Abstract
Type IV pilus (T4P) systems are complex molecular machines that polymerize major pilin proteins into thin filaments displayed on bacterial surfaces. Pilus functions require rapid extension and depolymerization of the pilus, powered by the assembly and retraction ATPases, respectively. A set of low abundance minor pilins influences pilus dynamics by unknown mechanisms. The Vibrio cholerae toxin-coregulated pilus (TCP) is among the simplest of the T4P systems, having a single minor pilin TcpB and lacking a retraction ATPase. Here we show that TcpB, like its homolog CofB, initiates pilus assembly. TcpB co-localizes with the pili but at extremely low levels, equivalent to one subunit per pilus. We used a micropillars assay to demonstrate that TCP are retractile despite the absence of a retraction ATPase, and that retraction relies on TcpB, as a V. cholerae tcpB Glu5Val mutant is fully piliated but does not induce micropillars movements. This mutant is impaired in TCP-mediated autoagglutination and TcpF secretion, consistent with retraction being required for these functions. We propose that TcpB initiates pilus retraction by incorporating into the growing pilus in a Glu5-dependent manner, which stalls assembly and triggers processive disassembly. These results provide a framework for understanding filament dynamics in more complex T4P systems and the closely related Type II secretion system.
Author Summary
Bacterial pathogens utilize a number of highly complex and sophisticated molecular systems to colonize their hosts and alter them, creating customized niches in which to reproduce. One such system is the Type IV pilus system, made up of dozens of proteins that form a macromolecular machine to polymerize small pilin proteins into long thin filaments that are displayed on the bacterial surface. These pili have a remarkable array of functions that rely on their ability to (i) adhere to many substrates, including host cell surfaces, pili from nearby bacteria, DNA and bacterial viruses (bacteriophage), and (ii) to depolymerize or retract, which pulls the bacteria along mucosal surfaces, pulls them close together in protective aggregates, and can even draw in substrates like DNA and bacteriophage for nutrition and genetic variation. For most Type IV pilus systems, retraction is an energy-driven process facilitated by a retraction ATPase. We show here that in the simplest of the Type IV pilus systems, the Vibrio cholerae toxin-coregulated pilus, a pilin-like protein initiates pilus retraction by what appears to be mechanical rather than enzymatic means. Our results provide a framework for understanding more complex Type IV pili and the related Type II secretion systems, which represent targets for novel highly specific antibiotics.
Introduction
Vibrio cholerae is a Gram-negative bacterial pathogen that causes the human diarrheal disease cholera, which afflicts millions of people each year [1]. Cholera is marked by copious watery diarrhea that can lead to dehydration, shock, organ failure and death as little as 24 hours after infection [2]. The severe diarrhea is caused by cholera toxin, an ADP ribosylating enzyme that is transported from the cytoplasm across the inner membrane via the Sec machinery and from the periplasm across the outer membrane via the Type II secretion (T2S) system [1, 3–5]. Colonization of the small intestine by V. cholerae requires a second virulence factor, the toxin co-regulated pilus (TCP), which self-associates to induce bacterial aggregation and microcolony formation [6–8]. In addition to building a pilus filament, the TCP assembly apparatus acts like a T2S system, exporting a soluble protein, TcpF, across the outer membrane [9, 10]. TCP is also the primary receptor for the lysogenic bacteriophage CTXϕ, which carries the cholera toxin genes ctxAB and is responsible for converting V. cholerae from a harmless marine microbe to a deadly human pathogen causing pandemic disease [11]. Understanding TCP biology is essential for understanding V. cholerae pathogenesis and designing prevention and treatment strategies for cholera disease.
TCP are members of the Type IV pilus (T4P) class, which are expressed by many Gram-negative bacteria as well as some Gram-positive species and archaea [12–16]. T4P perform a myriad of functions including adhesion, microcolony formation, phage and DNA uptake, twitching motility and protein secretion. They are polymers comprised primarily of a single protein subunit, the major pilin. X-ray crystal structures of full length major pilins reveal a small protein with a curved ~53-residue α-helical spine, α1, embedded via its C-terminal half, α1C, in an antiparallel β-sheet within the globular C-terminal domain of the protein [14, 17–20]. The N-terminal half of α1, α1N, is an exposed mostly hydrophobic stalk with an invariant glutamate at position 5. Pilin subunits are arranged in the pilus filament in a helical array with the α1Ns associated in the hydrophobic core of the pilus and Glu5 positioned to neutralize the positively charged N-terminal amine (N1+) of an adjacent subunit [18, 21, 22]. Two T4P sub-classes have emerged, Type IVa (T4a) and Type IVb (T4b). The T4b pili are present on enteric bacteria such as V. cholerae, enterotoxigenic Escherichia coli (ETEC), enteropathogenic E. coli (EPEC) and Salmonella Typhi as well as the Flp pili and R64 thin pilus [23, 24]. The T4b pili are distinguished from the T4a pili of Myxococcus xanthus, Pseudomonas aeruginosa and the pathogenic Neisseria in having fewer assembly components, all of which are encoded on a single gene cluster, a longer signal peptide and mature region for the major pilin and a distinct connectivity for the β-sheet in the globular domain [14].
Polymerization of the major pilin subunits into surface-displayed pili is accomplished by an assembly machinery that spans the bacterial envelope. This machinery is comprised of as few as 10 proteins to 30 or more components [16]. Despite recent major structural advances [25, 26] our understanding of T4P assembly remains incomplete. Major pilins are synthesized as prepilins, which are translocated across the inner membrane and are simultaneously processed by a dedicated prepilin signal peptidase that removes the signal peptide, leaving the subunits anchored in the inner membrane via their hydrophobic α1N [27–30]. Pilin subunits are thought to dock into the base of a growing pilus, which allows α1 to transition seamlessly from the acyl phase of the inner membrane to the hydrophobic core of the pilus where it is surrounded by its neighboring α1s [18]. The docked pilin is prevented from diffusing away from the pilus by extrusion of the growing pilus a short distance of 8–10 Å out of the membrane, equivalent to the axial rise per subunit [18, 21, 22], which opens up a gap around the base of the filament for the next subunit to dock. Polymerization is powered by ATP hydrolysis by the assembly ATPase located on the cytoplasmic side of the inner membrane [31–34]. Efficient pilus assembly requires the highly conserved Glu5, as amino acid substitutions at this position substantially reduce or eliminate pilus assembly [22, 35–37]. We have proposed that an electrostatic attraction between Glu5 on the incoming pilin subunit and the positively-charged N-terminal amino group on the terminal subunit in the growing pilus (N1+) contributes to subunit docking by neutralizing these charges in the acyl phase of the lipid bilayer prior to their extrusion from the membrane as part of the pilus [18, 22]. A bridge of electron density is observed connecting the N-terminal α-helices in the recent ~ 6 Å reconstruction of the Neisseria meningitidis T4P cryo-electron microscopy reconstruction, supporting the existence of a Glu5:N1+ salt bridge in the assembled pilus [21].
In Gram-negative bacteria T4P filaments grow across the periplasm and through the outer membrane via a complex conduit comprised of an outer membrane secretin channel and a network of proteins that connect this channel to the inner membrane assembly platform [38–41]. In addition to the assembly ATPase, most T4P systems possess a second “retraction” ATPase, often called PilT, which is required to depolymerize or retract the pili by an unknown mechanism [33, 42–46]. Retraction allows twitching motility, which pulls the bacteria along surfaces, and can draw bound substances like DNA and bacteriophage into the cell [47–53]. Retraction has been demonstrated, both directly and indirectly for P. aeruginosa, Neisseria gonorrhoeae, M. xanthus and Streptococcus sanguinis T4P by total internal reflection fluorescence microscopy, optical tweezers and elastic micropillars assays [52, 54–61]. N. gonorrhoeae pili can retract at an astonishing rate of ~1 μm/sec, equivalent to removing ~1000 pilin subunits per second. N. gonorrhoeae and M. xanthus T4P retract with forces of 100–150 pN in a PilT-dependent manner and thus PilT is considered to be the strongest molecular motor known [58]. Pilus retraction likely occurs by reversing pilus assembly, whereby subunits translocate from the base of the growing pilus into the inner membrane.
The T4P system is structurally and functionally related to the T2S system, whereby major “pseudopilins” assemble into a pseudopilus that grows from the inner membrane through the periplasm, driven by an assembly ATPase [4, 62–64]. While T4P are long thin filaments displayed on the bacterial surface, T2S pseudopili normally do not grow across the outer membrane: they are thought to form periplasmic pilus stubs that are rapidly assembled and disassembled, producing a piston-like motion to extrude substrates through the secretin channel [64–68]. Pseudopili can, however, be induced to grow into long surface-displayed pili when the major pseudopilin or the entire T2S operon are overexpressed [69, 70]. The T2S major pilins resemble Type IV pilins, having a hydrophobic N-terminus with Glu5 and a canonical pilin fold [71]. Despite the model for a dynamic pseudopilus, T2S systems lack a retraction ATPase to facilitate depolymerization.
In addition to the major pilin, the T4P and T2S systems each possess several minor pilins, pilin-like proteins that share N-terminal sequence homology and are structurally similar to their corresponding major pilins but are much less abundant in the cell. Minor pilins are known to be involved in T4P and T2S dynamics and functions but a mechanistic understanding is still lacking. Four core minor pilins, GspH, GspI, GspJ and GspK, are common to both systems, though their names differ depending on the system and bacterial species. T4P GspHIJK orthologs in P. aeruginosa, N. gonorrhoeae and EPEC are required for pilus assembly in an otherwise wild type (WT) background [72–74], but some pili are produced in minor pilin mutants that also lack the retraction ATPase, PilT [75–78]. Thus, it has been proposed that the minor pilins function as effectors of pilus homeostasis in the presence of the retraction ATPase rather than as essential initiators of pilus assembly [76]. Some T4P systems also possess one or more non-core minor pilins that are dispensable for pilus assembly but are required for pilus functions, including PilE in P. aeruginosa [77], PilV in N. gonorrhoeae [79] and ComP, PilX and PilV in N. meningitidis [80–86]. The T4P minor pilins localize to the pilus fraction and are shown by immunogold labeling and transmission electron microscopy (immunogold TEM) in some cases to be incorporated into the pilus [74, 82]. Structural studies on these minor pilins reveal the canonical pilin fold, consistent with their ability to incorporate into the pilus filament [77, 80, 82, 87–89].
Like the minor pilins of the T4P system, the T2S system minor pilins function in initiation of filament assembly [68, 70, 90–93]. Glu5 is conserved in most T2S minor pilins and was shown for at least one of these, PulH of Klebsiella oxytoca, to be necessary for secretion [94]. ETEC GspI, GspJ and GspK were crystallized in a ternary complex that is proposed to assemble at the tip of the pseudopilus [4, 95], consistent with a role in initiating pseudopilus assembly. However this minor pilin tip complex may also prevent growth of the pseudopilus through the secretin channel. The largest of the ETEC minor pseudopilins, GspK, has a bulky α-helical domain inserted between the first two strands of the pilin domain β-sheet. This large protein at the tip of the pilus may provide a steric or chemical signal that stalls pseudopilus assembly upon contact with the secretin channel [95]. ETEC GspK is a member of the GspK family of pilin-like proteins that are larger than their corresponding minor pilins and lack a position 5 glutamate [96]. Consistent with the proposed role in pseudopilus growth arrest, the GspK minor pilin XcpX was shown to control pseudopilus length in a P. aeruginosa T2S mutant overexpressing the major pseudopilin XcpT: longer more abundant pseudopili were produced in this strain when xcpX was deleted whereas fewer pseudopili were produced when xcpX was overexpressed [97]. Blocking passage of the pseudopilus through the secretin channel may trigger disassembly, producing a piston-like motion that extrudes protein substrates across the secretin channel independent of a retraction ATPase.
The closely related V. cholerae TCP and the ETEC CFA/III T4b pili provide comparatively simple systems in which to study the role of the minor pilins in filament dynamics as their assembly machinery has fewer than a dozen components, with only a single minor pilin. The functions associated with these pili—secretion, aggregation and phage transduction—suggest dynamic filaments that rapidly assemble and retract, as seen for the more complex T2S and T4P systems, yet they, like the T2S systems, lack a retraction ATPase. We show here that V. cholerae TCP are indeed retractile and that retraction is induced by the single minor pilin, TcpB. We further show that TcpB, like its ETEC counterpart CofB [98, 99], induces pilus assembly. Efficient TCP retraction but not assembly requires the conserved position 5 glutamate, consistent with TcpB having both a tip-associated localization, like the GspK family pseudopilins, but also incorporating into the growing pilus in the place of a major pilin. Our findings in the T4b pilus system provide important insights for understanding pilus dynamics in the more complex T4P and T2S systems.
Results
V. cholerae TcpB is a pilin-like protein
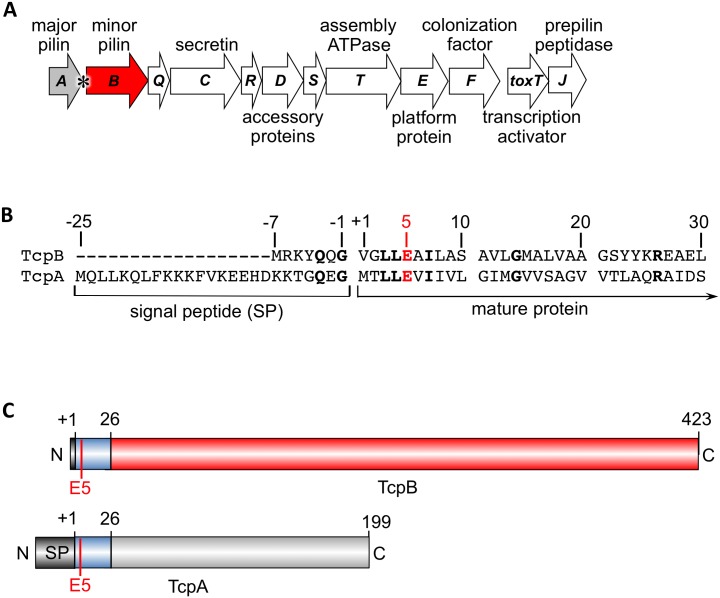
TcpB is encoded in the tcp operon immediately downstream of the tcpA gene encoding the major pilin subunit, TcpA (Fig 1A). TcpB is predicted to have a 7-amino acid signal peptide, which is considerably shorter than the 25-residue signal peptide of TcpA, and a 423 amino acid mature protein (46 kDa), which is much larger than the 199-residue TcpA (20 kDa, Fig 1B and 1C). The TcpB mature N-terminus shares amino acid sequence similarity with the N-terminal 25-residue polymerization domain of TcpA, including the conserved Glu5 (Fig 1B), but differs from TcpA in the C-terminal region. Importantly, tcpA has a putative rho-independent transcription termination site at its 3’ end (Fig 1A) [100], allowing it to be transcribed at higher levels than tcpB and other genes in the tcp operon. Apart from TcpA, TcpB is the only pilin-like protein encoded in this operon.
Fig 1. V. cholerae tcp operon and comparison of the minor pilin TcpB and the major pilin TcpA.
(A) tcp operon with the major pilin gene tcpA colored grey and the minor pilin gene tcpB colored red. The putative rho-independent transcription terminator is indicated with an asterisk. (B) Amino acid sequence alignment between the signal peptide and the N-terminal 30 residues of TcpB and TcpA (NCBI accession, CAA45456 and CAA45455, respectively). (C) Schematic of TcpB and TcpA pre-proteins.
TcpB is required for efficient pilus production
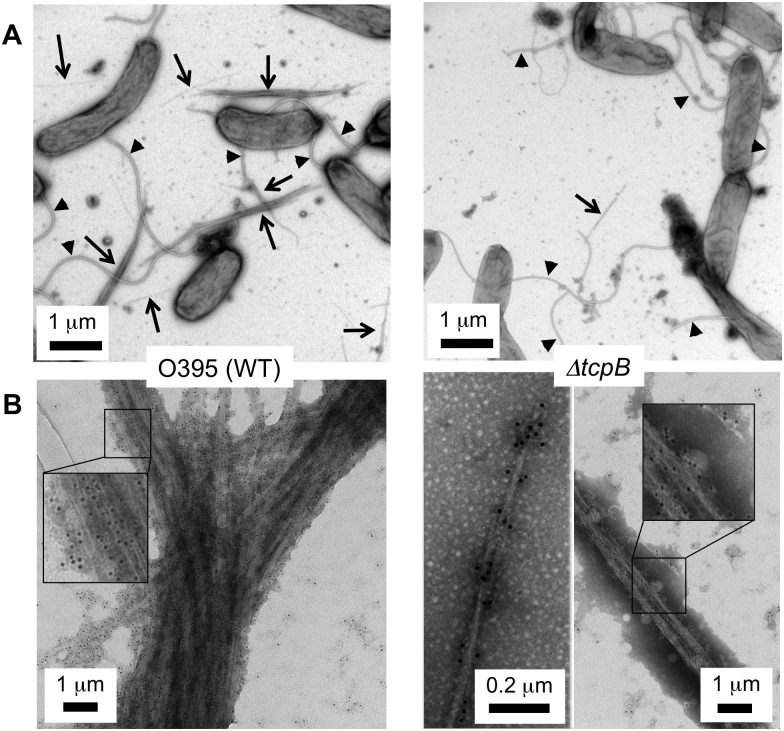
Previous reports indicate that TcpB is essential for TCP production [101–103]. We show here that the ΔtcpB strain can in fact assemble pili but at very low levels. Low magnification TEM images show abundant thick TCP bundles for V. cholerae WT strain O395, whereas few TCP are observed for the ΔtcpB mutant (Fig 2A). Nonetheless, individual TCP filaments are morphologically indistinguishable between the two strains. Pili from the WT and ΔtcpB strains were probed with polyclonal anti-TcpA primary antibodies and gold-labeled secondary antibodies and imaged by TEM. Though much less abundant, the ΔtcpB pili observed by TEM are gold labeled (Fig 2B), confirming that they are indeed TCP.
Fig 2. TCP are produced in very low numbers in the V. cholerae ΔtcpB strain.
(A) The left TEM image is representative of WT O395, in which TCP bundles are abundant, indicated by arrows. In contrast, very few TCP bundles are observed for the ΔtcpB strain. One small bundle is shown in the image on the right. Flagella are indicated with arrowheads. (B) TEM images of TCP labeled with anti-TcpA primary antibody and gold-labeled secondary antibody. The gold particles are 6 nm in diameter. A section of each image is magnified in the inset to show the gold particles attached to the pili.
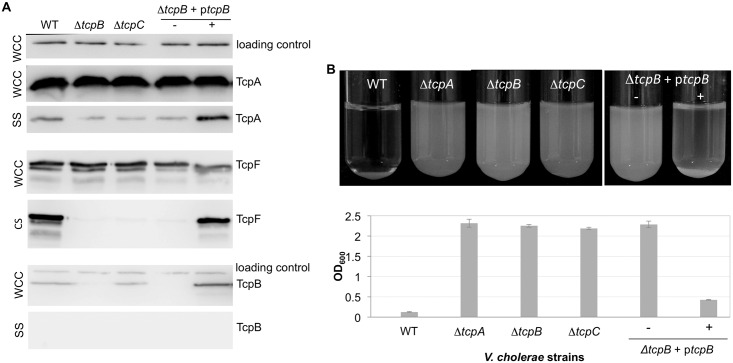
To evaluate whether TcpB influences pilus production by acting at the level of TcpA expression and/or stability, V. cholerae overnight cultures were examined by SDS-PAGE and immunoblotting. TcpA levels in whole cell culture samples (WCC), which represent the total protein within the cells and in the supernatant, are comparable in the WT O395 and ΔtcpB strains (Fig 3A), suggesting that TcpB is required for pilus assembly rather than for optimal expression and stability of TcpA. We also looked at the levels of TcpA in the culture supernatant after homogenizing the cells to shear off their pili (sheared cell supernatant, SS). Immunoblotting of the SS fraction with anti-TcpA antibodies showed low levels of TcpA for WT V. cholerae compared to the WCC fraction. This is likely due to the tendency of TCP to bundle, which makes them difficult to separate from the cells as the bundles pellet with the cells even at low g-forces. Nonetheless, TcpA levels were further reduced in the ΔtcpB mutant, comparable to that of a ΔtcpC mutant lacking a functional secretin channel. TcpA levels in the ΔtcpB SS fraction were restored when TcpB was expressed on a plasmid, ptcpB, from a rhamnose-inducible promoter PRHA (Fig 3A). Taken together, the immunoblots and TEM results suggest that TcpB, like the closely related ETEC CofB and other minor pilins [76, 78, 92, 98, 104], is involved in initiating pilus assembly.
Fig 3. V. cholerae requires TcpB for efficient pilus assembly and functions.
(A) Immunoblots of V. cholerae whole cell culture (WCC), sheared culture supernatant (SS) and cleared culture supernatant (CS) fractions probed with antibodies against TcpA, TcpF and TcpB, as indicated by the labeled protein bands. SS for the TcpA and TcpB blots was prepared by homogenizing the cells to shear the pili then removing intact cells by centrifugation. The SS fractions were topped up with PBS to a volume matching that of the WCC, and equal volumes (25 ul) of each sample were loaded onto the gel. TcpA in the SS fraction of the ΔtcpB and ΔtcpC mutants may represent contamination from cell membranes due to the shearing method or membrane blebbing due to high levels of TcpA that accumulate in the inner membrane. Secreted TcpF is detected in the CS fraction, obtained by removing V. cholerae cells by centrifugation and filtration without shearing. TcpF often appears as a doublet in immunoblots. The lower band of the doublet appears to be a proteolysed form of TcpF lacking the N-terminal ~25 amino acids. This segment is not resolved in the TcpF crystal structure suggesting it is flexible, which may explain its protease sensitivity [10]. The ΔtcpB strain is complemented with ptcpB without (-) and with (+) 0.001% rhamnose inducer. The loading control is an unknown ~60 kDa protein present in the WCC fraction and detected by the Strep-Tactin-HRP conjugate. (B) V. cholerae autoagglutination as a function of TcpB expression. Cell cultures are shown after overnight growth followed by 15 min stationary incubation at room temperature (top panel). The OD600 of the culture supernatant after the cells have settled is plotted for each sample in the bottom panel. The more complete the autoagglutination the lower the OD600 value. Values are averaged for 3 experiments; error bars represent standard deviations.
TCP-mediated functions of TcpF secretion and bacterial autoagglutination were analyzed for the ΔtcpB mutant and complemented strain. TcpF secretion is assayed by immunoblotting cleared culture supernatant (CS), in which overnight V. cholerae cultures are centrifuged and filtered to remove bacteria. TcpF secretion is abrogated in the ΔtcpB strain, comparable to that of the ΔtcpC strain, and is restored when TcpB is expressed ectopically (Fig 3A). Autoagglutination in overnight cultures can be assessed visually by examining the size and compactness of the macroscopic aggregates and the clarity of the supernatant, and more quantitatively by measuring the optical density (600 nm) of the culture supernatant, where the more complete the autoagglutination the lower the OD600 (Fig 3B). This assay serves as an in vitro readout for V. cholerae microcolony formation [8, 35]. Like TcpF secretion, autoagglutination is disrupted in the ΔtcpB strain and restored in the tcpB-complemented strain. The loss of pilus functions in the ΔtcpB mutant is consistent with a disruption of TCP assembly.
Because TcpB is homologous to TcpA in its N-terminal segment, and to ETEC CofB, which has a pilin domain [98], this minor pilin may incorporate into the pilus filament, most likely at its tip, consistent with its role in initiating pilus assembly. To test if TcpB co-localizes with TCP, WCC and SS fractions were immunoblotted with anti-TcpB antibody. As expected, TcpB is detected in the WCC fraction of WT and ΔtcpC strains and in the ΔtcpB+ptcpB strain, but not in the ΔtcpB strain (Fig 3A). While the intensities of the TcpA and TcpB bands cannot be directly compared as they are detected by different antibodies, the differences are nonetheless consistent with TcpB being produced in much lower levels than TcpA, as expected for a minor pilin. No TcpB was detected in the SS fraction for any of the strains (Fig 3A). However, based on the high ratio of TcpA:TcpB in the WCC fraction it is unlikely that TcpB would be detected in the SS fraction as so little TcpA is present in this fraction.
TcpB is a minor component of the pilus
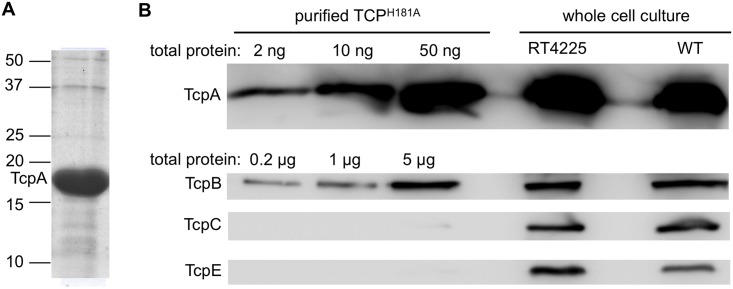
Since the immunoblots for TcpA and TcpB from sheared supernatant fractions were inconclusive regarding co-localization of TcpB with the pili, we looked for TcpB in a more concentrated pilus sample. The V. cholerae mutant, RT4225, has a His181Ala substitution in the major pilin TcpA [35]. TCPH181A pili can readily be purified by ammonium sulfate precipitation of the overnight culture supernatant [8, 22, 105]. This is likely because the pili don’t bundle and thus do not precipitate with the cells, and because they grow unusually long and fall off the bacteria into the culture supernatant during overnight growth. We can load much larger quantities of purified TCPH181A onto a gel (Fig 4A) compared to our sheared supernatant fractions from WT V. cholerae, which increases our chance of detecting the low abundance TcpB. Purified TCPH181A were examined by SDS-PAGE and immunoblotting with anti-TcpA and TcpB antibodies. TcpB was detected in the TCPH181A pilus preparation when loaded in much larger amounts than that required to visualize the major pilin TcpA (Fig 4B), indicating that it is indeed a minor component of the pilus. V. cholerae RT4225 produces TcpA and TcpB in levels comparable to that of WT V. cholerae O395, as shown by immunoblots of whole cell culture (Fig 4B). To ensure that the TcpB present in the TCPH181A preparation is not a contaminant of the V. cholerae envelope, this fraction was probed with antibodies against membrane components of the TCP assembly machinery: the outer membrane protein TcpC and the inner membrane protein TcpE. These proteins are almost undetectable even at the highest TCPH181A concentration loaded. These results indicate that TcpB co-localizes with the pilus but is present in much lower levels than TcpA, consistent with it being a minor pilin.
Fig 4. Co-localization of TcpB with purified TCPH181A.
(A) Coomassie-stained SDS gel of TCPH181A purified from V. cholerae RT4225, which has a mutation encoding a His181Ala substitution in the major pilin, TcpA. The protein concentration of the solution was found by spectrophotometry (A280) to be 10 mg/ml. The lane was overloaded with 90 μg of protein to show the purity, estimated at >95%. The positions of mass markers are indicated. The protein band at ~50 kDa is not likely to be TcpB (46 kDa), which based on the TcpA:TcpB stoichiometry determination (see below) is present in quantities too small to detect by Coomassie stain. (B) Immunoblots of pilins and pilus assembly components in purified TCPH181A and whole cell culture. For the purified TCPH181A samples, the total amount of protein loaded is indicated; for the whole cell culture samples, 25 μl was loaded into each well. Blots were probed with antibodies against TcpA and TcpB; TcpC and TcpE were also probed to assess contamination from outer and inner membranes, respectively. All blots were developed for 2 minutes.
To determine the TcpA:TcpB stoichiometry in the TCPH181A fraction, each protein was quantified by comparing the intensities of the immunoblot bands with those of purified recombinant N-terminally truncated (ΔN-) TcpA and TcpB of known concentrations using densitometry. Band densities of known amounts of ΔN-TcpA and ΔN-TcpB were normalized and their averages were plotted; these plots were used to determine the amount of TcpA and TcpB in the TCPH181A sample based on their densities (S1A and S1B Fig). The TCPH181A sample was found by densitometry to contain 12±1 mg/ml of TcpA and 4±1 μg/ml of TcpB, giving a mass ratio of 3000:1 and a molar ratio of ~ 7000:1 TcpA:TcpB. WT TCP bundles are typically ~ 2–3 μm in length, whereas individual TCPH181A pili appear to be longer but cannot be measured precisely as they extend past the edges of TEM images at magnifications at which they can be resolved (S1C Fig). Based on TCP having an axial rise of 8.4 Å [22], a ~ 6 μm-long pilus would be comprised of ~ 7000 TcpA subunits and only a single TcpB molecule. Since TcpB is involved in initiating pilus assembly we reason that this single minor pilin is located at the pilus tip. Interestingly, despite the very small amount of TcpB in the purified pilus fraction, TcpB is readily detected in whole cell culture by immunoblots (Figs 3A and 4B), indicating that more TcpB is made than is incorporated into the pili.
V. cholerae TCP are retractile
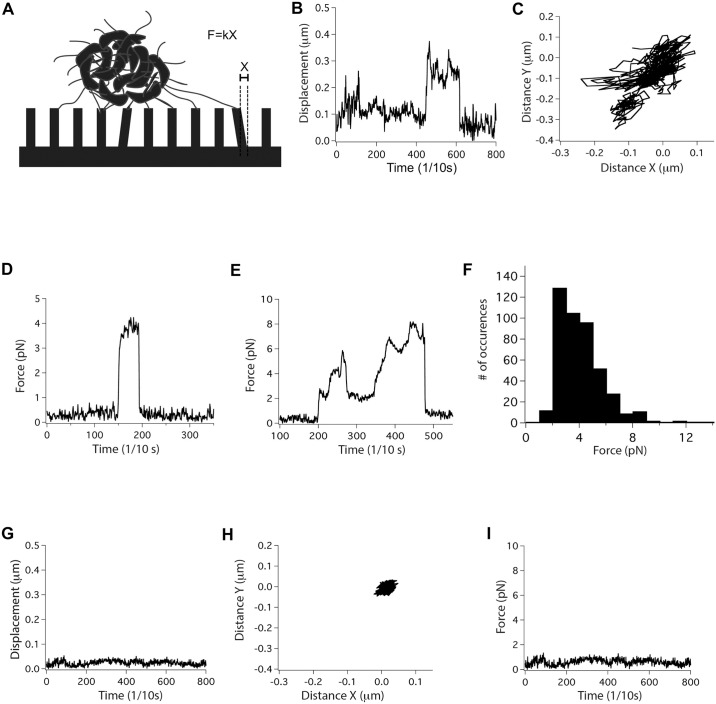
We proposed previously that TCP might be retractile despite lacking a retraction ATPase, based on their functions in TcpF secretion, autoagglutination and phage uptake [106]. We wondered if TcpB might be involved in this process in addition to its role in initiating pilus assembly. Our hypothesis is that TcpB induces pilus retraction by randomly incorporating into a growing pilus in place of TcpA and stalling assembly, causing the pilus to spontaneously disassemble. Stalling could occur by TcpB blocking addition of subsequent major pilins into the growing pilus or by preventing passage of the pilus through the secretin channel, as was proposed for the ETEC minor pseudopilin GspK [95]. To test if TCP are indeed retractile we performed an elastic micropillars assay, which has been used to demonstrate and quantify retraction events for T4P of N. gonorrhoeae [54, 107] and Streptococcus sanguinis [57]. Bacterial microcolonies are placed on an array of elastic micropillars and the movements of the tips of the micropillars under and adjacent to a microcolony are observed by differential interference contrast microscopy (Fig 5A). To assess TCP retraction in V. cholerae we employed a mutant lacking the flagellin gene, ΔflaA, to eliminate non-pilus-mediated micropillar movement that might result from flagellar rotation. Movements of micropillars directly under and within a few microns of microcolonies were observed in the direction of the microcolonies, consistent with TCP binding and retraction (Fig 5B and 5C and S1 and S2 Movies). The force exerted on the micropillars, F, is the product of the calibrated elastic spring constant of the micropillars, k, and the displacement, X, of micropillars from their resting position (Fig 5A, 5D–5F). Although many micropillars motions were recorded, only those movements whereby the micropillars returned to their resting position were included in the force calculations. Micropillars motions occur in single pulls lasting a few seconds (Fig 5D) and also in more complex pulls that last tens of seconds (Fig 5E). Such complex patterns showing different local force maxima and plateaus could be explained by the pulling of multiple pili, perhaps within TCP bundles, as seen for N. gonorrhoeae T4P [54]. This study does not distinguish between movements induced by individual pili and pilus bundles. The average force for recorded pulling events is 4 ± 1.7 pN (n = 447, Fig 5F), which is substantially weaker than the 50–100 pN forces observed for individual T4P from N. gonorrhoeae and S. sanguinis [54, 57, 58, 107].
Fig 5. Micropillars assay demonstrates that TCP are retractile.
(A) Illustration of the micropillars assay set-up. The force of retraction, F, is the product of the spring constant of the micropillars, k (here k = 25 pN/μm), and the displacement of a micropillar from its resting position, X. (B-F) Data plots from micropillars assays for V. cholerae ΔflaA. (B) Displacement of a single micropillar under a bacterial aggregate over time, measured from its resting position. The displacement is representative of hundreds of micropillars motions analyzed for several hours of recordings. (C) Projection in X and Y of the movement of the micropillars shown in (B) over the same period of time. (D, E) Representative values for force exerted on a single micropillar as a function of time for two separate retraction events. (F) Histogram of the force distribution for recorded TCP retraction events for V. cholerae ΔflaA. (G-I) Representative results for V. cholerae ΔtcpB/ΔflaA showing (G) the displacement of a single micropillar over time, which is not detected above noise level, (H) the projection in X and Y of the movement of the micropillars shown in (G), and (I) the calculated force exerted on the same micropillar over the same period of time.
Next, we examined the V. cholerae ΔtcpB/ΔflaA strain, which produces very few pili, to demonstrate that the micropillars movements observed for V. cholerae ΔflaA are pilus-mediated. Since this strain does not aggregate, these bacteria were treated with anti-lipopolysaccharide antibody prior to the micropillars assay to promote cell-cell aggregation and microcolony formation to better mimic the V. cholerae ΔflaA conditions. No micropillar movements were observed under these artificially induced V. cholerae ΔtcpB/ΔflaA microcolonies (Fig 5G–5I and S3 Movie). These results confirm that the micropillars movements observed for the V. cholerae ΔflaA strain are pilus-dependent, supporting our hypothesis that TCP are retractile despite lacking a retraction ATPase.
TcpB Glu5 is not required for pilus assembly but is required for pilus-mediated functions
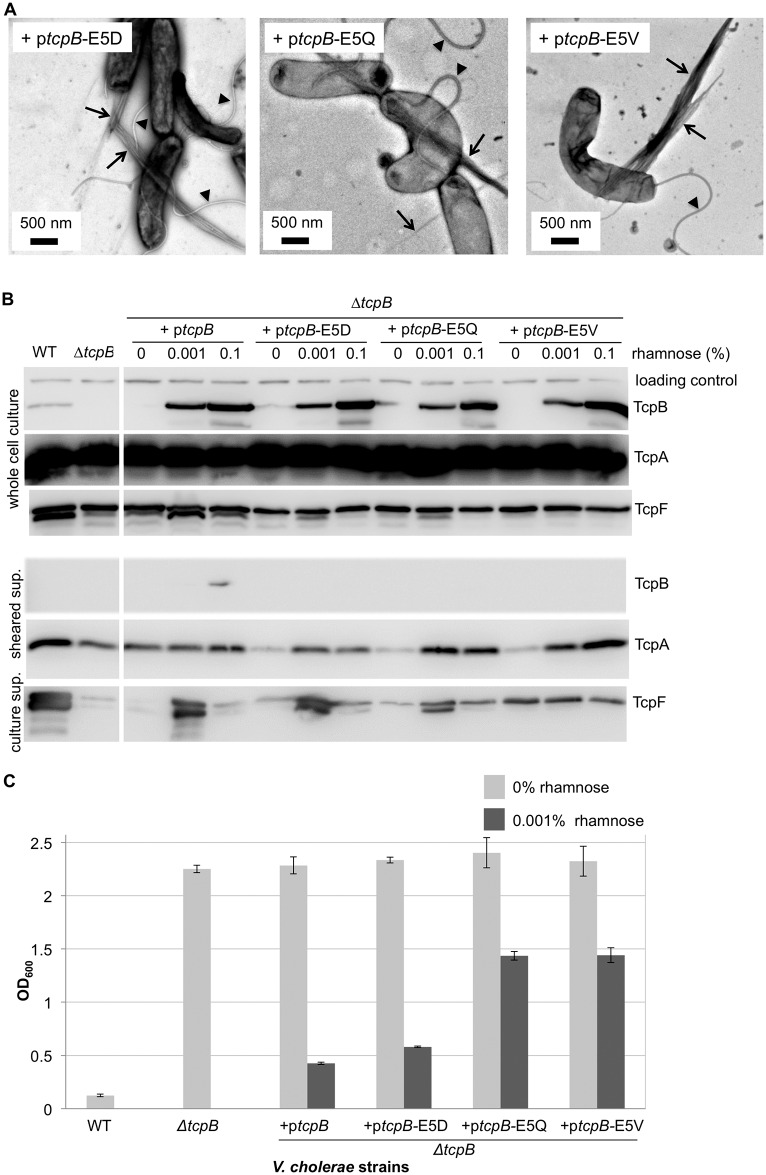
To determine whether or not TcpB influences pilus retraction it was necessary to identify a V. cholerae tcpB mutant capable of assembling TCP at WT levels, but not retracting them. We showed previously that the conserved Glu5 of major pilin TcpA is necessary for efficient TCP assembly [22]. We reasoned that TcpB should not require Glu5 to initiate pilus assembly as it would be the first subunit in the pilus, but Glu5 would be necessary for TcpB to incorporate into the growing pilus to initiate retraction. Retraction-deficient TCP would be unable to pull bacteria into compact microcolonies, to produce a piston-like motion to secrete TcpF, or to draw CTXϕ phage into the bacterium. To test the role of TcpB Glu5 in pilus assembly, retraction and functions we generated a series of tcpB mutants with position 5 substitutions and examined their ability to rescue pilus assembly and functions in a ΔtcpB mutant. Expression of the TcpB Glu5 variants in a ΔtcpB mutant restored pilus assembly: these strains produce WT or better levels of TCP, as assessed by TEM (Fig 6A). However, pilus-mediated TcpF secretion and autoagglutination are impaired in the complemented strains (Fig 6B and 6C). The degree of functional impairment depends on the nature of the Glu5 substitution: the ΔtcpB mutant complemented with the most conserved Glu5 substitution to the negatively charged aspartate (E5D) showed a slight reduction in both TcpF secretion and autoagglutination compared to the WT strain; the Glu5Gln (E5Q) variant, which results in loss of the negative charge but retains polarity, showed poor TcpF secretion and autoagglutination; and the Glu5Val (E5V) variant, which has a hydrophobic side chain at position 5, was substantially impaired for TcpF secretion and autoagglutination. These differences cannot be attributed to defective TcpB expression for the Glu5 variants, as TcpB-E5X levels are comparable to the WT rescue and in all cases are higher than for endogenous expression in the WT O395 strain (Fig 6B). Gao et al. showed that the V. cholerae chromosomal Glu5Val mutant is deficient in autoagglutination, TcpF secretion and CTXϕ transduction [102]. Thus, TcpB Glu5 is not required for pilus assembly but is required for optimal pilus functions.
Fig 6. Complementation of V. cholerae ΔtcpB with tcpB-E5V mutants results in impaired autoagglutination and TcpF secretion without disrupting pilus assembly.
(A) TEM images of V. cholerae ΔtcpB complemented with tcpB mutants encoding Glu5 substitutions show abundant pilus production. Arrows point to TCP bundles and arrowheads point to flagella. (B) Immunoblots of V. cholerae fractions for the ΔtcpB strain complemented with WT tcpB or tcpB mutants. TcpF secretion is disrupted for the TcpB Glu5 variants despite them producing TcpB levels comparable to that of the WT tcpB-complemented ΔtcpB strain. (C) Autoagglutination is impaired in the V. cholerae ΔtcpB strain rescued with TcpB Glu5 mutants. The lower the OD600 values the more complete the autoagglutination. Values are averaged for three replicates; error bars represent standard deviations.
Interestingly, while low levels of TcpB expression (induction with 0.001% rhamnose) restored TcpF secretion to varying degrees depending on the nature of the Glu5 substitution, high levels (0.1% rhamnose) were detrimental for both the WT and the mutant tcpB complementation (Fig 6B). Induction with 0.001% rhamnose apparently provides the optimal level of TcpB expression for TCP functions. TCP bundles were quantified for both expression levels, revealing a substantial reduction in piliation for cells induced with 0.1% rhamnose, compared to those induced with 0.001% rhamnose (see Fig 7B). Thus, overexpression of TcpB disrupts pilus assembly, which impacts pilus functions. Even lower levels of rhamnose were tested for the complementation of the ΔtcpB strain with WT ptcpB, but these did not result in improved autoagglutination levels (S2 Fig). Together these results imply that a precise TcpA:TcpB stoichiometry must be attained for optimal pilus assembly and functions.
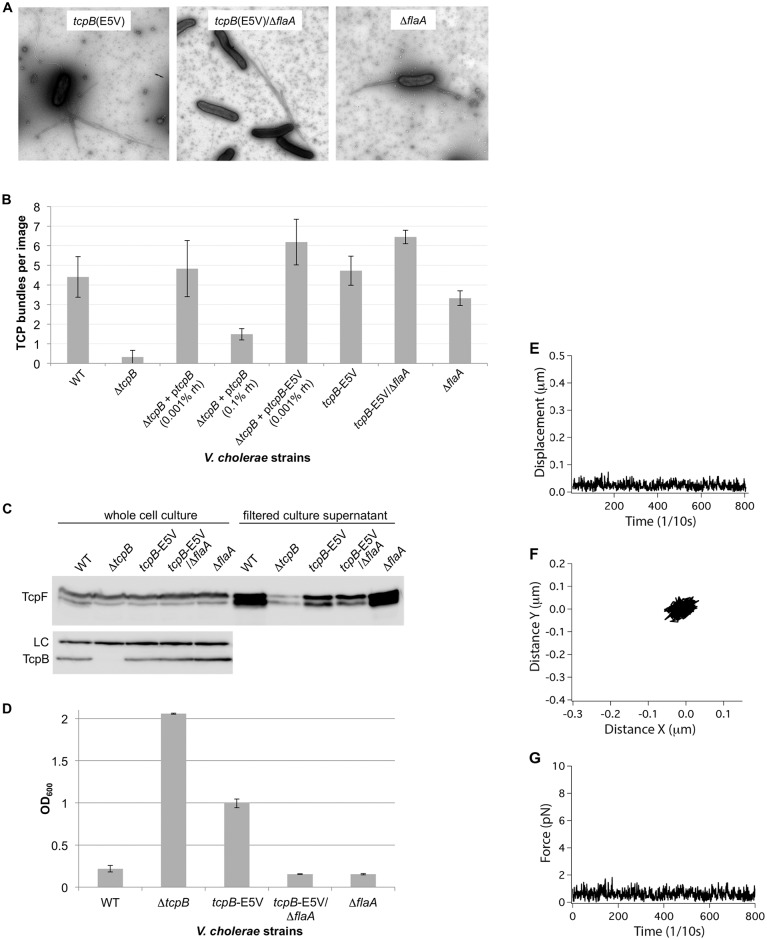
Fig 7. Autoagglutination, TcpF secretion and retraction but not TCP assembly are impaired in a chromosomal tcpB-E5V V. cholerae mutant.
(A) TEM images of piliated V. cholerae chromosomal tcpB-E5V/flaA mutants. (B) TCP bundles in TEM images were counted to quantify pilus expression in the various V. cholerae strains. Sixteen or more low magnification (1850X) images were counted for three growth replicates for each strain; error bars represent standard deviations. (C) Immunoblots of TcpF and TcpB in whole cell culture and cleared culture supernatant for tcpB-E5V/flaA mutants. The Glu5Val substitution disrupts TcpF secretion without affecting TcpB production. (D) The Glu5Val substitution in TcpB disrupts autoagglutination, which is restored in the tcpB-E5V/flaA double mutant. The lower the OD600 values the more complete the autoagglutination. Values are averaged for three replicates; error bars represent standard deviations. (E-G) Micropillars assay data plots for V. cholerae tcpB-E5V/ΔflaA showing (E) the displacement of a representative micropillar as a function of time, which is not detected above noise level, (F) the projection in X and Y of the movement of this same micropillar over time, and (G) the force exerted on this micropillar over the same time period.
Efficient TCP retraction requires TcpB Glu5
We hypothesized that the TcpB Glu5 variants are defective in inducing TCP retraction, explaining their impaired autoagglutination and TcpF secretion. To test this hypothesis we performed the micropillars assay on the V. cholerae tcpB-E5V mutant, which has the most severe functional defects. For these experiments we introduced the mutation encoding the Glu5Val substitution into the tcpB gene within the V. cholerae large chromosome via allelic exchange. Consistent with our results with ectopically expressed TcpB-E5V in the ΔtcpB strain (Fig 6A–6C), the chromosomal tcpB-E5V mutant produces pili that match WT TCP in morphology, bundling and quantity, as assessed by TEM (Fig 7A and 7B), but is impaired for both TcpF secretion and autoagglutination (Fig 7C and 7D). Interestingly, further deletion of flaA in the tcpB-E5V strain to eliminate flagella and non-pilus-mediated motility restored autoagglutination, though TcpF secretion remained impaired. Autoagglutination was also restored in another double mutant, tcpB-E5V/ΔmotAB, which is non-motile but does produce flagella (S3 Fig). Micropillars pulling events were very infrequent for the tcpB-E5V/ΔflaA mutant (Fig 7E–7G and S4 Movie) despite it being piliated at WT levels (Fig 7A). Thus, the single amino acid substitution in TcpB, Glu5Val, appears to disrupt TCP retraction without disrupting TcpB-mediated pilus assembly. We propose that V. cholerae autoagglutination and microcolony formation are driven by dynamic cycles of elongation and retraction of TCP. Initially, weak complementary interactions among extended pili bring the bacteria in contact with one another [8], followed by pilus retraction to form tight aggregates. Without retraction the forces of flagellar rotation will disrupt the pilus:pilus interactions and prevent aggregation. Conversely, without flagellar rotation, weak pilus:pilus interactions may persist long enough for inefficient TcpB-E5V-mediated retraction or spontaneous non-TcpB-mediated retraction to occur, allowing the tcpB-E5V/ΔflaA mutant to autoagglutinate during overnight growth.
Discussion
Our results represent an important conceptual advance in understanding pilus assembly and retraction and the role of the minor pilins in pilus dynamics for both the T4P and T2S pseudopilus systems. First, we show that the single minor pilin of the V. cholerae TCP system, TcpB, is required for efficient pilus assembly. Second, TCP are retractile despite lacking a retraction ATPase. Third, TCP retraction is induced by the minor pilin TcpB. Fourth, TcpB-mediated pilus retraction but not assembly requires TcpB-Glu5. Finally, retraction facilitates TCP-mediated processes of autoagglutination and TcpF secretion.
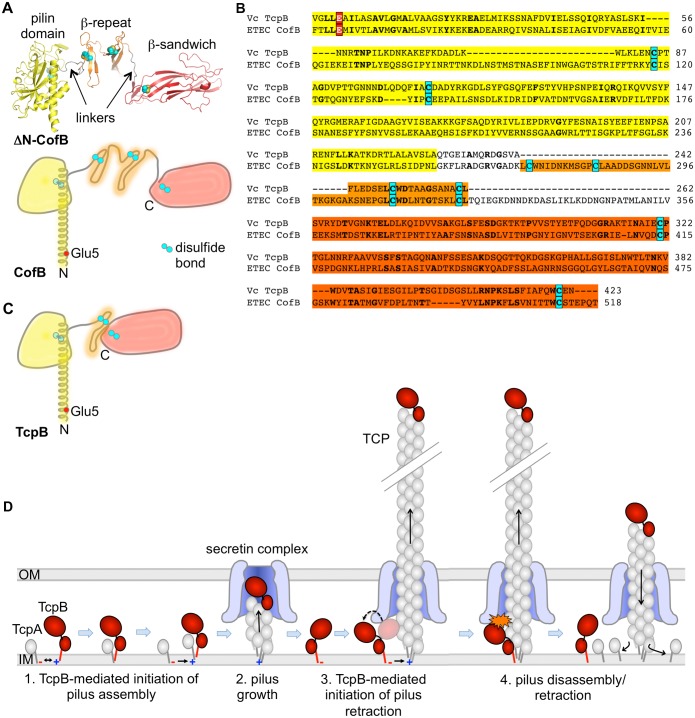
The role of the single minor pilin in initiating pilus assembly was shown previously in the CFA/III and Longus T4b pilus systems of ETEC [98, 99, 108], which are closely related to V. cholerae TCP in sequence, structure, gene synteny and pilus functions [98, 109]. We proposed that the ETEC minor pilin CofB forms the first subunit in the growing pilus and is thus tip-associated [98]. CofB has a well-defined pilin domain that resembles its corresponding major pilin, CofA, in fold and size, but has an extended C-terminal region comprised of a β-repeat domain followed by a β-sandwich domain, which are connected by flexible linkers (Fig 8A) [98, 99]. Each domain is stabilized by one or more disulfide bonds. While sequence similarity between CofB and TcpB is low, alignment of the conserved N-terminal α1s and 6 cysteines reveals additional shared residues (Fig 8B) and suggests a similar structure for TcpB, but lacking the first β-repeat sub-domain and having only a single linker (Fig 8C). The tip of the pilus is the only location that this bulky pilin could occupy without undergoing a conformational change and without having the C-terminus protrude from the pilus. As tip-associated pilins TcpB and CofB could recruit major pilins and/or trigger opening of the outer membrane secretin channel to allow passage of the pilus to the cell surface (Fig 8D). While we have not directly demonstrated that TcpB is tip-associated, we show here that it co-localizes with purified TCP with approximately one molecule per pilus, consistent with this minor pilin incorporating into the pilus tip to initiate pilus assembly rather than acting at the level of the trans-envelope pilus biogenesis machinery. It is important to note that TcpB is not essential for pilus assembly as small amounts of pili are observed in the ΔtcpB mutant. It may be that the major pilin can itself nucleate TCP assembly and/or trigger opening of the secretin channel but does so inefficiently.
Fig 8. Comparison of TcpB and the ETEC minor pilin CofB and model for TcpB-mediated assembly and retraction.
(A) Crystal structure of N-terminally truncated CofB (4QS4 [98]) shown in cartoon representation (top), and as a schematic (bottom) with the full N-terminal α-helix. Cysteines are colored cyan. (B) Amino acid sequence alignment of TcpB and CofB (NCBI Accession BAB62898). Alignment was first performed using Clustal Omega [110] then adjusted to align the cysteines. Identical residues are shown in boldface type. The conserved Glu5 (red with white text) and cysteines (cyan) are indicated. Discrete domains are shaded based on the coloring of CofB shown in (A). (C) Proposed schematic of the TcpB structure based on sequence alignment with CofB. (D) Model for TcpB-mediated initiation of pilus assembly and retraction. TcpB is represented as a pilin domain (red stick and small oval) with an additional C-terminal domain (large oval). Incorporation of TcpB into the growing pilus may block passage of the pilus through the secretin complex as shown (Steps 3, 4) or may prevent further incorporation of TcpA. If pilus assembly cannot proceed, the pilin subunits will melt back into the membrane, one subunit at a time, retracting the pilus.
We show here that the non-motile but piliated V. cholerae strain ΔflaA induces micropillars movements, whereas the non-piliated ΔtcpB/ΔflaA strain does not, implying that micropillars movements are pilus-mediated. T4P-mediated micropillars movements have been demonstrated for N. gonorrhoeae and S. sanguinis [54, 57, 111], and T4P retraction has been demonstrated by optical tweezers experiments for N. gonorrhoeae and M. xanthus [52, 54, 56, 58, 59, 61]. Piliation is not sufficient for micropillars pulling as strain tcpB-E5V/ΔflaA makes better than WT levels of pili but micropillars movements were rarely observed for this strain, demonstrating a role for Glu5 in TCP retraction but not assembly. An alternative explanation for the lack of micropillars movements in tcpB-E5V/ΔflaA, that the Glu5Val substitution destabilizes the pili causing them to break off the cells, was ruled out based on the following observations: (i) TCPTcpB-E5V bundles are more likely than WT TCP to be found by TEM attached to cells; (ii) TCPTcpB-E5V levels in the overnight culture supernatant are approximately the same as TCPWT; and (iii) TCPTcpB-E5V bundles tend to be longer than those of TCPWT, consistent with efficient pilus growth but inefficient retraction.
Our results with the tcpB-E5V strain, together with those of Gao et al. [102] show that pilus production alone is insufficient to mediate TCP functions of autoagglutination, TcpF secretion and CTXϕ uptake. Impaired functions in the tcpB-E5V strain correlate with lack of retraction, suggesting that TCP retraction is central to all of these processes: to produce the very tight, spherical aggregates observed for autoagglutinated V. cholerae in vitro and V. cholerae microcolonies in the intestines [35, 112]; to produce a piston-like motion to efficiently secrete TcpF; and to draw the filamentous CTXϕ into the periplasm. While the non-piliated ΔtcpB mutant is completely deficient in autoagglutination, TcpF secretion and phage uptake, the piliated tcpB-E5V mutant is able to perform these functions, though at substantially reduced levels, suggesting that pilus retraction occurs in this strain, perhaps inefficiently in a TcpB-E5V dependent manner, or by spontaneous TcpB-independent retraction. Nonetheless, it was surprising that the non-motile tcpB-E5V/ΔflaA mutant autoagglutinates as well as the WT strain. This observation highlights the disruptive force of flagellar rotation, which works against autoagglutination and microcolony formation. However TcpF secretion requires efficient retraction whether the cells are motile or not. It stands to reason that a pilus undergoing rapid cycles of elongation and retraction would extrude substrate more efficiently, whereas a pilus that grows long and then retracts would be more effective in establishing bacterial aggregates. TCP retract with forces averaging ~4 pN, enabling V. cholerae to move against moderate hydrodynamic flows to form compact aggregates and to draw phage into the periplasm. These forces are considerably weaker than the >100 pN forces exerted by N. gonorrhoeae and M. xanthus T4P [54, 56, 58], which perform twitching motility in addition to aggregation and phage uptake. The N. gonorrhoeae and M. xanthus T4P systems possess a retraction ATPase, which may influence the force, rate and frequency of the retraction events.
How might this minor pilin perform dual antagonistic roles in pilus biogenesis? We propose that TcpB incorporates at the pilus tip to initiate pilus assembly and remains at the pilus tip as the filament grows. Additionally TcpB incorporates transiently into the growing pilus where it causes assembly to stall, triggering disassembly or retraction. TcpB anchored in the inner membrane would occasionally insert into the growing pilus via its pilin domain, in place of TcpA (Fig 8D). The flexible linker would allow the bulky C-terminal domain to move out of the way of the pilin domain, forcing it onto the pilus surface where it would interfere with passage of the pilus through the secretin complex or perhaps prevent addition of the next major pilin, effectively stalling pilus assembly. Stalling would prevent incremental extrusion of the pilus so TcpB would eventually diffuse back into the inner membrane. As a result, the pilus would collapse a short distance into the membrane, whereupon the next pilin subunit would diffuse away, etc., disassembling the pilus one subunit at a time. The frequency of retraction events would depend on the molar ratio of TcpA to TcpB. This ratio would define the average length of the pilus: the more TcpB present the more frequent the retraction events and the fewer and shorter the pili. Our model infers that pilus retraction occurs as a consequence of the dynamic instability of the growing pilus filament, which requires energy from ATP hydrolysis to assemble but disassembles spontaneously, melting into the inner membrane one subunit at a time when assembly is interrupted, in this case by TcpB. This processive melting mechanism has been proposed by others [53], but was ruled out for T4P systems possessing a retraction ATPase as retraction visibly slows when ATP is depleted, suggesting an active role for the retraction ATPase [58]. While the mechanism of PilT-mediated retraction is not well understood, it clearly differs markedly from TcpB-mediated retraction, which, rather than utilizing chemical energy from ATP hydrolysis, appears to be a passive mechanical process that exploits the potential energy stored in the pilus.
Such a model explains why Glu5 is necessary for TcpB to induce pilus retraction but not pilus assembly. Docking of pilin subunits into a growing pilus is driven in part by charge complementarity between the N-terminal amine on the terminal subunit in the pilus and Glu5 on the incoming pilin (Fig 8D). As the first pilin subunit in the pilus, TcpB would need a positively charged N-terminus to neutralize the negatively charged Glu5 on an incoming TcpA, but would not itself need Glu5. However, to incorporate into the growing pilus TcpB would need Glu5 to neutralize N+ on the terminal TcpA at the base of the filament. Because TcpB-E5V can incorporate at the pilus tip it can initiate pilus assembly, thus the tcpB-E5V mutant is piliated, but TcpB-E5V cannot efficiently incorporate into the growing pilus to induce retraction, explaining the lack of micropillars motions and aberrant functions for this strain.
The sequence and functional similarities between V. cholerae TcpB and ETEC CofB suggest that the highly homologous ETEC minor pilins CofB and LngB also induce retraction of their respective T4b pili, CFA/III and Longus, respectively, to mediate aggregation and secretion. It is remarkable that a single minor pilin accomplishes both initiation and retraction, whereas four or more minor pilins work together to prime and in some cases to arrest pilus assembly in the more complex T4a pilus and T2S systems [74, 76, 77, 92, 93]. The extended C-terminal regions of the V. cholerae and ETEC minor pilins likely impart specialized functions accomplished by multiple minor pilins in the more complex systems, such as recruiting major pilins to initiate pilus assembly and/or opening the secretin channel. CofB, and by inference, TcpB, have flexible structures plus Glu5 that allow them to (i) incorporate at the tip of the pilus to initiate assembly, (ii) to pass through the secretin complex when tip-associated, and (iii) to incorporate into the growing pilus to trigger retraction. In contrast, the T2S minor pilin GspK is bulky but rigid and has a hydrophobic amino acid at position 5. While GspK in complex with GspHIJ appears to initiate pseudopilus assembly, its rigid structure might prevent its passage through the secretin complex [95], preventing growth of this pilus across the outer membrane, and triggering retraction. Rapid cycles of polymerization and retraction would serve to efficiently extrude substrates across the outer membrane. Importantly, its bulk, rigidity and lack of Glu5 would prevent GspK from incorporating at any site other than at the tip of the pilus. GspK is the only minor pilin of the GspHIJK cluster that lacks a glutamate at position 5. In fact, for most T2S and T4a pilus systems, as well as the EPEC T4b system of bundle-forming pili, only one of the four or five minor pilins has hydrophobic residue at position 5, the others having Glu5. In the T2S systems and the Gram positive T4P systems this minor pilin is also substantially larger than its corresponding major pilin, typical of GspK family pilins, and is encoded at the 3’ end of the minor pilin gene cluster. These features may represent signatures to identify minor pilins that form the first subunit in a nascent filament. Interestingly, for two of the minor pilins of the K. oxytoca T2S system, Glu5 has been implicated in interactions with inner membrane components of the pseudopilus assembly machinery, demonstrating a critical and complex role for this residue in filament formation [94].
The minimalist V. cholerae and ETEC T4b systems may represent precursors for other T4P and T2S systems. In these systems all genes necessary for T4b pilus assembly are clustered on a single operon. The V. cholerae tcp operon is present on a mobile genetic element, the Vibrio pathogenicity island [7], and the ETEC cof operon and lng operons are present on large virulence plasmids [113, 114]. These bacteria could pass their pilus operons to other bacteria during gut infections. Additional minor pilin genes likely arose via gene duplication. The T2S system appears more closely related to the V. cholerae and ETEC T4b pilus systems as T2S genes are also located on a single operon, they lack a retraction ATPase and they encode a large GspK family subunit. V. cholerae TcpB and ETEC CofB are large like the GspK family pilins but they have a flexible C-terminal region and Glu5, allowing them to incorporate at any point during pilus assembly and induce retraction. Our results suggest that the T4b pili, like the T4a pili and T2S pseudopili, are dynamic filaments that rapidly assemble and disassemble to perform their multiple and diverse functions, with each system having evolved distinct mechanisms to facilitate pilus dynamics. The V. cholerae minor pilin TcpB represents a simple but elegant multifunctional protein responsible for TCP dynamics and functions.
Materials and Methods
Bacterial strains and growth media
Bacterial strains used in this study are described in Table 1. All strains were maintained at -80°C in lysogeny broth (LB) containing 20% glycerol (v v-1). V. cholerae strains were grown under classical, TCP-expressing conditions of LB (starting pH of 6.5) at 30°C with aeration as previously described [7, 115]. Where appropriate, strains were grown with antibiotics at the following final concentrations: ampicillin (Ap) 100 μg/ml, gentamicin (Gm) 30 μg/ml, kanamycin (Km) 45 μg/ml, and streptomycin (Sm) 100 μg/ml, spectomycin (Sp), tetracyclin (Tet). All DNA manipulations were performed using standard molecular and genetic techniques [116, 117]. Expression of tcpB and tcpB mutants from ptcpB was induced using rhamnose at the indicated concentrations.
Table 1. List of bacterial strains, plasmids and primers.
| Reagent | Description or nucleotide sequence | Source / Reference |
|---|---|---|
| Strains | ||
| V. cholerae O395 | Classical O1, Ogawa, Sm | [7] |
| V. cholerae RT4031 | ΔtcpA, major pilin deletion strain | [35] |
| V. cholerae RT4368 | ΔtcpB, minor pilin deletion strain | [103] |
| V. cholerae RT4369 | ΔtcpC secretin deletion strain | [103] |
| V. cholerae RT4634 | TcpBE5V | [102] |
| V. cholerae TJK189 | Flagella minus O395 ΔflaA | [118] |
| V. cholerae CF12 | ΔflaA, ΔtcpB | This study |
| V. cholerae YG1257 | ΔflaA, TcpBE5V | This study |
| E. coli DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK− mK+), λ– | Life Technologies |
| E. coli SHuffle® T7 Express | MiniF lysY (CmR) / fhuA2 lacZ::T7 gene1 [lon] ompT ahpC gal λatt::pNEB3-r1-cDsbC (SpR, lacIq)ΔtrxB sulA11 R(mcr-73::miniTn10—TetS)2 [dcm] R(zgb-210::Tn10—TetS) endA1 Δgor Δ(mcrC-mrr)114::IS10 | New England Biolabs |
| Plasmids | ||
| pBAD22 | pMB1, Para promoter, araC, ApR | ATCC |
| pJMA10 | pBAD22 derivative; araC replaced with PrhaB. Bla, ApR | [102] |
| pJMA10.1 | pJMA10 with NcoI site removed | This study |
| ptcpB | pJMA10.1 containing the tcpB gene | This study |
| ptcpBE5D | ptcpB Glu5Asp | This study |
| ptcpBE5Q | ptcpB Glu5Gln | This study |
| ptcpBE5V | ptcpB Glu5Val | This study |
| pET15b-tcpA | pET-15b expressing TcpA29-199, ApR | [17] |
| pET15b-tcpB | pET-15b expressing TcpB25-423 | This study |
| pTK20 | pKAS32 ΔflaA; ApR | [118] |
| pTK2 | pKAS32 ΔtcpB; ApR | [118] |
| pTRNS101 | pKAS32 tcpB; ApR | [102] |
| pCHG006 | pKAS32 tcpB:E5V; ApR | [102] |
| pKAS32 | pGP704 rpsL; ApR | [119] |
| Primers | ||
| ABN26 | GATCGGGCGCCTGAGCCTCGGTTGTGTGGTG | This study |
| ABN27 | GATCGCCATGGTGAATTCCTCCTGAATTTCATTACGACCAGTC | This study |
| pJMA10-NcoDEL-F-NheI | TACTATCTTCAAAGCCACATTCGGTCGAAA | This study |
| pJMA10-NcoDel-R-KpnI | CGGGGTACCTTCCTCCTGAATTTCATTACGACCAGTCTAA | This study |
| VC-TcpB-F-KpnI | CGGGGTACCATGAGAAAATACCAACAAGGTGTCG | This study |
| VC-TcpB-R-XbaI | ATATATTCTAGATTAATTTTCACACCATTGAAACGCTATAAAA | This study |
| VC-tcpB(E5D)-F-KpnI | CGGGGTACCATGAGAAAATACCAACAAGGTGTC GGATTATTGGATGCG | This study |
| VC-TcpB(E5Q)F-KpnI | CGGGGTACCATGAGAAAATACCAACAAGGTGTCGGATTATTGCAGGCG | This study |
| VC-TcpB(E5V)F-KpnI | CGGGGTACCATGAGAAAATACCAACAAGGTGTCGGATTATTGGTGGCG | This study |
| TcpB-R-142-XbaI | ATATATTCTAGATTATTGTTTTATTTGTCGTTGAATTTCCG | This study |
| TcpB-R-217-XbaI | ATATATTCTAGATTACGAAAATTTTCTTTTAAAAGCGACGAAA | This study |
| TcpB-R-228-XbaI | ATATATTCTAGATTAAGCAAGACTGACCGCCAACG | This study |
| TcpB25-vc-fpcr-nde1 | GGAATTCCATATGAAGCGGGAAGCTGAACTCATGATTAAA | This study |
| TcpB25-vc-rpcr-bamh1 | CGCGGATCCTTAATTTTCACACCATTGAAACGCT | This study |
Plasmid and strain construction
Plasmids used in this study are listed in Table 1. Expression vector pJMA10 is derived from pBAD22 and constructed as follows. Primers ABN26/27 were used to amplify the rhamnose promoter PRHA from E. coli BL21 chromosomal DNA. The PCR amplified region was then cloned into pBAD22-TOPO (ATCC), replacing the araC promoter region to generate pJMA10. This vector was further modified to remove the endogenous NcoI restriction digest sequence from the multiple cloning site (MCS) to prevent a possible frameshift mutation. Primers NcoDEL-F-NheI and NcoDEL-R-KpnI were used to PCR amplify a 1.7 kb fragment of pJMA10 upstream of the MCS that contains a unique NheI restriction site. The fragment downstream of the MCS was directly digested from the purified plasmid. Both fragments were digested with restriction enzymes NheI and KpnI, and ligated together using T4 DNA ligase (New England Biolabs) to generate the corrected plasmid pJMA10.1. Both pJMA10 and pJMA10.1 were verified by DNA sequencing.
The vector expressing the minor pilin TcpB was derived from expression vector pJMA10.1, which contains an ApR marker. The gene fragment encoding TcpB was PCR amplified with primers TcpB-F-KpnI/-R-XbaI from V. cholerae WT strain O395 genomic DNA. The PCR product was purified, digested with KpnI and XbaI, and ligated into pJMA10.1 at the KpnI/XbaI restriction sites using T4 DNA ligase. The construct ptcpB was verified by DNA sequencing and transformed into V. cholerae strains for complementation testing. Pilin expression was induced using rhamnose at indicated concentrations.
The conserved Glu5 residue in TcpB was changed to Asp, Gln and Val on ptcpB to generate ptcpBE5D, ptcpBE5Q and ptcpBE5V respectively. Forward primers TcpB-E5D/E5Q/E5V-F-KpnI were used individually with reverse primer TcpB-R-XbaI to PCR amplify from ptcpB the tcpB gene fragments encoding the corresponding amino acid substitutions E5D, E5Q and E5V. PCR products were purified and subcloned into pJMA10.1 as previously described. All plasmids were verified by DNA sequencing and transformed into V. cholerae ΔtcpB and cells were grown on LB-Sm/Ap plates.
Generation of the tcpB-E5V chromosomal mutations
The ΔtcpB/flaA and tcpB-E5V/ΔflaA chromosomal mutants were generated by allelic exchange. pTRNS101 encoding tcpB-E5V [102] and pTK2 encoding tcpB with a central deletion [103] were used to introduce the E5V substitution and the ΔtcpB deletion into the V. cholerae ΔflaA strain TJK189 [118] by allelic exchange [119].
Immunoblotting
Fractions of overnight cell cultures were analyzed by SDS-PAGE and immunoblotting to compare TcpA, TcpB and TcpF production in the various V. cholerae strains and conditions. Whole cell culture (WCC) fractions used to assess total protein were obtained by resuspending overnight cultures by vortexing. Sheared supernatant (SS) fractions were obtained by homogenizing the resuspended cells using an IKA ULTRA-TURRAX T8.01 disperser (IKA-Werke) at the maximum setting for 20 seconds [8]. Cell debris was removed by centrifugation at 13,000 x g for 30 minutes at RT using a bench top microfuge, with the supernatant representing the SS fraction. For analysis of secreted TcpF in the culture supernatant (CS), overnight cultures were centrifuged at 3000 x g for 10 minutes at RT to remove the cells and the culture supernatant containing secreted TcpF was filtered through a 0.22 μm syringe-drive filter (Pall) to remove remaining cells. Each fraction was mixed with Laemmli sample buffer (60 mM Tris pH 6.8, 5% 2-mercaptoethanol, 2% SDS, 10% glycerol, 0.02% bromophenol blue) and boiled for 10 minutes prior to loading 25 μl onto 15% SDS polyacrylamide gels. The PageRuler Unstained Protein Ladder (Fermentas) was included for mass markers. Proteins were transferred onto polyvinylidene difluoride (PVDF) membrane (Bio-Rad) at 4°C in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) with a wet transfer apparatus (Bio-Rad). The membrane was blocked for 1 hour at RT with non-fat dried milk (5% w/v) in Tris-buffered saline with 0.1% Tween. Proteins were detected with rabbit polyclonal antisera raised against TcpA peptide 174–199 [120], TcpB peptide 64–78 (Pacific Immunology) and mouse monoclonal antisera for TcpF [10]. Goat-anti-rabbit or goat-anti-mouse secondary antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch) were used to bind the primary antibody. The protein mass markers were detected with Strep-Tactin-HRP conjugate (IBA). Immunoblots were visualized by enhanced chemiluminescence using the SuperSignal West Pico substrate (Thermo Fisher Scientific). Immunoblots were digitized using the FujiFilm LAS 4000 imager.
Expression and purification of ΔN-TcpA and ΔN-TcpB
Expression and purification of ΔN-TcpA displaying an N-terminal hexahistidine tag (His) and linker are described elsewhere [17]. The gene fragment encoding ΔN-TcpB (residues 25–423) was PCR amplified with primers TcpB25-vc-fpcr-nde1 and TcpB25-vc-rpcr-BamH1 from V. cholerae WT strain O395 genomic DNA, followed by digestion with restriction enzymes NdeI and BamHI. The product was ligated into expression vector pET15b (Novagen), which encodes a His-tag and linker with a thrombin cleavage site. The construct was transformed into SHuffle T7 Express lysY Competent E. coli (New England Biolabs) by heat shock. The cells were grown in LB broth supplemented with ampicillin (100 μg/ml) to OD600 of 0.1 in a shaking incubator 37°C, and protein expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG, final concentration to 0.1 mM). The temperature was reduced to 14°C and cells were grown for a further 20 hours and harvested by centrifugation (5000 x g, 30 min, 4°C). The culture supernatant was discarded and the cell pellet was resuspended in lysis buffer (50 mM Na2HPO4/NaH2PO4 pH 7.4, 20 mM Tris HCl pH 7.4, 100 mM NaCl, EDTA-free protease inhibitor cocktail [Roche], and 1 mg/ml lysozyme) and gently stirred at room temperature for an hour. Cells were lysed by sonication and cell debris was removed by centrifugation at 40,000 x g for 1 hour at 4°C.
Purification was carried out at 4°C. The cell lysate was filtered through 0.4 μm polyethersulfone membrane and loaded onto a Ni-NTA column pre-equilibrated with wash buffer (50 mM Na2HPO4/NaH2PO4 pH 7.4, 30 mM imidazole and 100 mM NaCl). After washing the column with 10 column volumes of wash buffer, the N-terminal His tag was cleaved off of the TcpB bound to the Ni-NTA column by thrombin digestion and the protein was eluted with elution buffer (50 mM Na2HPO4/NaH2PO4 pH 7.4, 250 mM imidazole, 100 mM NaCl). The protein was further purified by size exclusion chromatography using a HiPrep 26/60 Sephacryl S-100 HR column (GE Healthcare) in buffer containing 20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.5 mM EDTA, 0.5 mM EGTA and concentrated to 10 mg/ml using a stirred-cell concentrator (Millipore). The purity was estimated to be >95%.
Purification of TCPH181A
V. cholerae strain RT4225 expressing a mutation in tcpA encoding a His181Ala substitution [35], was grown on a LB plate containing ampicillin (LB/Ap). A single colony was used to inoculate 1 ml of LB/Ap, which was incubated at 37°C on a rotary shaker for 30 min. The cell culture was diluted to OD600 = 0.01 and 400 μl was used to inoculate 200 ml of LB, pH 6.5, 0.4 mM IPTG, 100 μg/ml Ap in a 2 L-flask, which was incubated at 30°C for 18 hours shaking at 250 rpm. Four ml of 0.5 M EDTA (pH 8) and 2 ml of 0.1 M histidine-HCl were added to the culture, which was centrifuged at 5000 xg for 15 minutes at 4°C to pellet cells. The supernatant was centrifuged again at 5000 xg for 10 minutes at 4°C to pellet residual cells. After transferring the supernatant to new tubes, solid ammonium sulfate (AmSO4) was added to 10% saturation and the solution was incubated at 4°C for 2 hours on a rocker. The solution was centrifuged at 10,000 xg for 30 minutes at 4°C, and AmSO4 was added to 30% saturation. The solution was incubated at 4°C for one hour on the rocker then centrifuged at 10,000 xg for 30 min at 4°C to pellet the pili. The pellet was resuspended in 200 μl of PBS containing 10 mM EDTA and dialyzed exhaustively using a 3500 kDa molecular weight cut-off membrane against precooled PBS, 10 mM EDTA.
Autoagglutination assay
Cells were grown in 2 mL LB with antibiotics (streptomycin, ampicillin as required) for 2 hours at 37°C with rotation and cell concentrations were normalized to OD600 of 0.01. Normalized cultures were diluted 1/500 in 3 mL LB (pH 6.5) and grown overnight on a Ferris wheel rotator at 30°C with antibiotics and rhamnose as required. Overnight cultures were allowed to settle at room temperature for 15 minutes, after which the autoagglutination phenotype was assessed visually and the OD600 of the culture supernatant was measured. Experiments were performed in triplicate.
Quantification of TcpA and TcpB in TCPH181A
ΔN-TcpA and ΔN-TcpB concentrations were determined by UV absorbance (260 nm) using a Nanodrop spectrophotometer (Thermo Fisher Scientific). Known amounts of each protein (standards) and fixed volumes of purified TCPH181A were separated on a 15% SDS polyacrylamide gel. Proteins were transferred onto PVDF membrane and immunoblotted using anti-TcpA and anti-TcpB antibodies. Protein bands were scanned with a FujiFilm LAS4000 imager and analyzed using ImageJ [121]. Band densities of the ΔN-TcpA and ΔN-TcpB standards were normalized to the densest standard band in that blot to make averaging possible. The averages of relative densities from two replicates were plotted against their protein amounts and the plot was used to interpolate the amount of native TcpA or TcpB present in the purified TCPH181A sample.
Negative staining and immunogold TEM imaging
V. cholerae cells were grown under TCP-expressing conditions. Carbon-coated grids (CF-300, Electron Microscopy Science) were inverted on top of a 20 μL drop of sample on Parafilm and incubated for 10 minutes. Following incubation, grids were transferred to drops of Tris-buffered saline with 0.1% Tween (TBST). Grids were then stained with 3% uranyl acetate and imaged on a Hitachi 8100 STEM operating at 120 kEv.
For immunogold labeled samples, grids were inverted on top of a 25 μl drop of overnight cell culture on Parafilm in a humidified chamber to prevent evaporation and incubated at 30°C for 10 min. Following incubation, grids were transferred to drops of fixative (4% paraformaldehyde/0.2% glutaraldehyde in 0.2 M sodium cacodylate pH 7.4) for 1 hour. The grids were washed with Tris-buffered saline with 0.1% Tween (TBST) and blocked for 1 hour in bovine serine albumin (BSA) in TBST. Samples were incubated with primary rabbit antibody raised against TcpA (peptide 174–199 [120], 1:50 dilution in TBST 1% BSA), washed in TBST, and then incubated for 30 minutes with gold-conjugated anti-rabbit secondary antibody (1:60 dilution in TBST 1% BSA) (Jackson Immunoresearch, Electron Microscopy Sciences). The final wash in TBST was followed by staining with 3% uranyl acetate. Samples were imaged on a Hitachi 8100 STEM at 200 kV with the exception of Fig 2B, which was imaged on a JEOL 100CX TEM at 100 kV.
Micropillars assays
Micropillars assays were performed as previously reported [107] using PoMPs (Polyacrylamide MicroPillars). Briefly, microfabricated silica molds were inverted onto 15 μL of a mixture of 20% polyacrylamide and 0.2% Bis-acrylamide with 1/10 volume of ammonium persulfate and 1/1000 volume of TEMED on activated 25 mm diameter round coverslips (Warner Instruments). Reticulation of the hydrogel and removal of the silica mold yielded a hexagonal array of polymerized micropillars spaced 3 μm center to center with a spring constant (k) of 25 +/- 4 pN/μm. The micropillars hydrogel was activated with sulfo-SANPAH crosslinker (Invitrogen) according to the manufacturer's recommendations, then the micropillars were treated with poly-L-lysine (30 μg/ml in phosphate buffered saline) for one hour at 37°C. After 3 washes in water the micropillars were coated for one hour with a 1/10 (V/V) solution of 20 nm carboxylated beads in water (Molecular Probes). After two washes with water and one with phosphate buffered saline, the coverslips supporting the micropillars were mounted at the bottom of an observation chamber (Attofluor chamber, Invitrogen). Two mls LB (pH 8.5) and 5 μL suspension of V. cholerae overnight culture was added to the chamber and incubated at 30°C for one hour. Ten Hz movies of the pillars tips were recorded using a 60X objective on an inverted microscope with an environmental chamber at 30°C (TiEclipse, Nikon). Positions of the micropillar tips were analyzed in the movies using a custom-designed cross-correlation algorithm [122]. The position of a non-deflected micropillar away from any colony was used as a reference to compensate for any global movement of the chamber. The forces exerted on the micropillars were calculated based of the displacement of the pillars and their calibrated spring constant using a commercial software package Matlab (Mathworks Inc. Natick, MA) and Igor Pro (WaveMetrics, Inc., Lake Oswego, OR).
The V. cholerae suspensions used to perform the micropillar assays were prepared as follows: ΔflaA, ΔtcpB/ΔflaA and tcpB-E5V/ΔflaA strains were streaked onto LB agar plates and grown for 24 hours at 37°C, a single colony was touched with a 2 mm sterilized loop and the bacteria attached to the loop were transferred to 1 ml of LB (pH 6.5). Ten μl of this solution was used to inoculate 3 ml LB (pH 6.5) in a culture tube, which was rotated at 30°C for 12 to 15 hours. For V. cholerae strain ΔtcpB/ΔflaA, 1 μl of anti-lipopolysaccharide antibody was added to induce bacterial aggregation and the suspension was rotated an additional 30 minutes prior to the micropillar assay.
Supporting Information
(A) Top panel: Immunoblot of known amounts of recombinant N-terminally truncated TcpA (ΔN-TcpA) and purified TCPH181A from V. cholerae RT4225 probed with anti-TcpA antibody. Bottom panel: graph of the ΔN-TcpA average band densities, determined by ImageJ analysis [121] from two replicates, plotted against the known protein amounts. From this graph the concentration of TcpA in purified TCPH181A was found to be 12±1 mg/ml (SEM, n = 3) or 0.59 mM. The error bars are standard deviations. (B) Top panel: Immunoblot of known amounts of recombinant N-terminally truncated TcpB (ΔN-TcpB) and purified TCPH181 probed with anti-TcpB antibody. Bottom panel: graph of the ΔN-TcpB average band densities, determined by ImageJ analysis [121] from three replicates, plotted against the known protein amounts. From this graph the TcpB concentration in purified TCPH181A was found to be 4±1 μg/ml (SEM, n = 4) or 86 nM. Thus, the stoichiometric ratio of TcpA:TcpB in TCPH181A is ~ 7000:1. (C) TEM image of purified TCPH181A and their dimensions.
(TIF)
TcpB expression and autoagglutination levels were assayed in the V. cholerae ΔtcpB strain complemented with ptcpB and induced with varying rhamnose concentrations (shown as %, w/v). (A) Immunoblot of V. cholerae whole cell cultures (WCC) probed with anti-TcpB antibodies. The loading control is an unknown ~60 kDa protein present in the WCC fraction and detected by the Strep-Tactin-HRP conjugate. (B) Autoagglutination levels. The more complete the autoagglutination the lower the OD600 value. Values are averaged for 3 experiments; error bars represent standard deviations. Autoagglutination is closest to that of WT V. cholerae O395 with 0.001% rhamnose.
(TIF)
The tcpB-E5V/ΔmotAB double mutant produces flagella and autoagglutinates at levels approaching WT, supporting the idea that the gain of autoagglutination observed for the tcpB-E5V/ΔflaA double mutant, as compared to that of the poorly autoagglutinating tcpB-E5V single mutant, is due to loss of motility rather than loss of flagella.
(TIF)
Differential contrast movie of the micropillars assay of V. cholerae ΔflaA. The average distance between pillars is 3 μm center to center and the full length of the movie is 4 min.
(ZIP)
Differential contrast movie of the micropillars assay of V. cholera ΔflaA. The average distance between pillars is 3 μm center to center and the full length of the movie is 6.5 min.
(ZIP)
Differential contrast movie of the micropillars assay of V. cholerae ΔtcpB/ΔflaA. The average distance between pillars is 3 μm center to center and the full length of the movie is 4 min.
(ZIP)
Differential contrast movie of the micropillars assay of V. cholerae tcpB-E5V/ΔflaA. The average distance between pillars is 3 μm center to center and the full length of the movie is 6.5 min.
(ZIP)
Acknowledgments
TEM images were collected at the Dartmouth College Electron Microscope Facility and the SFU Nanoimaging Facility, part of the 4D LABS shared facilities supported by the Canada Foundation for Innovation (CFI), British Columbia Knowledge Development Fund, Western Economic Diversification Canada and Simon Fraser University. We thank L. Howard for assistance with TEM, Kurt Piepenbrink (U. Maryland) for suggesting the method to determine TcpA:TcpB ratios, and Ed Egelman (U. Virginia) for providing feedback on the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding for this research was provided by grant MOP-125959 from the Canadian Institutes of Health Research (http://www.cihr-irsc.gc.ca/e/193.html) and grants AI116566, AI025096, AI007363 and AI007519 from the National Institutes of Health (https://www.nih.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kaper JB, Morris JG Jr., Levine MM. Cholera. Clin Microbiol Rev. 1995;8(1):48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gangarosa EJ, Mosley WH. The epidemiologic basis of cholera control. Bulletin of the Pan American Health Organization. 1974;8(3):189–97. Epub 1974/01/01. [PubMed] [Google Scholar]
- 3.Davis BM, Lawson EH, Sandkvist M, Ali A, Sozhamannan S, Waldor MK. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXphi. Science. 2000;288(5464):333–5. [DOI] [PubMed] [Google Scholar]
- 4.Korotkov KV, Sandkvist M, Hol WG. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10(5):336–51. Epub 2012/04/03. 10.1038/nrmicro2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez J, Holmgren J. Cholera toxin structure, gene regulation and pathophysiological and immunological aspects. Cell Mol Life Sci. 2008;65(9):1347–60. Epub 2008/02/19. 10.1007/s00018-008-7496-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168(4):1487–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84(9):2833–7. Epub 1987/05/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim MS, Ng D, Zong Z, Arvai AS, Taylor RK, Tainer JA, et al. Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation. Mol Microbiol. 2010;77(3):755–70. 10.1111/j.1365-2958.2010.07244.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirn TJ, Taylor RK. TcpF is a soluble colonization factor and protective antigen secreted by El Tor and classical O1 and O139 Vibrio cholerae serogroups. Infect Immun. 2005;73(8):4461–70. 10.1128/IAI.73.8.4461-4470.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Megli CJ, Yuen AS, Kolappan S, Richardson MR, Dharmasena MN, Krebs SJ, et al. Crystal structure of the Vibrio cholerae colonization factor TcpF and identification of a functional immunogenic site. J Mol Biol. 2011;409(2):146–58. Epub 2011/03/29. 10.1016/j.jmb.2011.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272(5270):1910–4. [DOI] [PubMed] [Google Scholar]
- 12.Melville S, Craig L. Type IV pili in Gram-positive bacteria. Microbiol Mol Biol Rev. 2013;77(3):323–41. 10.1128/MMBR.00063-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry JL, Pelicic V. Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS microbiology reviews. 2015;39(1):134–54. Epub 2015/03/21. 10.1093/femsre/fuu001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craig L, Li J. Type IV pili: paradoxes in form and function. Curr Opin Struct Biol. 2008;18(2):267–77. Epub 2008/02/06. 10.1016/j.sbi.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albers SV, Jarrell KF. The archaellum: how Archaea swim. Frontiers Microbiology. 2015;6:23. Epub 2015/02/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev. 2012;76(4):740–72. Epub 2012/12/04. 10.1128/MMBR.00035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11(5):1139–50. [DOI] [PubMed] [Google Scholar]
- 18.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, et al. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006;23(5):651–62. Epub 2006/09/05. 10.1016/j.molcel.2006.07.004 [DOI] [PubMed] [Google Scholar]
- 19.Hartung S, Arvai AS, Wood T, Kolappan S, Shin DS, Craig L, et al. Ultra-high resolution and full-length pilin structures with insights for filament assembly, pathogenic functions and vaccine potential. J Biol Chem. 2011;286(51):44254–65. 10.1074/jbc.M111.297242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tainer JA. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature. 1995;378(6552):32–8. 10.1038/378032a0 [DOI] [PubMed] [Google Scholar]
- 21.Kolappan S, Coureuil M, Xiong Y, Nassif X, Egelman E, Craig L. Structure of the Neisseria meningitidis Type IV pilus. Nat Commun. 2016;7:13015 10.1038/ncomms13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Egelman EH, Craig L. Structure of the Vibrio cholerae Type IVb pilus and stability comparison with the Neisseria gonorrhoeae Type IVa pilus. J Mol Biol. 2012;418(1–2):47–64. Epub 2012/03/01. 10.1016/j.jmb.2012.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2(5):363–78. 10.1038/nrmicro885 [DOI] [PubMed] [Google Scholar]
- 24.Pelicic V. Type IV pili: e pluribus unum? Mol Microbiol. 2008;68(4):827–37. Epub 2008/04/11. 10.1111/j.1365-2958.2008.06197.x [DOI] [PubMed] [Google Scholar]
- 25.Chang YW, Rettberg LA, Treuner-Lange A, Iwasa J, Sogaard-Andersen L, Jensen GJ. Architecture of the type IVa pilus machine. Science. 2016;351(6278):aad2001 Epub 2016/03/12. 10.1126/science.aad2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gold VA, Salzer R, Averhoff B, Kuhlbrandt W. Structure of a type IV pilus machinery in the open and closed state. eLife. 2015;4. Epub 2015/05/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufman MR, Shaw CE, Jones ID, Taylor RK. Biogenesis and regulation of the Vibrio cholerae toxin-coregulated pilus: analogies to other virulence factor secretory systems. Gene. 1993;126(1):43–9. [DOI] [PubMed] [Google Scholar]
- 28.Nunn DN, Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA. 1991;88(8):3281–5. Epub 1991/04/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strom MS, Lory S. Structure-function and biogenesis of the type IV pili. Ann Rev Microbiol. 1993;47:565–96. [DOI] [PubMed] [Google Scholar]
- 30.Morand PC, Bille E, Morelle S, Eugene E, Beretti JL, Wolfgang M, et al. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 2004;23(9):2009–17. Epub 2004/04/23. 10.1038/sj.emboj.7600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anantha RP, Stone KD, Donnenberg MS. Effects of bfp mutations on biogenesis of functional enteropathogenic Escherichia coli type IV pili. J Bacteriol. 2000;182(9):2498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tripathi SA, Taylor RK. Membrane association and multimerization of TcpT, the cognate ATPase ortholog of the Vibrio cholerae toxin-coregulated-pilus biogenesis apparatus. J Bacteriol. 2007;189(12):4401–9. 10.1128/JB.00008-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32(1):1–10. [DOI] [PubMed] [Google Scholar]
- 34.Wolfgang M, Park HS, Hayes SF, van Putten JP, Koomey M. Suppression of an absolute defect in Type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1998;95(25):14973–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol. 2000;35(4):896–910. [DOI] [PubMed] [Google Scholar]
- 36.Pasloske BL, Paranchych W. The expression of mutant pilins in Pseudomonas aeruginosa: fifth position glutamate affects pilin methylation. Mol Microbiol. 1988;2(4):489–95. [DOI] [PubMed] [Google Scholar]
- 37.Strom MS, Lory S. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J Biol Chem. 1991;266(3):1656–64. [PubMed] [Google Scholar]
- 38.Berry JL, Phelan MM, Collins RF, Adomavicius T, Tonjum T, Frye SA, et al. Structure and assembly of a trans-periplasmic channel for type IV pili in Neisseria meningitidis. PLoS Pathog. 2012;8(9):e1002923 Epub 2012/10/03. 10.1371/journal.ppat.1002923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karuppiah V, Derrick JP. Structure of the PilM-PilN inner membrane Type IV pilus biogenesis complex from Thermus thermophilus. J Biol Chem. 2011;286(27):24434–42. Epub 2011/05/21. 10.1074/jbc.M111.243535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leighton TL, Buensuceso RN, Howell PL, Burrows LL. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ Microbiol. 2015;17(11):4148–63. Epub 2015/03/27. 10.1111/1462-2920.12849 [DOI] [PubMed] [Google Scholar]
- 41.Leighton TL, Dayalani N, Sampaleanu LM, Howell PL, Burrows LL. Novel role for PilNO in Type IV pilus retraction revealed by alignment subcomplex mutations. J Bacteriol. 2015;197(13):2229–38. Epub 2015/04/29. 10.1128/JB.00220-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Misic AM, Satyshur KA, Forest KT. P. aeruginosa PilT structures with and without nucleotide reveal a dynamic Type IV pilus retraction motor. J Mol Biol. 2010;400(5):1011–21. Epub 2010/07/03. 10.1016/j.jmb.2010.05.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitchurch CB, Hobbs M, Livingston SP, Krishnapillai V, Mattick JS. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101(1):33–44. [DOI] [PubMed] [Google Scholar]
- 44.Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29(1):321–30. [DOI] [PubMed] [Google Scholar]
- 45.Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 2007;26(3):878–90. 10.1038/sj.emboj.7601544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anantha RP, Stone KD, Donnenberg MS. Role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells. Infect Immun. 1998;66(1):122–31. Epub 1998/01/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aas FE, Wolfgang M, Frye S, Dunham S, Lovold C, Koomey M. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol Microbiol. 2002;46(3):749–60. [DOI] [PubMed] [Google Scholar]
- 48.Burrows LL. Pseudomonas aeruginosa twitching motility: Type IV pili in action. Ann Rev Microbiol. 2012;66:493–520. Epub 2012/07/04. [DOI] [PubMed] [Google Scholar]
- 49.Chiang P, Habash M, Burrows LL. Disparate subcellular localization patterns of Pseudomonas aeruginosa Type IV pilus ATPases involved in twitching motility. J Bacteriol. 2005;187(3):829–39. Epub 2005/01/22. 10.1128/JB.187.3.829-839.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser D. Bacterial motility: how do pili pull? Curr Biol. 2000;10(21):R777–80. [DOI] [PubMed] [Google Scholar]
- 51.Mattick JS. Type IV pili and twitching motility. Ann Rev Microbiol. 2002;56:289–314. [DOI] [PubMed] [Google Scholar]
- 52.Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407(6800):98–102. 10.1038/35024105 [DOI] [PubMed] [Google Scholar]
- 53.Merz AJ, Forest KT. Bacterial surface motility: slime trails, grappling hooks and nozzles. Curr Biol. 2002;12(8):R297–303. [DOI] [PubMed] [Google Scholar]
- 54.Biais N, Ladoux B, Higashi D, So M, Sheetz M. Cooperative retraction of bundled Type IV pili enables nanonewton force generation. PLoS Biol. 2008;6(4):e87 Epub 2008/04/18. 10.1371/journal.pbio.0060087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Biais N, Higashi DL, Brujic J, So M, Sheetz MP. Force-dependent polymorphism in type IV pili reveals hidden epitopes. Proc Natl Acad Sci USA. 2010;107(25):11358–63. Epub 2010/06/11. 10.1073/pnas.0911328107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clausen M, Jakovljevic V, Sogaard-Andersen L, Maier B. High-force generation is a conserved property of Type IV pilus systems. J Bacteriol. 2009;191(14):4633–8. Epub 2009/05/12. 10.1128/JB.00396-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gurung I, Spielman I, Davies MR, Lala R, Gaustad P, Biais N, et al. Functional analysis of an unusual type IV pilus in the Gram-positive Streptococcus sanguinis. Mol Microbiol. 2015. Epub 2015/10/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maier B, Potter L, So M, Seifert HS, Sheetz MP. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci USA. 2002;99(25):16012–7. 10.1073/pnas.242523299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maier B, Wong GC. How bacteria use Type IV pili machinery on surfaces. Trends Microbiol. 2015;23(12):775–88. Epub 2015/10/27. 10.1016/j.tim.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 60.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA. 2001;98(12):6901–4. 10.1073/pnas.121171698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaburdaev V, Biais N, Schmiedeberg M, Eriksson J, Jonsson AB, Sheetz MP, et al. Uncovering the mechanism of trapping and cell orientation during Neisseria gonorrhoeae twitching motility. Biophys J. 2014;107(7):1523–31. Epub 2014/10/09. 10.1016/j.bpj.2014.07.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nivaskumar M, Francetic O. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta. 2014;1843(8):1568–77. Epub 2014/01/07. 10.1016/j.bbamcr.2013.12.020 [DOI] [PubMed] [Google Scholar]
- 63.Pugsley AP, Francetic O, Possot OM, Sauvonnet N, Hardie KR. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in Gram-negative bacteria—a review. Gene. 1997;192(1):13–9. Epub 1997/06/11. [DOI] [PubMed] [Google Scholar]
- 64.Sandkvist M. Type II secretion and pathogenesis. Infect Immun. 2001;69(6):3523–35. 10.1128/IAI.69.6.3523-3535.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Douzi B, Ball G, Cambillau C, Tegoni M, Voulhoux R. Deciphering the Xcp Pseudomonas aeruginosa type II secretion machinery through multiple interactions with substrates. J Biol Chem. 2011;286(47):40792–801. Epub 2011/09/29. 10.1074/jbc.M111.294843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nunn D. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 1999;9(10):402–8. [DOI] [PubMed] [Google Scholar]
- 67.Shevchik VE, Robert-Baudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 1997;16(11):3007–16. Epub 1997/06/02. 10.1093/emboj/16.11.3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vignon G, Kohler R, Larquet E, Giroux S, Prevost MC, Roux P, et al. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J Bacteriol. 2003;185(11):3416–28. 10.1128/JB.185.11.3416-3428.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Durand E, Bernadac A, Ball G, Lazdunski A, Sturgis JN, Filloux A. Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J Bacteriol. 2003;185(9):2749–58. 10.1128/JB.185.9.2749-2758.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sauvonnet N, Vignon G, Pugsley AP, Gounon P. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 2000;19(10):2221–8. 10.1093/emboj/19.10.2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McLaughlin LS, Haft RJ, Forest KT. Structural insights into the Type II secretion nanomachine. Curr Opin Struct Biol. 2012;22(2):208–16. Epub 2012/03/20. 10.1016/j.sbi.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alm RA, Mattick JS. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol Microbiol. 1995;16(3):485–96. Epub 1995/05/01. [DOI] [PubMed] [Google Scholar]
- 73.Alm RA, Mattick JS. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178(13):3809–17. Epub 1996/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martinez de la Pena CF, De Masi L, Nisa S, Mulvey G, Tong J, Donnenberg MS, et al. BfpI, BfpJ, and BfpK minor pilins are important for the function and biogenesis of bundle-forming pili expressed by enteropathogenic Escherichia coli. J Bacteriol. 2015. Epub 2015/12/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Giltner CL, Habash M, Burrows LL. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J Mol Biol. 2010;398(3):444–61. Epub 2010/03/27. 10.1016/j.jmb.2010.03.028 [DOI] [PubMed] [Google Scholar]
- 76.Winther-Larsen HC, Wolfgang M, Dunham S, van Putten JP, Dorward D, Lovold C, et al. A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol Microbiol. 2005;56(4):903–17. Epub 2005/04/28. 10.1111/j.1365-2958.2005.04591.x [DOI] [PubMed] [Google Scholar]
- 77.Nguyen Y, Sugiman-Marangos S, Harvey H, Bell SD, Charlton CL, Junop MS, et al. Pseudomonas aeruginosa Minor pilins prime Type IVa pilus assembly and promote surface display of the PilY1 adhesin. J Biol Chem. 2014. Epub 2014/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carbonnelle E, Helaine S, Nassif X, Pelicic V. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol. 2006;61(6):1510–22. 10.1111/j.1365-2958.2006.05341.x [DOI] [PubMed] [Google Scholar]
- 79.Winther-Larsen HC, Hegge FT, Wolfgang M, Hayes SF, van Putten JP, Koomey M. Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc Natl Acad Sci USA. 2001;98(26):15276–81. 10.1073/pnas.261574998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, et al. Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci USA. 2013;110(8):3065–70. Epub 2013/02/07. 10.1073/pnas.1218832110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Helaine S, Carbonnelle E, Prouvensier L, Beretti JL, Nassif X, Pelicic V. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol. 2005;55(1):65–77. 10.1111/j.1365-2958.2004.04372.x [DOI] [PubMed] [Google Scholar]
- 82.Helaine S, Dyer DH, Nassif X, Pelicic V, Forest KT. 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of Type IV pili. Proc Natl Acad Sci USA. 2007;104:15888–93. 10.1073/pnas.0707581104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bernard SC, Simpson N, Join-Lambert O, Federici C, Laran-Chich MP, Maissa N, et al. Pathogenic Neisseria meningitidis utilizes CD147 for vascular colonization. Nat Med. 2014;20(7):725–31. Epub 2014/06/02. 10.1038/nm.3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brissac T, Mikaty G, Dumenil G, Coureuil M, Nassif X. The meningococcal minor pilin PilX is responsible for type IV pilus conformational changes associated with signaling to endothelial cells. Infect Immun. 2012;80(9):3297–306. Epub 2012/07/11. 10.1128/IAI.00369-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller F, Phan G, Brissac T, Bouchiat C, Lioux G, Nassif X, et al. The hypervariable region of meningococcal major pilin PilE controls the host cell response via antigenic variation. mBio. 2014;5(1):e01024–13. Epub 2014/02/13. 10.1128/mBio.01024-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Imhaus AF, Dumenil G. The number of Neisseria meningitidis Type IV pili determines host cell interaction. EMBO J. 2014;33(16):1767–83. Epub 2014/05/28. 10.15252/embj.201488031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karuppiah V, Thistlethwaite A, Derrick JP. Structures of type IV pilins from Thermus thermophilus demonstrate similarities with type II secretion system pseudopilins. J Struct Biol. 2016. Epub 2016/09/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen Y, Harvey H, Sugiman-Marangos S, Bell SD, Buensuceso RN, Junop MS, et al. Structural and functional studies of the Pseudomonas aeruginosa minor pilin, PilE. J Biol Chem. 2015;290(44):26856–65. Epub 2015/09/12. 10.1074/jbc.M115.683334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piepenbrink KH, Maldarelli GA, Martinez de la Pena CF, Dingle TC, Mulvey GL, Lee A, et al. Structural and evolutionary analyses show unique stabilization strategies in the type IV pili of Clostridium difficile. Structure. 2015;23(2):385–96. Epub 2015/01/21. 10.1016/j.str.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Douzi B, Durand E, Bernard C, Alphonse S, Cambillau C, Filloux A, et al. The XcpV/GspI pseudopilin has a central role in the assembly of a quaternary complex within the T2SS pseudopilus. J Biol Chem. 2009;284(50):34580–9. Epub 2009/10/16. 10.1074/jbc.M109.042366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuo WW, Kuo HW, Cheng CC, Lai HL, Chen LY. Roles of the minor pseudopilins, XpsH, XpsI and XpsJ, in the formation of XpsG-containing pseudopilus in Xanthomonas campestris pv. campestris. J Biomed Sci. 2005;12(4):587–99. Epub 2005/08/04. 10.1007/s11373-005-7372-3 [DOI] [PubMed] [Google Scholar]
- 92.Cisneros DA, Bond PJ, Pugsley AP, Campos M, Francetic O. Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J. 2012;31(4):1041–53. Epub 2011/12/14. 10.1038/emboj.2011.454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cisneros DA, Pehau-Arnaudet G, Francetic O. Heterologous assembly of type IV pili by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol Microbiol. 2012;86(4):805–18. Epub 2012/09/26. 10.1111/mmi.12033 [DOI] [PubMed] [Google Scholar]
- 94.Nivaskumar M, Santos-Moreno J, Malosse C, Nadeau N, Chamot-Rooke J, Tran Van Nhieu G, et al. Pseudopilin residue E5 is essential for recruitment by the type 2 secretion system assembly platform. Mol Microbiol. 2016. Epub 2016/06/05. [DOI] [PubMed] [Google Scholar]
- 95.Korotkov KV, Hol WG. Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat Struct Mol Biol. 2008;15(5):462–8. 10.1038/nsmb.1426 [DOI] [PubMed] [Google Scholar]
- 96.Bleves S, Voulhoux R, Michel G, Lazdunski A, Tommassen J, Filloux A. The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family). Mol Microbiol. 1998;27(1):31–40. Epub 1998/02/18. [DOI] [PubMed] [Google Scholar]
- 97.Durand E, Michel G, Voulhoux R, Kurner J, Bernadac A, Filloux A. XcpX controls biogenesis of the Pseudomonas aeruginosa XcpT-containing pseudopilus. J Biol Chem. 2005;280(36):31378–89. Epub 2005/07/14. 10.1074/jbc.M505812200 [DOI] [PubMed] [Google Scholar]
- 98.Kolappan S, Ng D, Yang G, Harn T, Craig L. Crystal structure of the minor pilin CofB, the Initiator of CFA/III pilus assembly in enterotoxigenic Escherichia coli. J Biol Chem. 2015;290(43):25805–18. Epub 2015/09/02. 10.1074/jbc.M115.676106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kawahara K, Oki H, Fukakusa S, Yoshida T, Imai T, Maruno T, et al. Homo-trimeric structure of the type IVb minor pilin CofB suggests mechanism of CFA/III pilus assembly in human enterotoxigenic Escherichia coli. J Mol Biol. 2016. Epub 2016/02/16. [DOI] [PubMed] [Google Scholar]
- 100.Brown RC, Taylor RK. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16(3):425–39. Epub 1995/05/01. [DOI] [PubMed] [Google Scholar]
- 101.Fullner KJ, Mekalanos JJ. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect Immun. 1999;67(3):1393–404. Epub 1999/02/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gao Y, Hauke CA, Marles JM, Taylor RK. Effects of tcpB mutations on biogenesis and function of TCP, the Type IVb pilus of Vibrio cholerae. J Bacteriol. 2016. Epub 2016/08/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kirn TJ, Bose N, Taylor RK. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol Microbiol. 2003;49(1):81–92. [DOI] [PubMed] [Google Scholar]
- 104.Nguyen Y, Jackson SG, Aidoo F, Junop M, Burrows LL. Structural characterization of novel Pseudomonas aeruginosa Type IV pilins. J Mol Biol. 2010;395(3):491–503. Epub 2009/11/10. 10.1016/j.jmb.2009.10.070 [DOI] [PubMed] [Google Scholar]
- 105.Li J, Lim MS, Li S, Brock M, Pique ME, Woods VL Jr., et al. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure. 2008;16(1):137–48. 10.1016/j.str.2007.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ford CG, Kolappan S, Phan HT, Waldor MK, Winther-Larsen HC, Craig L. Crystal structures of a CTXphi pIII domain unbound and in complex with a Vibrio cholerae TolA domain reveal novel interaction interfaces. J Biol Chem. 2012;287(43):36258–72. Epub 2012/09/04. 10.1074/jbc.M112.403386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Biais N, Higashi D, So M, Ladoux B. Techniques to measure pilus retraction forces. Methods Mol Biol. 2012;799:197–216. Epub 2011/10/14. 10.1007/978-1-61779-346-2_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saldana-Ahuactzi Z, Rodea GE, Cruz-Cordova A, Rodriguez-Ramirez V, Espinosa-Mazariego K, Gonzalez-Montalvo MA, et al. Effects of lng mutations on LngA expression, processing, and CS21 assembly in enterotoxigenic Escherichia coli E9034A. Front Microbiol. 2016;7:1201 Epub 2016/08/19. 10.3389/fmicb.2016.01201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yuen AS, Kolappan S, Ng D, Craig L. Structure and secretion of CofJ, a putative colonization factor of enterotoxigenic Escherichia coli. Mol Microbiol. 2013;90(4):898–918. Epub 2013/10/11. 10.1111/mmi.12407 [DOI] [PubMed] [Google Scholar]
- 110.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539 Epub 2011/10/13. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Higashi DL, Biais N, Weyand NJ, Agellon A, Sisko JL, Brown LM, et al. N. elongata produces type IV pili that mediate interspecies gene transfer with N. gonorrhoeae. PloS one. 2011;6(6):e21373 Epub 2011/07/07. 10.1371/journal.pone.0021373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Krebs SJ, Kirn TJ, Taylor RK. Genetic mapping of secretion and functional determinants of the Vibrio cholerae TcpF colonization factor. J Bacteriol. 2009;191(11):3665–76. Epub 2009/03/24. 10.1128/JB.01724-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Giron JA, Levine MM, Kaper JB. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol Microbiol. 1994;12(1):71–82. [DOI] [PubMed] [Google Scholar]
- 114.Taniguchi T, Akeda Y, Haba A, Yasuda Y, Yamamoto K, Honda T, et al. Gene cluster for assembly of pilus colonization factor antigen III of enterotoxigenic Escherichia coli. Infect Immun. 2001;69(9):5864–73. Epub 2001/08/14. 10.1128/IAI.69.9.5864-5873.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gardel CL, Mekalanos JJ. Regulation of cholera toxin by temperature, pH, and osmolarity. Methods Enzymol. 1994;235:517–26. Epub 1994/01/01. [DOI] [PubMed] [Google Scholar]
- 116.Burnett LC, Lunn G, Coico R. Biosafety: guidelines for working with pathogenic and infectious microorganisms. Curr Prot Microbiol. Biosafety: Guidelines for working with pathogenic and infectious microorganisms: John Wiley and Sons Inc.; 2009. [Google Scholar]
- 117.Maniatis T, Fritsch E, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 118.Martinez RM, Jude BA, Kirn TJ, Skorupski K, Taylor RK. Role of FlgT in anchoring the flagellum of Vibrio cholerae. J Bacteriol. 2010;192(8):2085–92. Epub 2010/02/16. 10.1128/JB.01562-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169(1):47–52. [DOI] [PubMed] [Google Scholar]
- 120.Sun D, Seyer JM, Kovari I, Sumrada RA, Taylor RK. Localization of protective epitopes within the pilin subunit of the Vibrio cholerae toxin-coregulated pilus. Infect Immun. 1991;59:114–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Meth. 2012;9(7):671–5. Epub 2012/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gelles J, Schnapp BJ, Sheetz MP. Tracking kinesin-driven movements with nanometre-scale precision. Nature. 1988;331(6155):450–3. Epub 1988/02/04. 10.1038/331450a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Top panel: Immunoblot of known amounts of recombinant N-terminally truncated TcpA (ΔN-TcpA) and purified TCPH181A from V. cholerae RT4225 probed with anti-TcpA antibody. Bottom panel: graph of the ΔN-TcpA average band densities, determined by ImageJ analysis [121] from two replicates, plotted against the known protein amounts. From this graph the concentration of TcpA in purified TCPH181A was found to be 12±1 mg/ml (SEM, n = 3) or 0.59 mM. The error bars are standard deviations. (B) Top panel: Immunoblot of known amounts of recombinant N-terminally truncated TcpB (ΔN-TcpB) and purified TCPH181 probed with anti-TcpB antibody. Bottom panel: graph of the ΔN-TcpB average band densities, determined by ImageJ analysis [121] from three replicates, plotted against the known protein amounts. From this graph the TcpB concentration in purified TCPH181A was found to be 4±1 μg/ml (SEM, n = 4) or 86 nM. Thus, the stoichiometric ratio of TcpA:TcpB in TCPH181A is ~ 7000:1. (C) TEM image of purified TCPH181A and their dimensions.
(TIF)
TcpB expression and autoagglutination levels were assayed in the V. cholerae ΔtcpB strain complemented with ptcpB and induced with varying rhamnose concentrations (shown as %, w/v). (A) Immunoblot of V. cholerae whole cell cultures (WCC) probed with anti-TcpB antibodies. The loading control is an unknown ~60 kDa protein present in the WCC fraction and detected by the Strep-Tactin-HRP conjugate. (B) Autoagglutination levels. The more complete the autoagglutination the lower the OD600 value. Values are averaged for 3 experiments; error bars represent standard deviations. Autoagglutination is closest to that of WT V. cholerae O395 with 0.001% rhamnose.
(TIF)
The tcpB-E5V/ΔmotAB double mutant produces flagella and autoagglutinates at levels approaching WT, supporting the idea that the gain of autoagglutination observed for the tcpB-E5V/ΔflaA double mutant, as compared to that of the poorly autoagglutinating tcpB-E5V single mutant, is due to loss of motility rather than loss of flagella.
(TIF)
Differential contrast movie of the micropillars assay of V. cholerae ΔflaA. The average distance between pillars is 3 μm center to center and the full length of the movie is 4 min.
(ZIP)
Differential contrast movie of the micropillars assay of V. cholera ΔflaA. The average distance between pillars is 3 μm center to center and the full length of the movie is 6.5 min.
(ZIP)
Differential contrast movie of the micropillars assay of V. cholerae ΔtcpB/ΔflaA. The average distance between pillars is 3 μm center to center and the full length of the movie is 4 min.
(ZIP)
Differential contrast movie of the micropillars assay of V. cholerae tcpB-E5V/ΔflaA. The average distance between pillars is 3 μm center to center and the full length of the movie is 6.5 min.
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.