Abstract
Phosphorylation of phosphoenolpyruvate carboxylase (PEPc; EC 4.1.1.31) plays an important role in the control of central metabolism of higher plants. This phosphorylation is controlled largely at the level of expression of PEPc kinase (PPCK) genes. We have analyzed the expression of both PPCK genes and the PEPC genes that encode PEPc in soybean (Glycine max). Soybean contains at least four PPCK genes. We report the genomic and cDNA sequences of these genes and demonstrate the function of the gene products by in vitro expression and enzyme assays. For two of these genes, GmPPCK2 and GmPPCK3, transcript abundance is highest in nodules and is markedly influenced by supply of photosynthate from the shoots. One gene, GmPPCK4, is under robust circadian control in leaves but not in roots. Its transcript abundance peaks in the latter stages of subjective day, and its promoter contains a sequence very similar to the evening element found in Arabidopsis genes expressed at this time. We report the expression patterns of five PEPC genes, including one encoding a bacterial-type PEPc lacking the phosphorylation site of the plant-type PEPcs. The PEPc expression patterns do not match those of any of the PPCK genes, arguing against the existence of specific PEPc-PPCK expression partners. The PEPC and PPCK gene families in soybean are significantly more complex than previously understood.
Phosphoenolpyruvate carboxylase (PEPc; EC 4.1.1.31) catalyzes the carboxylation of phosphoenolpyruvate to form oxaloacetate and inorganic phosphate. It is a cytosolic enzyme that plays a wide range of roles in different tissues of higher plants (Andreo et al., 1987; Chollet et al., 1996; Vidal and Chollet, 1997). A photosynthetic isoform of PEPc catalyzes the primary fixation of CO2 in C4 and Crassulacean acid metabolism (CAM) plants. Other isoforms are thought to be responsible for different functions, including the replenishment of tricarboxylic acid cycle intermediates, pH control, and the provision of malate in guard cells, developing fruit and legume root nodules.
PEPc plays a central role in the metabolism that allows fixation of atmospheric N2 by bacteroids in legume root nodules. It provides the C4 dicarboxylates that are required by the bacteroids for energy generation and also the carbon skeletons that are needed for the subsequent assimilation of 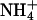 into amino acids (Vance et al., 1994). Like the enzyme from other plant tissues, nodule PEPc is an allosteric enzyme, inhibited by l-malate and activated by Glc-6-P (Schuller et al., 1990; Chollet et al., 1996). Superimposed on this, again like other plant PEPcs (Chollet et al., 1996; Nimmo, 2000), it is controlled by reversible phosphorylation of a single, highly conserved Ser residue close to the N terminus of the protein (Schuller and Werner, 1993; Zhang et al., 1995; Wadham et al., 1996; Zhang and Chollet, 1997). PEPc is more phosphorylated and less sensitive to inhibition by malate in nodules from illuminated plants than in nodules from stem-girdled or decapitated plants or plants placed in prolonged darkness (Zhang et al., 1995; Wadham et al., 1996). The effects of these treatments on the malate sensitivity of PEPc correlated with their effects on PEPc kinase (PPCK) activity. This led to the conclusion that soybean nodule PPCK is up-regulated reversibly by photosynthate and controls the phosphorylation state and activity of PEPc (Zhang and Chollet, 1997).
into amino acids (Vance et al., 1994). Like the enzyme from other plant tissues, nodule PEPc is an allosteric enzyme, inhibited by l-malate and activated by Glc-6-P (Schuller et al., 1990; Chollet et al., 1996). Superimposed on this, again like other plant PEPcs (Chollet et al., 1996; Nimmo, 2000), it is controlled by reversible phosphorylation of a single, highly conserved Ser residue close to the N terminus of the protein (Schuller and Werner, 1993; Zhang et al., 1995; Wadham et al., 1996; Zhang and Chollet, 1997). PEPc is more phosphorylated and less sensitive to inhibition by malate in nodules from illuminated plants than in nodules from stem-girdled or decapitated plants or plants placed in prolonged darkness (Zhang et al., 1995; Wadham et al., 1996). The effects of these treatments on the malate sensitivity of PEPc correlated with their effects on PEPc kinase (PPCK) activity. This led to the conclusion that soybean nodule PPCK is up-regulated reversibly by photosynthate and controls the phosphorylation state and activity of PEPc (Zhang and Chollet, 1997).
PPCK genes have been cloned recently from several higher plants (Hartwell et al., 1999a; Taybi et al., 2000; Tsuchida et al., 2001; Fontaine et al., 2002; Marsh et al., 2003). It is now clear that PPCK comprises a minimal Ser/Thr kinase domain, closely related to the catalytic domain of plant calcium-dependent protein kinases but lacking the N- and C-terminal extensions of these enzymes, and that PPCK activity is largely controlled at the level of PPCK gene expression. In C4 plants, the transcript abundance of the photosynthetic PPCK isoform is regulated via light, whereas in CAM plants, PPCK is under circadian control (Hartwell et al., 1999a, 1999b; Taybi et al., 2000; Tsuchida et al., 2001). In C3 plants, light and nitrogen modulate PPCK expression (Duff and Chollet, 1995; Li et al., 1996). Arabidopsis contains two PPCK genes that differ in their tissue expression patterns. This has led to the proposal that plants contain a small PPCK gene family, members of which may play specific metabolic roles (Nimmo, 2003).
In soybean, one PEPc gene (GmPEPC7) is highly and relatively specifically expressed in nodules. We therefore asked whether PPCK expression shows a similar pattern by testing the hypothesis that one PPCK gene is nodule specific and controlled by photosynthate. The recent work of Xu et al. (2003) suggests that this is the case. However, our data present a more complex picture: soybean contains at least four PPCK genes, of which two show nodule-enhanced expression that may be regulated by the availability of photosynthate. Additionally, the transcript abundance of one of the PPCK genes appears to be under circadian control in certain organs. To our knowledge, this is the first report of a C3 PPCK gene that is controlled in this manner.
RESULTS
Identification of PPCK Genes in Soybean
Potential PPCK genes were identified by searching soybean (Glycine max) expressed sequence tag (EST) databases with known plant PPCK amino acid sequences. Putative PPCK clones (GenBank accession nos. AI736847, AW0990717, and AW756453) were sequenced completely and found to have different 3′ untranslated regions (UTRs). The corresponding genes are termed GmPPCK1, GmPPCK2, and GmPPCK3. Gene-specific primers were designed, and 5′ RACE was used to obtain full-length sequences of GmPPCK1, GmPPCK2, and GmPPCK3. GmPPCK4 was identified from EST CA783260.
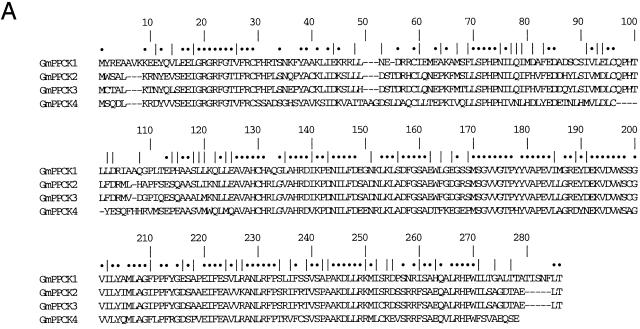
Adapter ligation PCR was used to obtain 5′ and 3′ sequences from genomic DNA. Full-length PCR products for all of the genes were obtained from genomic DNA and cDNA, and the sequences have been deposited in GenBank under accession numbers AY144180 to AY144185, AY568713, and AY568714. Figure 1A shows the alignment of the deduced amino acid sequences. These sequences are typical of plant PPCK genes. The encoded proteins comprise a Ser/Thr kinase catalytic domain similar to the catalytic region of plant calcium-dependent protein kinases but lacking the N terminal extensions, the auto-inhibitory region, and the Ca2+-binding EF hands of that family. The genomic sequences revealed a single intron (81–107 bp long) close to the 3′ end of the coding sequence, the position of which is conserved in all plant PPCK sequences identified to date (Nimmo, 2003). The GmPPCK3 sequence is unusual in not having an in-frame stop codon in the intron, unlike almost all other PPCK genes. Hence, unprocessed GmPPCK3 transcripts would encode a protein identical to GmPPCK3 over most of its length but with one amino acid alteration (the codon interrupted by the intron) and an insertion of 30 residues.
Figure 1.
Sequence analysis of the soybean PPCK genes. A, Clustal alignment of deduced amino acid sequences of the soybean PPCK genes. Dots and vertical lines denote identical and similar residues respectively. B, Phylogenetic analysis of higher plant PPCK genes based on full-length deduced amino acid sequences. Branch numbers indicate bootstrap support (1,000 replicates). Accession numbers are given in Marsh et al. (2003), Nimmo (2003), and this work; other sequences are our unpublished data.
Of the four GmPPCK sequences, GmPPCK2 and GmPPCK3 are the most similar. At the nucleotide level, GmPPCK2 and GmPPCK3 are 94% identical in the coding region and 81% identical in the 3′ UTR. GmPPCK1 is 71% and 70% identical to GmPPCK2 and GmPPCK3, respectively, in the coding region and 42% and 40% identical in the 3′ UTR. GmPPCK4 is more divergent, being only 66% and 67% identical to GmPPCK2 and GmPPCK3, respectively, in the coding region. The sequences available include, respectively, 555, 373, 812, and 645 bases upstream of the translation start site for GmPPCK1-4. No sequences similar to the nodulin consensus sequences 5′AAAGAT and 5′CTCTT, which are found in the promoter regions of leghemoglobin and other late-nodulin genes (Stougaard et al., 1987, 1990), were detected in the promoter sequences of GmPPCKs. However, some other putative elements were observed (see “Discussion”).
Figure 1B shows an unrooted tree derived from phylogenetic analysis of full-length PPCK amino acid sequences. As noted by Xu et al. (2003), GmPPCK1, GmPPCK2, GmPPCK3, and a PPCK from Lotus japonicus cluster together. Interestingly, however, GmPPCK4 is more closely related to PPCKs from tomato and potato (LePPCK2 and StPPCK2) than it is to any of the other three GmPPCKs. This is considered further in “Discussion.”
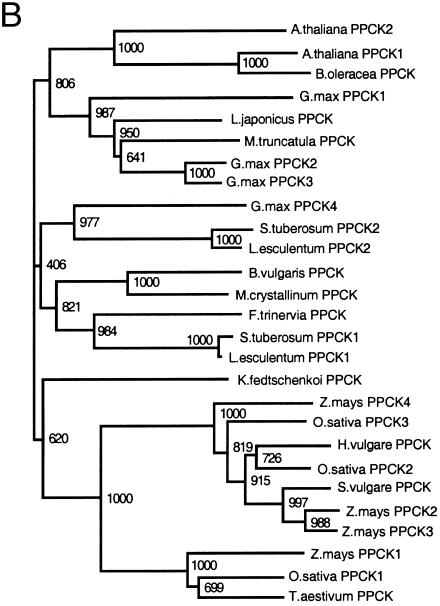
To test the function of the gene products, full-length cDNA sequences for each GmPPCK and the full length GmPPCK3 genomic DNA sequence were cloned behind a T7 promoter. The plasmids were linearized, transcribed, and translated, and the translation products were assayed for PPCK activity. Figure 2 shows the phosphor images of the 35S-labeled translation products separated by SDS-PAGE and of the 32P incorporated into purified PEPc in PPCK assays. All of the cDNA sequences gave rise to translation products of the expected size (31–32 kD), while the genomic GmPPCK3 sequence encoded a larger protein (34 kD) owing to the translation of the intron sequence. However, while the cDNA translation products were able to phosphorylate PEPc, confirming their identification as functional PPCKs, the product from the genomic GmPPCK3 sequence showed essentially no kinase activity.
Figure 2.
Functional analysis of full-length GmPPCK clones. A, Phosphor images of 35S-Met-labeled products from in vitro translation of RNA samples separated on 12.5% SDS-PAGE gel. B, Phosphor images of immunoprecipitated, 32P-labeled PEPc from assays of the PPCK activity of translation products separated on an 8% SDS-PAGE gel. Lanes are as follows: 1, GmPPCK1 cDNA; 2, GmPPCK2 cDNA; 3, GmPPCK3 genomic clone; 4, GmPPCK3 cDNA; 5, no RNA control; and 6, GmPPCK4 cDNA. The arrows indicate the two subunit types of K. fedtshenkoi PEPc (Nimmo et al., 1986).
Expression of PEPc and PPCK Genes in Soybean Tissues
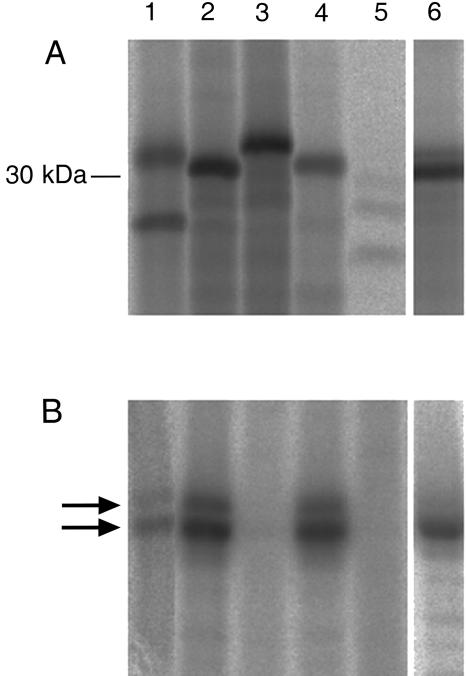
In Arabidopsis, PPCK genes are expressed only at low levels (Fontaine et al., 2002). Although PPCK transcripts in soybean nodules are sufficiently abundant to be detected by northern analysis (Xu et al., 2003), their expression in other tissues is lower. Therefore, the expression of the four PPCK sequences in soybean tissues was assessed using reverse transcription (RT)-PCR (Fig. 3A). GmPPCK1 shows highest expression in stem and root tissue and low levels of expression in all other tissues tested. GmPPCK2 and GmPPCK3 have very similar expression patterns. They are both preferentially expressed in nodule tissue, with lower expression levels in all other tissues. However, GmPPCK3 is more highly expressed in flowers than GmPPCK2. GmPPCK4 is also expressed in all tissues examined, with highest transcript levels in nodules, roots, and flowers.
Figure 3.
Expression patterns of GmPPCK and GmPEPC genes. This shows the products obtained from RT-PCR analysis of GmPPCK genes (27 cycles; A) and GmPEPC (27 cycles; B) genes from nodulated soybean plants. Ubiquitin was used as a control for equal loading. Tissue was collected 4 h into the light period.
The number and expression pattern of PEPc genes in soybean was also investigated. The presence of a nodule-enhanced PEPc isoform in legume root nodules is well documented (Pathirana et al., 1997; Suganuma et al., 1997; Hata et al., 1998). The nodule-enhanced gene in soybean is GmPEPC7. Three other PEPc genes have also been identified in soybean, GmPEPC1 (Vazquez-Tello et al., 1993), also termed GmPEPC15 (Hata et al., 1998), GmPEPC4 (Hata et al., 1998), and GmPEPC16 (Sugimoto et al., 1992). Results from northern-blotting experiments showed that GmPEPC1 and GmPEPC16 are expressed at low levels in the tissues examined (Sugimoto et al., 1992; Hata et al., 1998), while the full sequence and expression pattern of GmPEPC4 has not been investigated before due to its low level of similarity to other soybean PEPc isoforms (Hata et al., 1998). Based on the partial sequence (Hata et al., 1998) and using 5′ RACE, a full-length cDNA sequence of GmPEPC4 was obtained (GenBank accession no. AY563044).
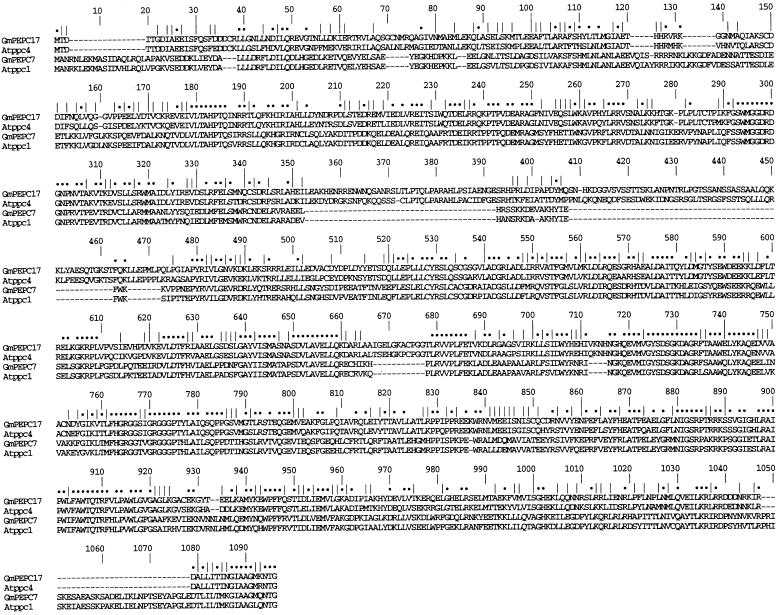
Querying of The Institute for Genomic Research soybean EST database (see http://www.tigr.org/) revealed a further three Tentative Consensus sequences with high sequence similarity to plant PEPc genes. Recently, it was shown that Arabidopsis contains a PEPc gene termed Atppc4 encoding a putative protein that resembles bacterial PEPcs much more than higher plant PEPcs; for example, it lacks the phosphorylation site of the plant PEPc genes (Sanchez and Cejudo, 2003). One soybean Tentative Consensus sequence, TC192656, closely resembles Atppc4. Using a degenerate 5′ primer based on the sequence of Atppc4 and a 3′ primer based on TC192656, we isolated an RT-PCR product termed GmPEPC17 and obtained full-length sequence by 5′ RACE (GenBank accession no. AY563043). The deduced amino acid sequence of GmPEPC17 is shown in Figure 4 aligned against the Arabidopsis homolog Atppc4, Atppc1, which is expressed in all organs (Sanchez and Cejudo, 2003), and GmPEPC7, which encodes the nodule-enhanced PEPc. As expected, GmPEPC17 is much more similar to Atppc4 than to the other soybean PEPc sequences and lacks the phosphorylation site. Figure 3B shows the expression patterns of GmPEPC7, GmPEPC1, GmPEPC4, GmPEPC16, GmPEPC17, and another Tentative Consensus sequence, TC178069, which seems to represent an additional PEPc gene. Our data confirm the preferential expression of GmPEPC7 in nodules and the broad but low-level expression patterns of GmPEPC16 and GmPEPC1, although GmPEPC1 is not detectably expressed in leaves. TC178069 showed a broad expression pattern, while both GmPEPC4 and GmPEPC17 were expressed mainly in aboveground organs.
Figure 4.
Sequence analysis of two soybean PEPC genes. Clustal alignment of deduced amino acid sequences of soybean and Arabidopsis PEPcs. Atppc1 (AJ532901) and Atppc4 (AJ532903) are from Sanchez and Cejudo (2003). GmPEPC17 and Atppc4 both encode bacterial-type PEPcs. Dots and vertical lines denote identical and similar residues, respectively.
Effect of Darkness and Detopping on PPCK Expression in Nodules
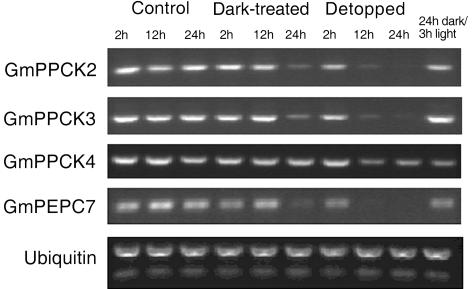
Several reports have shown that treatments that perturb the supply of photosynthate from the shoots, such as extended darkness, stem girdling, or detopping (removal of all aboveground organs), cause a decline in the phosphorylation state of nodule PEPc because of reduced PPCK activity (Zhang et al., 1995; Wadham et al., 1996; Zhang and Chollet, 1997). In order to examine the effects of light, soybean plants were subjected to 2, 12, or 24 h of darkness or 24 h darkness followed by 3 h of illumination. In other experiments, plants were detopped and tissue collected after 2, 12, and 24 h. Figure 5 shows the results of the RT-PCR experiments from these plants. The transcript abundance of both GmPPCK2 and GmPPCK3 shows a reduction in nodules as a result of reduced photosynthate supply from the shoots. The effect of detopping is much more dramatic than that of darkness, with transcripts barely detectable after 12 h. The effects of 24 h of darkness are reversible by 3 h of illumination. By contrast, the transcript abundance of GmPPCK4 shows little change in response to the treatments (Fig. 5), as does that of GmPPCK1 (data not shown). The expression of the nodule-enhanced PEPc gene, GmPEPC7, is greatly reduced after 24 h of darkness and detopping. This is in agreement with previously reported findings (Nakagawa et al., 2003a, 2003b) for both soybean and L. japonicus nodule-enhanced PEPc.
Figure 5.
Expression of soybean PPCK genes and GmPEPC7 in root nodules in response to darkness or shoot detopping. Nodulated soybean plants (5 weeks old) were used. Control plants were grown under normal light/dark conditions. All experiments were started 6 h into the light period. For dark treatments, plants were transferred to an identical environmental chamber under the same growth conditions but without lighting. For detopping, all aboveground organs were removed from below the point at which the cotyledons were attached. Gene-specific primers were used to obtain RT-PCR products (27 cycles). Ubiquitin was used as a control for equal loading.
GmPPCK4 Is under Circadian Control in Leaves But Not in Roots
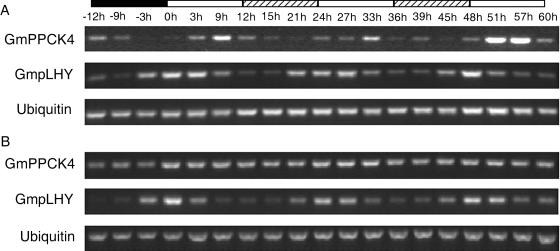
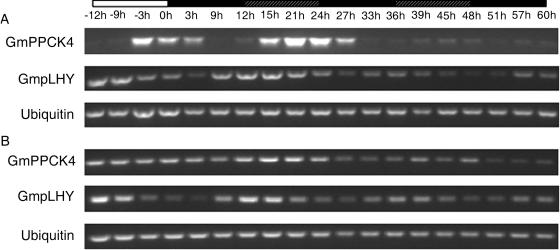
PPCK expression is under circadian control in leaves of the Crassulacean acid metabolism plants Kalanchoë fedtschenkoi and Mesembryanthemum crystallinum but not in Arabidopsis tissues (Hartwell et al., 1999a; Taybi et al., 2000; Fontaine et al., 2002). To investigate the situation in soybean, 2- to 3-week-old non-nodulated plants were transferred to continuous light (LL) and 22°C at the end of a normal dark period or to continuous dark (DD) and 22°C at the end of a normal light period. Leaf and root samples were taken over the following 60 h, and the expression of all four PPCK genes was assessed by RT-PCR. GmPPCK1-3 showed no signs of circadian control in either leaves or roots (data not shown). For GmPPCK2 and GmPPCK3, transcript abundance was consistently higher in LL than in the preceding dark period in leaves, while GmPPCK1 transcript abundance showed no systematic variation during the experiment. By contrast, GmPPCK4 was clearly under robust circadian control in leaves. In LL, transcript abundance was highest in the second half of subjective day but was low in subjective night (Fig. 6). GmPPCK4 transcript abundance also cycled in DD (Fig. 7), though the amplitude of the second peak in DD was lower than that of the first peak. In roots, however, GmPPCK4 transcript abundance was not under clear circadian control in either LL (Fig. 6) or DD (Fig. 7). Since such organ dependence of circadian control is unusual (see “Discussion”), we designed primers to amplify a partial-length soybean homolog of the Arabidopsis clock gene LHY, which is part of the central machinery of the clock (Alabadi et al., 2001), using tentative consensus sequence TC189020 (see http://www.tigr.org/). The soybean sequence (hereafter termed GmpLHY for Glycine max putative LHY) has been deposited in GenBank (AY568715). The putative soybean LHY is 80% identical to a LHY ortholog isolated from Phaseolus vulgaris (Kaldis et al., 2003) and 49% identical to the Arabidopsis LHY at the amino acid level. As expected from the behavior of Arabidopsis LHY (Schaffer et al., 1998), GmpLHY was expressed in a circadian fashion in both LL and DD in leaves and roots, with transcript abundance maximal early in subjective day (Figs. 6 and 7). As with GmPPCK4, the amplitude of the second peak in GmpLHY transcript abundance in DD was lower than that of the first peak.
Figure 6.
Expression of GmPPCK4 in constant light. The time points indicate hours in constant light. The black box indicates the preceding dark period, white boxes indicate subjective day, and the shaded boxes indicate subjective night. GmpLHY was used as a circadian control and ubiquitin as a control for equal loading. A, Expression in leaves. B, Expression in roots.
Figure 7.
Expression of GmPPCK4 in constant darkness. The time points indicate hours in constant darkness. The white box indicates the preceding light period, black boxes indicate subjective night, and the shaded boxes indicate subjective day. GmpLHY was used as a circadian control and ubiquitin as a control for equal loading. A, Expression in leaves. B, Expression in roots.
DISCUSSION
Our data substantially increase understanding of PEPC and PPCK genes in soybean and the relationship between them. We show that the PPCK gene family contains at least four members, and we present the sequence and expression pattern of two hitherto uncharacterized members. Our data show that, unexpectedly, expression of one member of the family, GmPPCK4, is under circadian control. We have been able to extend previous work by comparing the organ expression patterns of four PPCK and five PEPC genes. Moreover, we report the sequence and expression pattern of a bacterial-type PEPc gene in soybean similar to the recently reported Arabidopsis and rice genes.
Recently, Xu et al. (2003) reported the sequence and expression pattern for two soybean PPCK genes. These are a nodule-enhanced gene termed NE-PpcK, identical to GmPPCK2 in this work, and a more broadly expressed gene termed GmPpcK, identical to our GmPPCK1. They observed striking regulation of the NE-PpcK transcript abundance by photosynthate supply and suggested the existence of PpcK-Ppc expression partners exemplified by NePpcK and NE-Ppc in the soybean nodule. Our data show that the situation is appreciably more complex.
First, soybean contains not one but two PPCK genes, GmPPCK2 and GmPPCK3, that are primarily expressed in nodules and regulated by photosynthate. These genes are very similar at the nucleotide sequence level, and both are expressed in the later stages of nodule development (not illustrated), as reported for GmPPCK2 by Xu et al. (2003). While they show some minor differences in their tissue expression patterns (Fig. 3A), it is not possible to ascribe different functions to the two genes based on expression data. Second, we show for the first time, to our knowledge, the existence of another PPCK gene, GmPPCK4, that is expressed in nodules, although its expression does not seem to be affected by photosynthate supply from the shoots. It seems likely that the increase in nodule PPCK activity in response to photosynthate is mainly due to the expression of GmPPCK2 and GmPPCK3, but it is clearly important to assess how much of the nodule PPCK activity is contributed by these three genes (GmPPCK2-4) under different conditions. It is noteworthy that GmPPCK2 and GmPPCK3 are expressed in immature and mature leaves as well as in nodules, whereas GmPEPC7 is not. Hence, if the target of the GmPPCK2 and 3 in nodules is GmPEPC7, their targets must be different in leaf compared to nodule tissue. Furthermore, our data demonstrate the existence and expression of a bacterial-type PEPc gene in soybean. These bacterial-type PEPc isoforms lack the conserved Ser close to the N terminus that is the target of PPCK (Sanchez and Cejudo, 2003) and therefore are probably not regulated by phosphorylation. The transcripts of the different GmPEPC genes were detectable via RT-PCR after the same number of amplification cycles. Hence, it appears that the bacterial-type gene GmPEPC17 is expressed at a level comparable to those for the other GmPEPC genes. This is in contrast to Arabidopsis and rice, in which the bacterial-type genes Atppc4 and Osppc-b are expressed at a low level (Sanchez and Cejudo, 2003). A further contrast is evident in organ expression patterns; Atppc4 is expressed exclusively in siliques, flowers, and roots (Sanchez and Cejudo, 2003), whereas GmPEPC17 shows a much more uniform pattern of expression (Fig. 3B). This expression pattern offers no obvious clues to the function of the bacterial-type PEPc. However, we have noted the presence of several other potential homologs of Atppc4, Osppc-b, and GmPEPC17 in EST databases, and we conclude that bacterial-type PEPc genes are present in many plant species.
In our hands, GmPPCK1 is mainly expressed in roots and stems. This pattern is somewhat different from the data reported by Xu et al. (2003), in which the highest expression of GmPPCK1 was observed in seeds, flowers, and leaves. We therefore repeated our RT-PCR experiment with the primers reported by Xu et al. (2003). The results were very similar to those in Figure 3A. The differences between our data and those of Xu et al. (2003) may be due to growth conditions or a cultivar difference.
The new PPCK gene identified in this study, GmPPCK4, shows several interesting properties. It is the first PPCK gene shown to be under circadian control other than those in CAM species. Moreover, under constant conditions, transcript abundance cycles in leaves but not roots. To our knowledge, this is the first example of a plant gene where circadian control of expression is organ specific. Many studies of circadian rhythms in plants have involved use of whole seedlings, and organ specificity has rarely been assessed. However circadian-controlled expression of Arabidopsis PHYTOCHROME A has been observed in both leaves and roots, and similar observations have been made with CHALCONE SYNTHASE in aerial tissue and roots (Hall et al., 2001; Thain et al., 2002). As judged by the behavior of GmpLHY, the circadian clock operates in both roots and leaves of soybean in DD and LL. Many plant genes show robust rhythms of transcript abundance in LL that dampen rapidly in DD, as is the case for GmPPCK4 in leaves (Fig. 7). In tobacco, such behavior involves the ZGT gene that links the circadian oscillator to LHCB expression (Xu and Johnson, 2001). One explanation of our data is that machinery such as ZGT exists to connect the expression of GmPPCK4 to the clock in soybean leaves but not in roots.
Harmer et al. (2000) identified a nine-nucleotide evening element, AAAATATCT, that plays an important role in conferring rhythmic gene expression toward the end of the day in Arabidopsis. Since GmPPCK4 shows this pattern of expression, we analyzed the limited sequence information available for all four GmPPCK promoters. The GmPPCK4 promoter contains the sequence ATAATATCT, which differs from the evening element by just one base and includes the core seven-nucleotide sequence AATATCT noted by Xu and Johnson (2001). It also contains the sequence CACGTG, one version of the G-box core light-regulation motif (Chattopadhyay et al., 1998). None of the other GmPPCK promoters contain such a close match to the nine-nucleotide evening element or a G-box core, and it is possible that these features contribute to the control of GmPPCK4 expression.
Harmer et al. (2000) also noted that several genes whose products are involved in the consumption, translocation, or storage of sugars are clock controlled, with transcript abundance peaking in the late afternoon. They suggested that the circadian clock might play an important role in allocating sugar carbon to different pathways. This is consistent with the circadian control of GmPPCK4 in leaves and the potential role of the gene product in directing carbon flow toward biosynthesis.
The phylogenetic tree (Fig. 1B) shows that GmPPCK1-3 group with the PPCK sequences from two other legume species, L. japonicus and Medicago truncatula. The Lotus PPCK gene has been shown to have nodule-enhanced expression (Nakagawa et al., 2003a). While the Medicago PPCK has yet to be investigated, it is expressed in nodules, as two ESTs were isolated from nodule libraries. It may be that this group represents the nodule-enhanced PPCK isoforms for these plants. The placement of GmPPCK4 away from the other legume PPCK sequences is surprising. GmPPCK4 is more closely related to LePPCK2 and StPPCK2 than it is to other soybean PPCKs. The two kinase sequences from tomato and potato (LePPCK1 and 2, and StPPCK1 and 2) are also separate from each other in the tree. The placing of these sequences within the phylogenetic tree implies that duplication of an ancestral PPCK gene occurred significantly earlier during the evolution of the flowering plants than has been appreciated to date. Unlike the majority of PPCK genes, LePPCK2 and StPPCK2 contain a second intron that is subject to alternative splicing (Marsh et al., 2003). However, it is clear from the genomic sequence that GmPPCK4 contains only one intron. Expression of LePPCK2 is strongly induced during fruit ripening, but this does not correlate with changes in the levels of organic acids in the fruit, and the role of this gene is currently unknown.
As noted in the introduction, PEPc plays many different metabolic roles in plants, and the expression of PPCK genes plays an important role in regulating flux through PEPc. Hence, the existence of at least four soybean PPCK genes that differ in their organ expression patterns is, perhaps, not surprising. It seems likely that the expression of these genes will be controlled by several different factors and perhaps by different mechanisms. For example, at a gross level, GmPPCK2 and GmPPCK3 both respond to photosynthate. However, they may respond to different levels of photosynthate, in different cells, by different signaling pathways. In addition, there may be differences in the responses of the two genes to other factors, such as the availability of inorganic nutrients. Recently, Lodwig et al. (2003) have shown that an amino acid cycle is required for symbiotic nitrogen fixation in pea nodules. Whether such a cycle exists in soybean remains to be investigated, but the work of Lodwig et al. (2003) raises the possibility that the metabolic connection between plant and symbiont is more complicated than previously thought. This might account for the unexpected complexity of PEPc and PPCK expression in nodules. Clearly, more detailed studies of PPCK expression are required, including the analysis of promoter-reporter fusions. This will allow the analysis of expression at the cellular level and of the promoter elements involved in the circadian control of GmPPCK4.
MATERIALS AND METHODS
Plant Material
Nodulated soybean plants (Glycine max cv Williams 82) were grown in a perlite/sand mix in a controlled environment chamber (Microclima 1750; Snijders Scientific, Tilburg, Holland) at 22°C/18°C day/night temperature, 60% relative humidity, with a 16-h photoperiod and a light intensity of 400 μmol m−2 s−1. Plants were inoculated with Bradyrhizobium japonicum (strain 3402). Nodulated plants were watered daily with a nitrogen-free nutrient solution (Ryle et al., 1978). For the experiments in constant conditions, non-nodulated soybean plants were grown in a perlite/compost mix at 22°C, 60% relative humidity, with a 12-h photoperiod.
Nucleic Acid Isolation
All plant material was frozen under liquid nitrogen and stored at −80°C. Genomic DNA was isolated from leaf material using a DNA isolation kit (PUREgene DNA isolation kit; Gentra Systems, Minneapolis). Tissue for RNA extraction was collected from 5-week-old nodulated plants except for flowers, which were collected from 7- to 8-week-old plants. Non-nodulated plants for the time course experiments were harvested after 2 to 3 weeks. RNA was extracted using the Qiagen RNeasy kit (Qiagen USA, Valencia, CA) according to the manufacturer's protocol. Total RNA was DNase treated (DNA-free; Ambion, Huntingdon, UK) to remove contaminating genomic DNA. Concentration and purity were determined spectrophotometrically.
RT-PCR
cDNA was synthesized from 2.5 μg of total RNA as described previously (Fontaine et al., 2002). PCR reactions were performed with 2.5 μL of cDNA in a reaction mixture (25 μL) containing 0.5 μm of each primer and 1× ReddyMix PCR master mix with 1.5 mm MgCl2 (Abgene, Epsom, UK). For GmPPCK1, 2, and 3, a conserved forward primer (GAGTACGATGAGAAGGTTGATGTGT) was used with specific reverse primers designed to the 3′ UTR (GmPPCK1-R, ATCCGAAACTGAAACTCTTTAAAC; GmPPCK2-R, CACTACTACTACAGCATGTAGCTAC; and GmPPCK3-R, AAAATCTACAGAAGCGTGCTCTGTTCC). For GmPPCK4, gene-specific primers were designed in the coding region (GmPPCK4-F, CAAGATCGTTCAGCTTCTCTCC, and GmPPCK4-R, AAAAACACGAGTCGGAAACCT). For PEPc expression analysis, primers were designed to the 3′ UTR using the Primer3 program (Rozen and Skaletsky, 2000). Ubiquitin was used as a constitutive control (GmUb-F, GTACCCTCGCCGACTACAAC, and GmUb-R, GGGTCCTTCCATCCTCTAGC).
5′ RACE
The 5′ sequences of GmPPCK1, 2, 3, GmPEPC17, and GmPEPC4 were obtained using the GeneRacer RACE kit (Invitrogen, Carlsbad, CA).
Genomic PCR
Full-length genomic sequence for GmPPCK4 and promoter regions of GmPPCK1-4 were obtained using the Universal GenomeWalker kit (BD Biosciences, Erembodegem, Belgium). Primers were then designed to amplify the full-length GmPPCK4 sequence from cDNA.
Cloning
PCR products from genomic and cDNA were gel extracted, purified (Qiagen USA), and cloned using the TOPO TA cloning kit (Invitrogen). Universal primers were used for sequencing (MWG-Biotech, Ebersberg, Germany).
In Vitro Transcription and Translation
Full-length cDNA clones of GmPPCK1, 2, and 4, the full-length EST AW756453 (corresponding to GmPPCK3), and a full-length genomic clone of GmPPCK3 were used as templates for in vitro transcription, followed by translation as described previously (Hartwell et al., 1999a). Sequences cloned into the pcr4TOPO vector in the T7 orientation were transcribed using the T7 mMessage mMachine kit (Ambion). Transcribed RNA (1 μg) was translated in a rabbit reticulocyte lysate system (Amersham Biosciences, Piscataway, NJ) labeled with Redivue [35S]Met (Amersham Biosciences). Translation products were separated on SDS polyacrylamide gels and imaged using a Fuji (Tokyo) FLA-5000 phosphor imager.
Assay of PEPc Kinase Activity
The products of in vitro translation were assayed for PPCK activity as previously described (Hartwell et al., 1996). SDS polyacrylamide gels were analyzed using a Fuji FLA-5000 phosphor imager.
Sequence Alignment and Phylogenetic Analysis
Nucleotide alignments were performed using LALIGN at http://www.ch.embnet.org/. Deduced amino acid sequences were aligned using the ClustalX multiple sequence alignment program (version 1.63b; Thompson et al., 1997). A total of 1,000 bootstrap replicates were performed on the Neighbor-Joining tree. Trees were viewed using TreeView (Page, 1996).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY144180 to AY144185, AY568713, AY568714, AY563044, AY563043, and AY568715.
Acknowledgments
Soybean seeds were kindly provided by Prof. Brian Diers, University of Illinois, Urbana, and the Bradyrhizobium strain 3402 was kindly supplied by Dr. Frank Minchin, IGER, Aberystwyth, UK. We thank Dr. J. Hartwell for helpful discussion.
This work was supported by the Biotechnology and Biological Sciences Research Council (Ph.D. studentship support to S.S. and research support to H.G.N.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042762.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Andreo CS, Gonzalez DH, Iglesias AA (1987) Higher plant phosphoenolpyruvate carboxylase. FEBS Lett 213: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 273–298 [DOI] [PubMed] [Google Scholar]
- Duff SMG, Chollet R (1995) In vivo regulation of wheat-leaf phosphoenolpyruvate carboxylase by reversible phosphorylation. Plant Physiol 107: 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine V, Hartwell J, Jenkins GI, Nimmo GA, Nimmo HG (2002) Arabidopsis thaliana contains two phosphoenolpyruvate carboxylase kinase genes with different expression patterns. Plant Cell Environ 25: 115–122 [Google Scholar]
- Hall A, Kozma-Bognár L, Tóth R, Nagy F, Millar AJ (2001) Conditional circadian regulation of PHYTOCHROME A gene expression. Plant Physiol 127: 1808–1818 [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (1999. a) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20: 1–10 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Jenkins GI, Wilkins MB, Nimmo HG (1999. b) The light induction of maize phosphoenolpyruvate carboxylase kinase translatable mRNA requires transcription but not translation. Plant Cell Environ 22: 883–889 [Google Scholar]
- Hartwell J, Smith LH, Wilkins MB, Jenkins GI, Nimmo HG (1996) Higher plant phosphoenolpyruvate carboxylase kinase is regulated at the level of translatable mRNA in response to light or a circadian rhythm. Plant J 10: 1071–1078 [Google Scholar]
- Hata S, Izui K, Kouchi H (1998) Expression of a soybean nodule-enhanced phosphoenolpyruvate carboxylase gene that shows striking similarity to another gene for a house-keeping isoform. Plant J 13: 267–273 [DOI] [PubMed] [Google Scholar]
- Kaldis AD, Kousidis P, Kesanopoulos K, Prombona A (2003) Light and circadian regulation in the expression of LHY and Lhcb genes in Phaseolus vulgaris. Plant Mol Biol 52: 981–997 [DOI] [PubMed] [Google Scholar]
- Li B, Zhang XQ, Chollet R (1996) Phosphoenolpyruvate carboxylase kinase in tobacco leaves is activated by light in a similar but not identical way as in maize. Plant Physiol 111: 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwig EM, Hosie AHF, Bordes A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS (2003) Amino-acid cycling drives nitrogen fixation in the legume-rhizobium symbiosis. Nature 422: 722–726 [DOI] [PubMed] [Google Scholar]
- Marsh JT, Sullivan S, Hartwell J, Nimmo HG (2003) Structure and expression of phosphoenolpyruvate carboxylase kinase genes in solanaceae. A novel gene exhibits alternative splicing. Plant Physiol 133: 2021–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Izumi T, Banba M, Umehara Y, Kouchi H, Izui K, Hata S (2003. a) Characterization and expression analysis of genes encoding phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxylase kinase of Lotus japonicus, a model legume. Mol Plant Microbe Interact 16: 281–288 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Takane K, Sugimoto T, Izui K, Kouchi H, Hata S (2003. b) Regulatory regions and nuclear factors involved in nodule-enhanced expression of a soybean phosphoenolpyruvate carboxylase gene: implications for molecular evolution. Mol Genet Genomics 269: 163–172 [DOI] [PubMed] [Google Scholar]
- Nimmo GA, Nimmo HG, Hamilton ID, Fewson CA, Wilkins MB (1986) Purification of the phosphorylated night form and dephosphorylated day form of phosphoenolpyruvate carboxylase from Bryophyllum fedtshenkoi. Biochem J 239: 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo HG (2000) The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci 5: 75–80 [DOI] [PubMed] [Google Scholar]
- Nimmo HG (2003) Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch Biochem Biophys 414: 189–196 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pathirana MS, Samac DA, Roeven R, Yoshioka H, Vance CP, Gantt JS (1997) Analyses of phosphoenolpyruvate carboxylase gene structure and expression in alfalfa nodules. Plant J 12: 293–304 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HS (2000) Primer3 on the WWW for general users and for biologist programmers. In S Krawetz, S Misener, eds, Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed]
- Ryle GJA, Powell CE, Gordon AJ (1978) Effect of source of nitrogen on the growth of Fiskeby soybean: the carbon economy of whole plants. Ann Bot (Lond) 42: 637–648 [Google Scholar]
- Sanchez R, Cejudo FJ (2003) Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol 132: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schuller KA, Turpin DH, Plaxton WC (1990) Metabolite regulation of partially purified soybean nodule phosphoenolpyruvate carboxylase. Plant Physiol 94: 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller KA, Werner D (1993) Phosphorylation of soybean (Glycine max L.) nodule phosphoenolpyruvate carboxylase in vitro decreases sensitivity to inhibition by L-malate. Plant Physiol 101: 1267–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard J, Jorgensen JE, Christensen T, Kuhle A, Marcker KA (1990) Interdependence and nodule specificity of cis-acting regulatory elements in the soybean leghemoglobin-Lbc3 and N23 gene promoters. Mol Gen Genet 220: 353–360 [DOI] [PubMed] [Google Scholar]
- Stougaard J, Sandal NN, Gron A, Kuhle A, Marcker KA (1987) 5′ analysis of the soybean leghemoglobin Lbc3 gene-regulatory elements required for promoter activity and organ specificity. EMBO J 6: 3565–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma N, Okada Y, Kanayama Y (1997) Isolation of a cDNA for nodule-enhanced phosphoenolpyruvate carboxylase from pea and its expression in effective and plant-determined ineffective pea nodules. J Exp Bot 48: 1165–1173 [Google Scholar]
- Sugimoto T, Kawasaki T, Kato T, Whittier RF, Shibata D, Kawamura Y (1992) cDNA sequence and expression of a phosphoenolpyruvate carboxylase gene from soybean. Plant Mol Biol 20: 743–747 [DOI] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman JC (2000) A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in crassulacean acid metabolism-induced leaves of the common ice plant. Plant Physiol 123: 1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ (2002) The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol 130: 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida Y, Furumoto T, Izumida A, Hata S, Izui K (2001) Phosphoenolpyruvate carboxylase kinase involved in C4 photosynthesis in Flaveria trinervia: cDNA cloning and characterization. FEBS Lett 507: 318–322 [DOI] [PubMed] [Google Scholar]
- Vance CP, Gregerson RG, Robinson DL, Miller SS, Gnatt JS (1994) Primary assimilation of nitrogen in alfalfa nodules: molecular features of the enzymes involved. Plant Sci 101: 51–64 [Google Scholar]
- Vazquez-Tello A, Whittier RF, Kawasaki T, Sugimoto T, Kawamura Y, Shibata D (1993) Sequence of a soybean (Glycine max L.) phosphoenolpyruvate carboxylase cDNA. Plant Physiol 103: 1025–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal J, Chollet R (1997) Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci 2: 230–237 [Google Scholar]
- Wadham C, Winter H, Schuller KA (1996) Regulation of soybean nodule phosphoenolpyruvate carboxylase in vivo. Physiol Plant 97: 531–535 [Google Scholar]
- Xu W, Zhou Y, Chollet R (2003) Identification and expression of a soybean nodule-enhanced PEP-carboxylase kinase gene (NE-PpcK) that shows striking up-/down-regulation in vivo. Plant J 34: 441–452 [DOI] [PubMed] [Google Scholar]
- Xu Y, Johnson CH (2001) A clock- and light-regulated gene that links the circadian oscillator to LHCB gene expression. Plant Cell 13: 1411–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Chollet R (1997) Phosphoenolpyruvate carboxylase protein kinase from soybean root nodules: partial purification, characterization, and up/down-regulation by photosynthate supply from the shoots. Arch Biochem Biophys 343: 260–268 [DOI] [PubMed] [Google Scholar]
- Zhang XQ, Li B, Chollet R (1995) In vivo regulatory phosphorylation of soybean nodule phosphoenolpyruvate carboxylase. Plant Physiol 108: 1561–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]