-(γ-glutamyl)-polyamines (bis-PAs; Folk, 1980). The posttranslational cross-linking of proteins is one of the essential physiological processes involved in the stabilization of tissue and cellular matrices. High molecular protein networks can be generated by TGase catalysis when cross-links are formed between Gln and either Lys residues or PAs (bis-PA derivatives) thus forming bridges between different proteins. At present, both in eukaryotes and prokaryotes several TGases have been described exhibiting a large number of functions (Aeschlimann et al., 1998; Griffin et al., 2002; Lorand and Graham, 2003). Eight distinct TGase isoenzymes have been identified in different animal tissues, and some of these have been purified and characterized at molecular level (Aeschlimann et al., 1998; Griffin et al., 2002; Lorand and Graham, 2003).
TGases are classified in three structural families: (1) the papain-like TGases, (2) the protein disulfide isomerase-like TGases, and (3) the bacterial toxin TGases. TGases catalyze a variety of posttranslational protein modifications such as transamidation, Lys acylation, esterification, deamidation, and isopeptide cleavage (Lorand and Graham, 2003). Furthermore, some TGases are multifunctional, as they can act also as GTPases (Lee et al., 1989) and as protein disulfide isomerases (Hasegawa et al., 2003).
One typical feature of TGases is the fact that they exhibit a catalytic triad (Cys, His, and Asp), analogous to that of thiol-proteinases, and a critical Trp located upstream from the active center Cys, which is not involved in disulfide bridges (Weiss et al., 1998; Noguchi et al., 2001).
With few exceptions, the activity of TGases and in particular the correspondence to tissueTGase, is activated by calcium and can be inhibited by GTP. These ligands induce opposite structural protein modifications that result in the tightening or relaxing of the four tissueTGase domains (Griffin et al., 2002).
At present, several TGase activities have been detected both in higher and lower plants, supporting the presence of this type of enzyme. Indeed, this type of activity has been found in various cell compartments such as chloroplasts, mitochondria, cytoplasm, and cell walls (for review, see Serafini-Fracassini et al., 1995; Serafini-Fracassini and Del Duca, 2002). Although their roles are still poorly understood, these plant TGase activities, on the basis of the knowledge acquired from animal systems, are supposed to contribute to structural or conformational modification processes. In addition, it is not excluded that plant TGases may play a role in some organelle-specific metabolisms. In this respect, TGase substrates are found in chloroplasts in which light-dependent plastid TGase(s) may have a role in the photosynthesis or photo-protection reactions. In addition, plant TGase activities also appear to be related to growth (cell cycle, apical growth, and seedling growth), differentiation, programmed cell death, and stress (Serafini-Fracassini and Del Duca, 2002). Indeed, in the apical growth of germinating pollen, cytoskeleton proteins, such as tubulin and actin, were found to be conjugated with PAs (Del Duca et al., 1997).
However, the research on plant TGases has been greatly delayed so far because, although several purified plant protein extracts could establish a close association between their TGase activities with some purified proteins detected by SDS-PAGE, at present the identity of these proteins remains unknown. In addition, in the Arabidopsis, Zea mays, Oryza sativa, and Solanum tuberosum databases no DNA sequence has been found to share homology with animal TGases. This makes it difficult or even excludes the possibility to identify plant TGases by sequence comparison with other well-known animal TGases.
Recently, computational analysis has identified a gene in Arabidopsis, named AtPng1p, coding for a putative peptide N-glycanase (Suzuki et al., 2001). PNGases are de-N-glycosylating enzymes involved in the degradation of misfolded proteins. As amidases, they have a Cys residue that acts as a critical nucleophile for their enzymatic activity (Suzuki et al., 2002). In fact, the AtPng1p gene product has the Cys-His-Asp sequence typically present in the catalytic domain of TGases (Suzuki et al., 2001). To elucidate whether AtPng1p encodes a TGase protein, we have overexpressed its coding sequence in Escherichia coli and purified the recombinant protein by affinity chromatography for performing TGase enzymatic assays. These results demonstrate that AtPng1p conjugates PA primary amines to glutamyl residues of known TGase substrates and polymerizes bovine serum albumine (BSA) in a Ca2+-, pH-, and GTP-dependent manner. In addition, antibodies raised against the recombinant protein have allowed the identification of the AtPng1p gene product as a microsomal-associated protein.
Based on these results, the AtPng1p gene product could therefore be considered as a TGase. Although the existence of some TGase activities have been demonstrated so far in purified plant protein extracts, this is the first characterization to our knowledge of a known protein, AtPng1p, having TGase activity in plants.
RESULTS
AtPng1p Protein Contains the TGase Tripartite Domain
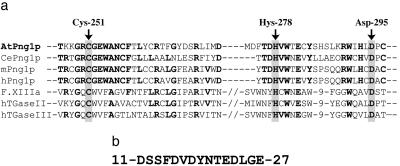
AtPng1p gene product is a 721-amino acid residue protein containing the tripartite domain typical of TGases. Alignment of the AtPng1p TGase domain with other well-characterized TGases is shown in Figure 1A. Thus, in common with other TGases and PNGases, AtPng1p contains two cysteinyl residues within the first domain, a histidyl residue within the second domain, and an aspartyl residue within the third TGase domain.
Figure 1.
ClustalW alignment of the tripartite TGase domain of AtPng1p. A, Alignment among AtPng1p tripartite domain with those of C. elegans (CePng1p; AAF74721), mouse (mPng1p; NP_067479), and human (hPng1p; AF250924.2) peptide N-glycanases, the human Factor XIIIa (NP_000120), human TGase II (NP_945189), and human TGase III (Q08188). Arrows indicate the amino acids Cys-His-Asp of the catalytic triad. Numbers indicate the AtPng1p amino acidic residue position. In bold, amino acid identity. B, N-terminal peptide signal sequence of AtPng1p: putative Tyr sulfation domain, which is associated to Golgi membranes (based on ScanProsite program prediction).
In addition to the tripartite domain, the ScanProsite program predicts in the N-terminal region of AtPng1p (Fig. 1B) a signal peptide which is typically associated with proteins that transit through (or are located in) the Golgi apparatus.
AtPng1p Is a Single, Ubiquitous, and Low-Expressed Gene
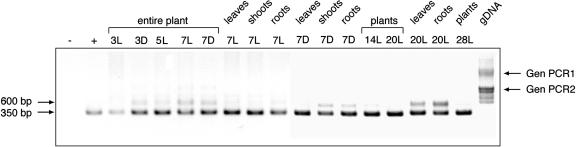
Based on database information, AtPng1p is located in chromosome 5 and contains 17 exons. When searching in Arabidopsis databases, no homologs of AtPng1p were found, indicating that it is a single gene. In addition, only a few expressed sequence tags arising from AtPng1p could be identified in databases so far, suggesting that it is a low-expressed gene. In fact, no mRNA accumulation could be detected by northern-blot analysis (results not shown). Therefore, we performed nested reverse transcription (RT)-PCR for identifying the AtPng1p mRNA accumulation (Yang and Marchand, 2002) in the entire plant, in different organs, growth stages, and light conditions. Plants were grown under light and dark or dark conditions for the days reported in Figure 2. In all conditions, the nested AtPng1p cDNA fragment of the expected size (350 bp) was amplified. In some cases, a fragment of 600 bp was also observed, which corresponds to the expected bands amplified from the first PCR step. Amplification of contaminant genomic DNA is excluded by the fact that using two couples of primers for the amplification of Arabidopsis genomic DNA, the first amplification product results in a 1,600-bp band (first couple of primers), whereas a fragment of 1,000 bp results from the second couple of primers, the difference being due to the size of the introns.
Figure 2.
Nested RT-PCR assay. Nested RT-PCR from Arabidopsis mRNA extracted from entire plants, leaves, shoots, and roots. 3, 5, 7, 14, 20, and 28 refer to the number of growing days. L refers to plants grown under 16-h-light/8-h-dark conditions. D refers to plants grown under dark conditions. An empty pEt-28a(+) vector was used as a negative control (−), whereas, the AtPng1p cDNA clone was used as a positive one (+). Gen PCR1 and Gen PCR2 indicate the 1,600 and 1,000-bp fragments, respectively, obtained from the first and second couple of primers using Arabidopsis genomic DNA (gDNA) as a template.
Recombinant AtPng1p Purification and Immunodetection
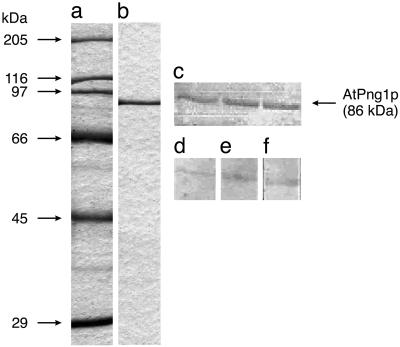
To elucidate whether AtPng1p encodes a TGase protein, we have overexpressed its coding sequence in E. coli and purified the recombinant protein by Ni2+-affinity chromatography. The 86-kD band obtained is shown in Figure 3B.
Figure 3.
Immunodetection of recombinant AtPng1p by western-blot analysis using different animal TGase antibodies. A, Molecular mass standards. B, Coomassie staining of AtPng1p purified with Ni2+-affinity column at 400 mm imidazol. C, Red Poinceau positive bands on nitrocellulose deriving from 10% (w/v) SDS-PAGE (1 μg protein). Immuno-staining with TGase II-Ab 3 and antibodies (D) for C. elegans TGase (E), rat prostatic gland TGase (F). Primary antibodies dilution: 1:1,000; secondary antibodies dilution 1:3,000.
As no plant protein was known as TGase until now, no specific plant antibodies were available, and therefore animal TGase antibodies have been used for the detection of TGases in plant extracts exhibiting TGases activities (Del Duca et al., 1994; Waffenschmidt et al., 1999; Dondini et al., 2001; Serafini Fracassini et al., 2002). Therefore, we have also tested whether the recombinant AtPng1p is detected by these animal antibodies (Fig. 3, D–F). As expected, our results indicate that AtPng1p protein is immuno-recognized also by the following animal TGase antibodies, TGase II-Ab 3 (Neomarker, Fremont, CA), anti-Caenorhabditis elegans TGase, anti-rat prostatic gland TGase, which is in agreement with previous results obtained with purified plant protein extracts having TGase activity.
Subcellular Immuno-Localization of AtPng1p
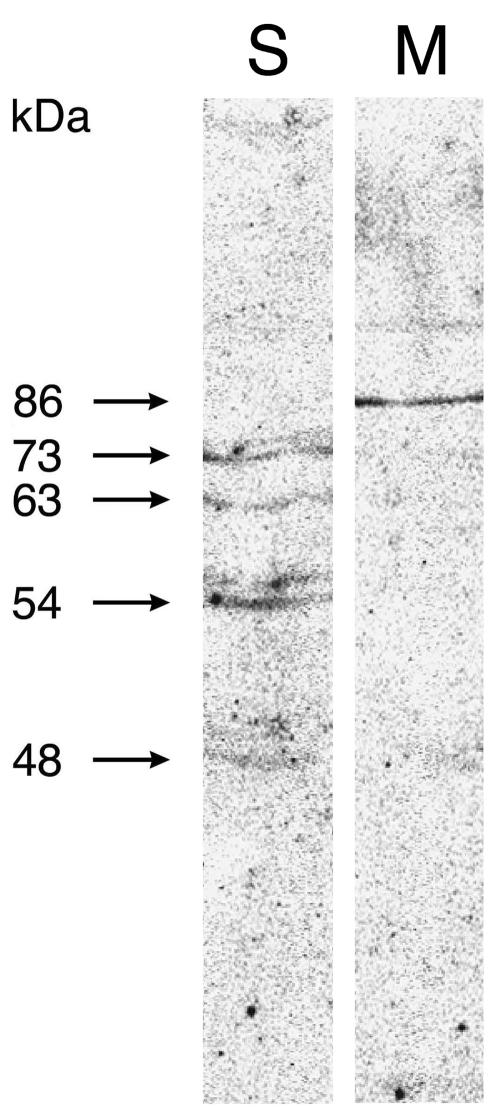
To elucidate more precisely how AtPng1p gene product is expressed in Arabidopsis and which is its subcellular localization, we have raised specific antibodies against the purified recombinant AtPng1p and used them to immuno-localize the protein (Fig. 4M). Western-blot analyses have detected AtPng1p protein (a single band of 86 kD) in the microsomal-enriched fraction only. This result is therefore in agreement with the ScanProsite program, which predicts AtPng1p gene product as a membrane-associated protein.
Figure 4.
Immunodetection of AtPng1p by western-blot analysis using AtPng1p antibodies. S refers to 20-d-old Arabidopsis soluble enriched fraction, whereas M refers to microsomal enriched fraction. We ran 100 μg of proteins in 10% (w/v) SDS-PAGE and blotted to a nitrocellulose membrane. The membranes were then treated as described in “Materials and Methods” and incubated with antibodies raised against AtPng1p (used in 1:1,000 dilution). The mass of proteins is indicated in kD.
Some others bands (showed as doublets) of lower molecular mass (73, 63, 54, and 48 kD, respectively) were identified in the cytosolic fraction (Fig. 4S), opening the question of whether these bands represent soluble fragments of AtPng1p protein produced by proteolytic degradation.
Enzymatic Activity Assays
Recombinant AtPng1p Protein Covalently Links PAs to Dimethylcasein
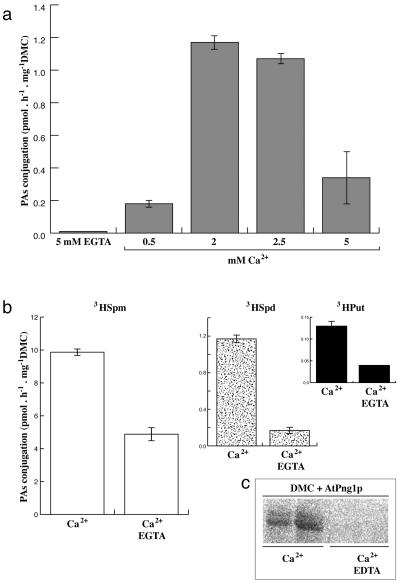
The capability of AtPng1p to incorporate primary amines of radiolabeled PAs or biotinilated-cadaverine in a well-known TGase substrate, such as dimethylcasein (DMC), was investigated. This substrate contains a large percentage of lysyl residues blocked by methylations and, therefore, the available reaction sites consist mainly of glutamyl residues. Figure 5A shows that recombinant AtPng1p is able to conjugate spermidine (Spd) to DMC, in a Ca2+-dependent manner, with a peak of incorporation at 2 to 2.5 mm Ca2+, and a dramatic fall at 5 mm. Furthermore, the enzyme preferentially catalyzes the conjugation of spermine (Spm): 10-fold more than Spd and 100-fold more than putrescine (Put; Fig. 5B). In addition, the Ca2+-dependent incorporation of PAs is inhibited by 5 mm EGTA (Fig. 5, A and B). The autoradiography obtained utilizing [3H]Spm shows that the labeling of DMC only occurs when Ca2+ is present in the reaction mixture; in fact the presence of EDTA inhibits the conjugation event (Fig. 5C).
Figure 5.
TGase conjugation assays. The assays were performed using PAs as substrates. A, [3H]Spd conjugation to DMC catalyzed by AtPng1p incubated at different Ca2+ molarity. B, [3H]Spm, -Spd, and -Put conjugation to DMC catalyzed by AtPng1p incubated with 2 mm Ca2+ and 10 mm DTT in the presence or absence of 5 mm EGTA. C, Autoradiography of the 15% (w/v) SDS-PAGE of the DMC conjugated to [3H]Spm by catalysis of the AtPng1p as in B, with or without 5 mm EDTA, performed by Fuji imaging [3H]plate. Means significantly differs (1% probability level, Student's t test).
Recombinant AtPng1p Protein Conjugates Spermine to Glutamyl Residues of DMC
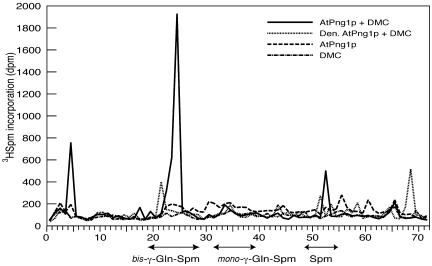
To confirm that AtPng1p protein conjugates the primary amine of [3H]Spm to Gln residues of DMC, γ-glutamyl-derivatives were investigated (Fig. 6). HPLC analysis shows the formation mainly of bis-γ-gln-Spm derivatives in the presence of 2 mm calcium. In addition, neither DMC alone nor AtPng1 protein alone or denatured with 10% (w/v) trichloroacetic acid (TCA) in the presence of substrate are able to generate γ-glutamyl-derivatives. Furthermore, the biotin-cadaverine assay using DMC as substrate also confirms the amine incorporation due to the action of AtPng1p protein only in the presence of calcium (results not shown).
Figure 6.
Glutamyl-derivatives assay. The conjugation of DMC to [3H]Spm produced by the action of AtPng1p were analyzed by HPLC. DMC alone, AtPng1p alone, and denatured AtPng1p incubated with DMC are used as negative control. Arrows indicate standard retention times for bis-γ-glutamyl-Spm and mono-γ-glutamyl-Spm obtained with DMC and guinea pig liver TGase.
Recombinant AtPng1p Protein Polymerizes BSA in a pH-Dependent Manner
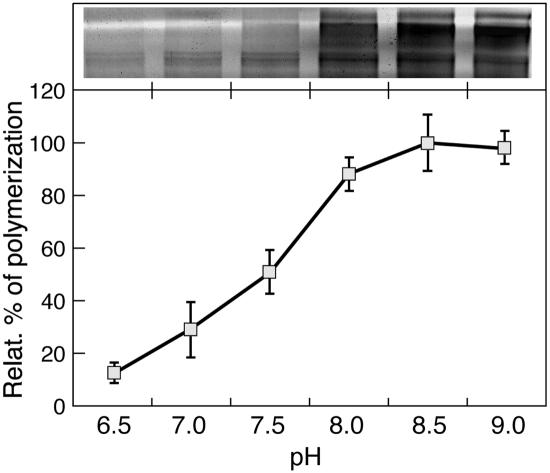
The effect of AtPng1p protein on BSA polymerization was assayed in the absence of PAs to avoid the competition with their primary amino-groups. To avoid an unspecific BSA aggregation, due to the thermal treatment, samples were incubated with 100 mm dithiotreitol (DTT) in the sample buffer prior to and after boiling. Then, samples were run in 10% (w/v) SDS-PAGE, and the effect of polymerization was visualized by Coomassie Blue staining (Fig. 7). These results show that polymerization increases starting at pH 7.5 with increasing values of pH, the maximum being reached at pH 8.5, demonstrating that AtPng1p protein polymerizes the substrate in a pH-dependent way.
Figure 7.
pH-dependent polymerization assay. Relative percentage of cross-linked BSA after incubation with AtPng1p at different pH values. After enzymatic assay, proteins were run in 10% (w/v) SDS-PAGE, and the polymers formation were then measured as O.D. (FLA 3000, Fuji, Milan) of the high molecular mass region (>250 kD) of the SDS-PAGE gel, as shown in the insert. The 100% refers to O.D. of the polymerized zone at pH 8.5. Assay condition: 20 mm Tris-HCl, 30°C, BSA/AtPng1p ratio = 12.5/1.
Recombinant AtPng1p Protein Polymerizes BSA in a Ca2+- and DTT-Dependent Manner
We have performed TGase assays using BSA and DMC as substrates to reach a better understanding of the polymerization phenomenon. After these assays, the samples were run in 10% (w/v) SDS-PAGE and the proteins were visualized by Coomassie Blue staining (Fig. 8).
Figure 8.
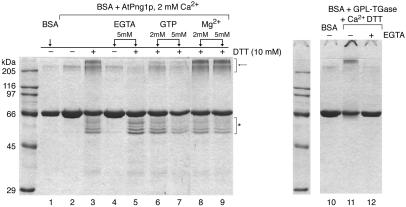
BSA polymerization assay. Left, Coomassie staining of 10% (w/v) SDS-PAGE of BSA after incubation with AtPng1p. Ca2+, DTT, EGTA, GTP, and Mg2+ were assayed on the cross-linking formation as indicated in the lanes. Assay condition: 40 mm MES-NaOH, pH 7.0, 30°C, BSA/AtPng1p ratio = 12.5/1. Right, Polymerization of BSA catalyzed by GPL-TGase. Assay condition: 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 30°C, BSA/GPL-TGase ratio = 12.5/1. Arrow, BSA polymers; asterisk, bands below BSA.
Although BSA rarely acts as a TGase substrate, our results show the formation of BSA polymers (>200 kD, indicated by arrow in the figure) through the action of the AtPng1p protein when calcium and DTT are simultaneously present in the reaction mixture (Fig. 8, left, lane 3, and compare with lane 2). In addition, this polymerization event is strongly inhibited when EGTA chelates the calcium in the absence or presence of DTT in the assayed reaction (lane 4 and 5, and compare with lane 3). Similar to the inhibition produced by EGTA, the addition of GTP strongly inhibits polymer formation (lanes 6 and 7, and compare with lane 3). However, the addition of Mg2+ to the reaction mixture does not interfere with the degree of polymerization (lanes 8 and 9, and compare with lane 3). Like magnesium, neither sodium nor potassium affects the polymerization of BSA (results not shown).
BSA can also be polymerized using commercial guinea pig liver transglutaminase (GPL-TGase) instead of AtPng1p protein. In this case, the polymerization is also Ca2+-dependent (Fig. 8, right, lane 11), in fact it is strongly inhibited when EGTA is added to the reaction mixture (lane 12).
In addition to the polymerization product, 5 to 6 bands of lower molecular mass, with respect to BSA, appear in the presence of the AtPng1p protein, calcium, and DTT (lane 3, and compare with lane 2, indicated with asterisk in the figure). Depletion of Ca by EGTA does not affect the formation of these bands (Fig. 8, right, lane 5, and compare with lane 3), suggesting that this event is DTT-dependent but not Ca2+-dependent. GTP shows a partial inhibition of this phenomenon only at a concentration of 5 mm (lane 6, and compare with lane 3).
At variance with polymerization, the generation of these slower bands appears to be specifically related to AtPng1p protein as no such bands are observed when the commercial GPL-TGase is used in the assays (Fig. 8, right, lane 11).
In addition to BSA, we have also tested whether DMC is a polymerization substrate for the AtPng1p protein and the commercial GPL-TGase. When the assays were performed in the presence of Ca2+ and DTT, DMC polymerized, although to a lower extent than BSA (results not shown). This phenomenon could indeed be interpreted by the fact that a high percentage of DMC Lys is blocked by methylation.
DISCUSSION
This paper reports the first known protein, AtPng1p, showing TGase activity in plants. Furthermore, as the protein catalyzes the formation of covalent linkages between primary amino-groups of PAs and glutamyl-residues in a Ca2+-dependent way (Figs. 5 and 6), we suggest that the tripartite Cys-His-Asp TGase domain of the AtPng1p protein (Fig. 1) may be implicated in this enzymatic process. However, whether one or more of these residues are critical for enzymatic activity is still unknown and directed mutagenesis should be performed to answer this question. In addition, AtPng1p protein recognizes as substrates and polymerizes BSA and DMC (Figs. 5, 6, and 8) in a Ca2+-, DTT-, GTP-, and pH-dependent way (Figs. 7 and 8), like GPL-TGase and other well-known animal TGases reported so far (Aeschlimann et al., 1998; Griffin et al., 2002; Lorand and Graham, 2003). The biochemical features exhibited by the AtPng1p gene product agree with previous results obtained with partially purified plant protein extracts showing TGase activity. In these different plants, the calcium-dependent TGase activity appears to be regulated by particular physiological conditions such as senescence (Serafini-Fracassini et al., 2002), light (Dondini et al., 2003), or salt stress (Dondini et al., 2001). However, the precise binding sites for calcium and GTP in the AtPng1p protein remain to be determined. In addition, this protein is immuno-recognized by three animal TGase antibodies (Fig. 3), a feature that is in agreement with those previously reported for plant extracts exhibiting TGase activity (Del Duca et al., 1994, 2000; Waffenschmidt et al., 1999; Dondini et al., 2001).
The fact that very few expressed sequence tags are found in databases arising from AtPng1p agrees with our difficulty detecting AtPng1p mRNA accumulation. This situation was solved by RT-PCR assays (Fig. 2), which demonstrated that this gene is ubiquitously expressed. The presence of the AtPng1p mRNA, even though low expressed, in all tissues, growth stages, and light conditions suggests that AtPng1p gene product could be a constitutive enzyme. Lilley et al. (1998) as well as Falcone et al. (1993) and Kang et al. (1998) reported the occurrence of TGase activities in several organs of the same plant.
Western-blot analysis also revealed that the amount of AtPng1p protein is low in the cell, in agreement with the low AtPng1p mRNA levels detected by RT-PCR. In fact, these low amounts of AtPng1p protein in the cell could explain why several laboratories have failed in isolating plant TGases over the years. To our knowledge, only Kang and Cho (1996) obtained the purification from soybean leaves of an 80-kD protein to homogeneity, so far unsequenced, showing TGase activity. Similarly, a 39-kD protein has been recently partially purified from a fraction of chloroplast thylakoids showing TGase activity (Della Mea et al., 2004). In addition, in the Arabidopsis genome, only AtPng1p appears to have the TGase Cys-His-Asp catalytic domain. This situation raises the question of whether Arabidopsis has a unique TGase enzyme. However, several TGase activities have been detected in plants so far, as above reported.
Interestingly, an 86-kD band corresponding to the AtPng1p gene product has been detected mainly in the microsomal protein fraction. This result is in agreement with the ScanProsite program, which predicts an N-terminal region typically associated to proteins that transits through (or are located in) the Golgi apparatus. However, a few other smaller bands were detected in the soluble protein fraction (Fig. 4). Therefore, although AtPng1p appears to be a single gene, these results could indicate that its product may undergo posttranslational modifications that could modulate its subcellular location. This possibility is in keeping with results obtained from TGases of animal origin, in which both soluble and plasma membrane-bound TGases have been identified. For example, tissue TGase is found at several intracellular sites including mitochondria, the nuclear membrane, and the nucleoplasm (Griffin et al., 2002; Lorand and Graham, 2003). Also in plants, some evidences show that more than one TGase form is present in the same tissue and probably in the same cell, differently compartmented (chloroplasts, mitochondria, cell wall, etc.; Falcone et al., 1993; Del Duca et al., 1994; Bernet et al., 1999; Votyakova et al., 1999; Waffenschmidt et al., 1999). In chloroplasts, there is a TGase activity associated with the stroma compartment and another activity associated to the thylakoid (Dondini et al., 2003).
In conclusion, although the Arabidopsis AtPng1p protein only shares a TGase domain (Suzuki et al., 2001) that could be compared to that of other well-known TGases, the biochemical and immunological features of the recombinant purified protein confirm its identification as a plant TGase. At present, the production of transgenic lines down-regulated for this gene is in progress to further study and to precisely determine the role(s) of AtPng1p in the plant.
Although the existence of some TGase activities have been demonstrated so far in plant protein extracts, this is the first report in which a known protein, AtPng1p, is characterized as a TGase enzyme in plants.
MATERIALS AND METHODS
Plant Material and Vectors
Arabidopsis L. cv Columbia was used. All plants were grown in greenhouse conditions (23°C) with 16 h light/8 h dark or only on dark conditions (light intensity, 55 μmol photons m−2 s−1).
The coding sequence of AtPng1p (2,163 bp; N96715) was obtained from the Arabidopsis G3B4T7 cDNA clone from TAIR database. At the beginning of this work, this clone was the longest one, but by sequence comparison with genomic DNA, it lacked the start Met and two other amino acid residues of the coding sequence. Therefore, during the course of the cloning step, we added these three amino acid residues prior to cloning this cDNA into the pET-28a (+) expression vector (Novagen, La Jolla, CA). The coding sequence was cloned in frame by adding the NdeI and SacI restriction sites.
This coding sequence was confirmed by another longer cDNA clone (GenBank accession no. AI993733) containing these three amino acid residues and the 5′ untranslated region that appeared later in databases.
All reagents were obtained from Sigma-Aldrich (St. Louis) unless otherwise specified.
Production of Recombinant AtPng1p Protein and Affinity Purification
The sequence of the upstream and downstream primers is:
5′-GCATATGGTTGCTCGGAAATTTGTCGTCCGCCATGAAG-3′ (upstream);
5′-CACGGAGCTCTCACTGGTGACTTCTG-3′ (downstream).
Underlined sequences refer to NdeI and SacI restriction sites, respectively, and correspond to the restriction sites used for cloning in-frame into pET-28a (+) expression vector. Heat shock transformed Escherichia coli (BL21 strain) were grown in 250 mL Luria-Bertani medium at 37°C. When the culture reached optical density (O.D., 600 nm) of 0.7, the production of recombinant protein was induced with 1 mm isopropil-β-d-thiogalactopyranoside and the culture incubated at 37°C for 3 h. Then, bacteria were centrifuged (3,000g) at 4°C and the pellet was manipulated according to manufacturer's conditions (Novagen). Finally, as the recombinant AtPng1p protein was present in the bacteria inclusion bodies, it was purified by Ni2+ affinity chromatography in the presence of 8 m urea (Novagen).
Nested RT-PCR
mRNAs deriving from different organs in different growth condition (as reported in Fig. 2) of Arabidopsis were isolated using standard procedures. The RNA was suspended in 50 μL of diethyl pyrocarbonate-treated distilled H2O, analyzed by gel electrophoresis, and quantified spectrophotometrically. Ten micrograms of RNA were used to synthesize cDNAs using oligo(dT) primers according to manufacturer's conditions (Invitrogen, Giuliano Milanese, Italy).
First round PCR was performed with 5 μL of cDNA in 30 cycles using specific primers (upstream primer sequence, 5′-GCAGTTTCTTCCCGCACTAGGC-3′; downstream primer sequence, 5′-CTTCTGTACAGATCGATGCTCCC-3′). Four microliters of first round PCR product were added to 46 μL of a second round master mix containing nested primers (upstream primer sequence, 5′-GCATTGCCAGTTGCATTGGACGC-3′; downstream primer sequence, 5′-CCACCATCGTTACTTCCTTCAAG-3′), followed by a further 30 cycles of amplification.
Preparation of Enriched Microsomal and Soluble Protein Fractions
Protein extracts were prepared from 20-d-old Arabidopsis plants, with all steps performed at 4°C. Four grams of whole 20-d-old plants were added to 30 mL ice-cold extraction buffer (0.25 m sorbitol, 50 mm Tris-HCl, pH 7.4, 3 mm EGTA, 1 mm EDTA supplemented immediately prior to use with 1 mm DTT, 10 mg/mL pepstatin, 0.5 mg/mL leupeptin, and 1 mm phenylmethylsulfonyl fluoride) and homogenized. The separation of microsomal and soluble proteins was then performed as already described (Terry and Williams, 2002).
In all cases, isolated proteins were frozen with liquid nitrogen and stored at −80°C prior to use.
Production of AtPng1p Antibodies
A quantity of 50 μg × 3 of affinity-purified AtPng1p protein were injected in chicken. Antibodies were recovered from 80 mL egg yolks 30 d after each injection. Each sample was suspended in 2 volumes of phosphate-buffered saline (PBS) 100 mm, pH 7.5, 5.25% (w/v) of polyethyleneglycol-8000, gently mixed for a period of 5 min, then centrifuged at 12,000g for 10 min at 4°C. The supernatant was recovered, filtered through two layers of Miracloth, incubated for 10 min at room temperature with 25 mL of PBS 100 mm, pH 7.5, 37.5% (w/v) of polyethyleneglycol-8000, then centrifuged at 12,000g for 15 min at 4°C. The pellet was recovered and resuspended in 25 mL 100 mm PBS, pH 7.5. Finally, antibodies were aliquoted and stored at −80°C.
Western-Blot Analyses
A total of 100 μg of Arabidopsis enriched microsomal and soluble proteins were boiled in SDS loading buffer, loaded onto a denaturing 10% (w/v) SDS-PAGE (Laemmli, 1970), and migrated in a 1× tris-borate-EDTA buffer at 25 mA for 2 h using a Bio-Rad Mini-protean II apparatus (Segrate, Italy). After protein migration, the gel was blotted to a nitrocellulose membrane (Amersham Biosciences, Buckinghamshire, UK) using a semidry Trans-Blot system (Bio-Rad). Incubation of membranes with AtPng1p antibodies was performed as previously described (Sambrook et al., 1989). Antibodies dilution was 1:1,000. Proteins were finally detected by chemiluminescence using goat antibodies raised against chicken immunoglobulins coupled with horse radish peroxidase (Amersham Biosciences).
Studies on cross-reactivity between AtPng1p and animal TGase antibodies was performed as above using: anti-TGase II-Ab 3 (Neomarker), anti-Caenorhabditis elegans TGase (kind gift of Prof. K. Mehta, Houston, TX), and anti-rat prostatic gland TGase (kind gift of Prof. C. Esposito, Salerno, Italy). Anti-mouse or anti-rabbit horse radish peroxidase conjugates (Sigma-Aldrich) were used as secondary antibodies. Proteins were detected using amino-ethilcarbazole tablets (Sigma-Aldrich).
TGase Assays
Prior to enzymatic assays, the recombinant purified AtPng1p protein was renatured according to Novagen protocols. Refolding was obtained by performing a multistep dialysis in 100 mm Tris-HCl, pH 8.5 buffer with a progressive depletion of urea under the presence of 5 mm Ca2+, 10 mm DTT, and 10% (w/v) Suc. Finally, with the same technique, samples were deprived of Ca2+ and DTT.
Radiometric Assay
Two micrograms of recombinant purified AtPng1 protein was assayed using 50 μg of DMC or BSA as substrates, in the presence of 2 μL of (740 kBq) [1,4(n)-3H]Put (specific activity 1.5 TBq mmol−1; NEN, Milan), 2 μL of [1,4(n)-3H]Spd (specific activity 1.5 TBq mmol−1; NEN), or 2 μL of [1,4(n)-3H]Spm (specific activity 1.5 TBq mmol−1; ICN, Segrate, Italy), 40 mm MES buffer, pH 7.0, 10 mm DTT, and 0 to 5 mm of CaCl2 in a final volume of 50 μL. When required, 5 mm EGTA or EDTA were added for the chelation of Ca2+. After 1 h at 30°C, assays were blocked with TCA containing 2 mm unlabeled Put (7% w/v final concentration) or with 2× SDS-PAGE loading buffer in the case of autoradiography. In the first case, the incorporation of the labeled amines was counted in a Beckman LS 6500 (Fullerton, CA) scintillation counter, as previously reported (Del Duca et al., 1994). In the second assay, proteins were separated by 10% (w/v) SDS-PAGE and stained with Coomassie-Blue G reagent (Hames and Rickwood, 1990). The gel was then dried, exposed at −80°C to an imaging plate BAS-TR2025 (Fuji, Milan), and revealed at different exposure times with FLA 3000 Laser System (Fuji).
Biotin-Labeled Cadaverine Incorporation Assay
The assay was carried out according to Lilley et al. (1997). Three micrograms of purified recombinant AtPng1p protein was incubated at pH 7.5 with immobilized DMC as a substrate, with or without 5 mm EDTA.
Identification of γ-Glutamyl-Polyamine Derivatives
After radiometric assays, TCA 7% (w/v) final concentration was added to block TGase activity. The pellet was washed at least three times with anhydrous diethyl-ether and proteolyzed according to the method previously described (Folk et al., 1980). γ-Glutamyl-PAs present in the TCA-insoluble fractions were separated by ion-exchange chromatography using a Jasco HPLC-system (4.5 mm × 90 mm column, packed with Ultropac 8 resin, Na+ form; Jasco Europe Srl, Milan) and the five-buffer system previously reported (Folk et al., 1980). The conjugated PAs were released from the ion-exchange fractions by acid hydrolysis and their identity determined by comparison with the corresponding retention times of glutamyl-PA standards.
Polymerization Assay
DMC and BSA were tested by the radiometric assay conditions with the exception of (1) the absence of PAs, (2) the use of 20 mm Tris-HCl for the pH curve, and (3) the incubation time (16 h). Then, an aliquot corresponding to 5 μg of substrate was separated by 10% or 15% (w/v) SDS-PAGE and further analyzed by densitometric assay.
Gel Densitometries
The densitometric analysis of stained and labeled hydrated gels was performed with the specific software Total Lab (Raytest, Milan) on the gel scanned by a FLA 3000 Laser System (Fuji) provided with an imaging plate ultrasensitive for tritium (BAS-TR 2025, Fuji).
Statistics
Quantitative determinations were repeated at least three times. All values are means with indicated ses. The Student's t test was used to compare means, as reported in the legends.
Statement
Upon request, all novel material described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AI993733.
Supplementary Material
Acknowledgments
We thank the Arabidopsis TAIR database for providing us with the AtPng1p cDNA clone used in this work. Authors are very indebted to Dr. J. Casas (CSIC) for producing antibodies against AtPng1p, Dr. M. Capellades (IBMB-CSIC) for the technical support in greenhouses for growing Arabidopsis, the DNA sequencing team of IBMB-CSIC of Barcelona, Mr. N. Mele (University of Bologna) for the images elaboration, and Professor A. Serafini-Fracassini, Emeritus of St. Andrews University, for scientific and language suggestions.
This work was supported by Ministero dell'Università e della Ricerca Scientifica e Tecnologica (FIRB 2001), by the ESF Transglutaminases Program and the COST 844 Transglutaminases in Apoptosis (grant to M.D.M.), and by the Spanish Government (Secretaría de Estado de Educación, Universidades e Investigación, support to D.S.-F.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042549.
References
- Aeschlimann D, Koeller MK, Allen-Hoffmann BL, Mosher DF (1998) Isolation of a cDNA encoding a novel member of the transglutaminase gene family from human keratinocytes. Detection and identification of transglutaminase gene products based on reverse transcription-polymerase chain reaction with degenerate primers. J Biol Chem 273: 3452–3460 [DOI] [PubMed] [Google Scholar]
- Bernet E, Claparols I, Dondini L, Santos MA, Serafini-Fracassini D, Torné JM (1999) Changes in polyamine content, arginine and ornithine decarboxilases and transglutaminase activities during light/dark phases of initial differentiation in maize calluses and their chloroplasts. Plant Physiol Biochem 37: 1–11 [Google Scholar]
- Del Duca S, Bregoli AM, Bergamini C, Serafini-Fracassini D (1997) Transglutaminase-catalyzed modification of cytoskeletal proteins by polyamines during the germination of Malus domestica pollen. Sex Plant Reprod 10: 89–95 [Google Scholar]
- Del Duca S, Dondini L, Della Mea M, Muñoz de Rueda P, Serafini-Fracassini D (2000) Factors affecting transglutaminase activity catalysing polyamine conjugation to endogenous substrates in the entire chloroplast. Plant Physiol Biochem 38: 429–439 [Google Scholar]
- Del Duca S, Tidu V, Bassi R, Serafini-Fracassini D, Esposito C (1994) Identification of transglutaminase activity and its substrates in isolated chloroplast of Helianthus tuberosus. Planta 193: 283–289 [Google Scholar]
- Della Mea M, Di Sandro A, Dondini L, Del Duca S, Vantini F, Bergamini C, Bassi R, Serafini Fracassini D (2004) A Zea mays 39-kDa thylakoidal transglutaminase catalyses the modification by polyamines of light-harvesting complex II in a light-dependent way. Planta DOI 10.1007/s00425-004-1280z [DOI] [PubMed]
- Dondini L, Bonazzi S, Bregoli AM, Del Duca S, Serafini-Fracassini D (2001) Adaptation of chloroplast transglutaminase to salt stress in a polyamine-deficient variant strain of Dunaliella salina. J Plant Physiol 158: 185–197 [Google Scholar]
- Dondini L, Del Duca S, Dall'Agata L, Bassi R, Gastaldelli M, Della Mea M, Di Sandro A, Claparols I, Serafini-Fracassini D (2003) Suborganellar localisation and effect of light on Helianthus tuberosus chloroplast transglutaminases and their substrates. Planta 217: 84–95 [DOI] [PubMed] [Google Scholar]
- Falcone P, Serafini-Fracassini D, Del Duca S (1993) Comparative studies of transglutaminase-like activity and substrates in different organs of Helianthus tuberosus. J Plant Physiol 142: 265–273 [Google Scholar]
- Folk JE (1980) Transglutaminases. Annu Rev Biochem 49: 517–531 [DOI] [PubMed] [Google Scholar]
- Folk JE, Park MH, Chung SI, Schrode J, Lester EP, Cooper HL (1980) Polyamines as physiological substrates for transglutaminase. J Biol Chem 255: 3695–3700 [PubMed] [Google Scholar]
- Griffin M, Casadio R, Bergamini CM (2002) Transglutaminases: nature's biological glues. Biochem J 368: 377–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames BD, Rickwood D (1990) One-dimensional polyacrylamide gel electrophoresis. In BD Hames, D Rickwood, eds, Gel Electrophoresis of Proteins. A Practical Approach. Oxford University Press, Oxford, pp 1–383
- Hasegawa G, Suwa M, Ichikawa Y, Ohtsuka T, Kumagai S, Kikuchi M, Sato Y, Saito Y (2003) A novel function of tissue-type transglutaminase: protein disulfide isomerase. Biochem J 373: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Cho YD (1996) Purification and properties of transaminase from soybean (Glycine max) leaves. Biochem Biophys Res Commun 223: 228–292 [DOI] [PubMed] [Google Scholar]
- Kang H, Lee GS, Cho YD (1998) Identification of glycinin in vivo as a polyamine-conjugated protein via a γ-glutamyl linkage. Biochem J 332: 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227: 680–683 [DOI] [PubMed] [Google Scholar]
- Lee KN, Birckbichler PJ, Patterson MK Jr (1989) GTP hydrolysis by guinea pig liver transglutaminase. Biochem Biophys Res Commun 162: 1370–1375 [DOI] [PubMed] [Google Scholar]
- Lilley G, Griffin M, Bonner PLR (1997) Assays for the measurement of tissue transglutaminase (type II) mediated protein crosslinking via epsilon-(gamma-glutamyl) lysine and N′,N′-bis (gamma-glutamyl) polyamine linkages using biotin labelled casein. J Biochem Biophys Methods 34: 31–43 [DOI] [PubMed] [Google Scholar]
- Lilley G, Skill J, Griffin M, Bonner PLR (1998) Detection of Ca2+ dependent transglutaminase activity in root and leaf tissue of monocotyledoneous and dicotyledoneous plants. Plant Physiol 117: 1115–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorand L, Graham RM (2003) Transglutaminases: cross-linking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol 4: 140–156 [DOI] [PubMed] [Google Scholar]
- Noguchi K, Ishikawa K, Yokoyama KI, Ohtsuka T, Nio N, Suzuki E (2001) Crystal structure of red sea bream transglutaminase. J Biol Chem 276: 12055–12059 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Gel electrophoresis of DNA. In C Nolan, ed, Molecular Cloning: A Laboratory Manual, Ed 2, Vol 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Serafini-Fracassini D, Del Duca S (2002) Biochemistry and function of plant transglutaminases. Minerva Biotecnol 14: 135–141 [Google Scholar]
- Serafini-Fracassini D, Del Duca S, Beninati S (1995) Plant transglutaminases. Phytochemistry 40: 355–365 [DOI] [PubMed] [Google Scholar]
- Serafini-Fracassini D, Del Duca S, Monti F, Poli F, Sacchetti G, Bregoli AM, Biondi S, Della Mea M (2002) Transglutaminase activity during senescence and programmed cell death in the corolla of tobacco (Nicotiana tabacum) flowers. Cell Death Differ 9: 309–321 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Park H, Anderson Till E, Lennarz WJ (2001) The PUB domain: a putative protein-protein interaction domain implicated in the ubiquitin-proteasome pathway. Biochem Biophys Res Commun 287: 1083–1087 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Park H, Lennarz WJ (2002) Cytoplasmic peptide:N-glycanase (PNGase) in eukaryotic cells: occurrence, primary structure, and potential functions. FASEB J 16: 635–641 [DOI] [PubMed] [Google Scholar]
- Terry MJ, Williams LE (2002) Fractionation of plant tissue for biochemical analyses. In PM Gilmartin and C Bowler, eds, Molecular Plant Biology. A Practical Approach, Ed 2. Oxford University Press, Oxford, pp 147–151
- Votyakova VT, Wallace HM, Dunbar B, Wilson SB (1999) The covalent attachment of polyamines to proteins in plant mitochondria. Eur J Biochem 260: 250–257 [DOI] [PubMed] [Google Scholar]
- Waffenschmidt S, Kusch T, Woessner JP (1999) A transglutaminase immunologically related to tissue transglutaminase catalyzes cross-linking of cell wall proteins in Chlamydomonas reinhardtii. Plant Physiol 121: 1003–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MS, Metzner HJ, Hilgenfeld R (1998) Two non-proline cis peptide bonds may be important for factor XIII function. FEBS Lett 423: 291–296 [DOI] [PubMed] [Google Scholar]
- Yang X, Marchand JE (2002) A sensitive technique to clone low abundance receptor transcripts from single microdissected tissue punches. Brain Res Brain Res Protoc 9: 135–146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.