Abstract
Li et al. [1] reported over ten thousand mismatches between mRNA and DNA sequences from the same individuals, which they attributed to previously unrecognized mechanisms of gene regulation. We found that at least 88% of these sequence mismatches can likely be explained by technical artifacts such as errors in mapping sequencing reads to a reference genome, sequencing errors, and genetic variation.
Li et al.[1] sequenced cDNA from lymphoblastoid cell lines derived from 27 individuals whose genomes have been sequenced at low coverage [2], and identified 10,210 sites of mismatches between an individual's mRNA and DNA sequences (RDD sites, for RNA-DNA difference). RDD sites included all possible combinations of sequence mismatches, and the authors validated a subset of these mismatches by additional assays. These observations were interpreted as evidence for novel mechanisms of gene regulation, analogous perhaps to A→I RNA editing [3].
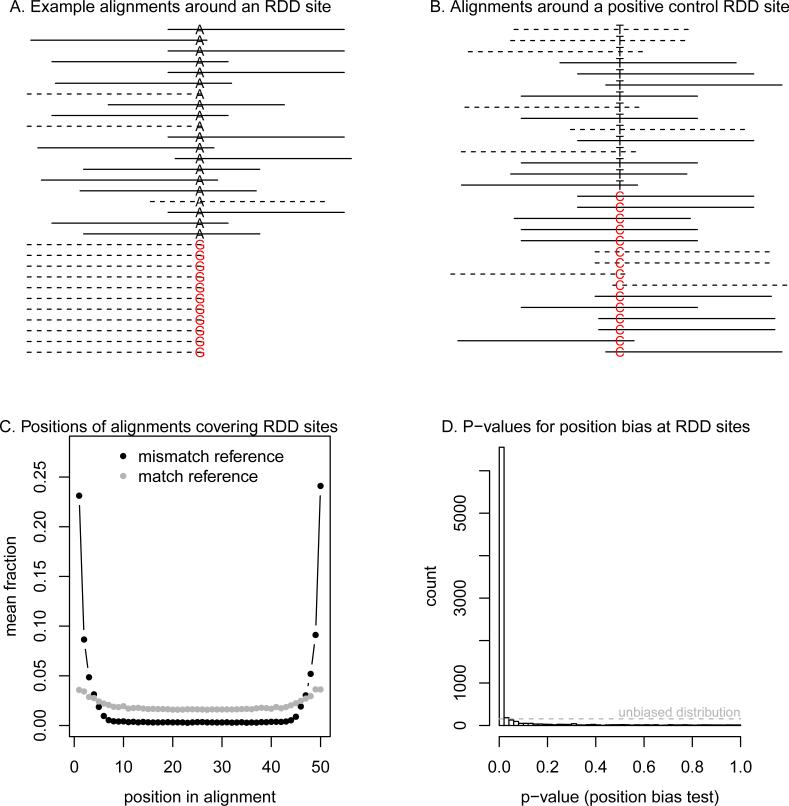
An alternative explanation is that some RDD sites are technical artifacts due to errors in mapping sequencing reads to a reference genome or systematic sequencing errors. To evaluate this possibility, we examined the sequence alignments used to call RDD sites (Supplementary Material). Visualizing these alignments revealed a number of anomalies. For example, at the RDD site presented in Figure 1A, all mismatches to the genome occur at the last base of reads aligned to the negative DNA strand. No such anomalies are seen in alignments around a positive control site (Figure 1B). The biases in the first example are consistent with several known issues that cause spurious differences between Illumina sequencing reads and a reference genome; these include read-mapping errors between paralogous genomic regions and around insertions and deletions [2; 4], as well as position and strand biases in the error rate of Illumina sequencing [5–7].
Figure 1. Identifying false positive RDD calls.
A. RNA-seq read alignments around an RDD call from Li et al. (2011). Plotted are the positions of read alignments to the genome surrounding the RDD site at chromosome 11, position 105,473,792. The solid lines show sequencing reads aligning to the (+) strand of the genome, and dotted lines are alignments to the (−) strand of the genome. At the center of the plot is the base corresponding to the RDD site; the reference base is in black, and the non-reference base is in red, and both are labeled with respect to the (+) DNA strand. Alignments have been organized such that the mismatches to the genome are at the bottom of the figure. For plotting, we randomly sampled 20 alignments that match the genome at the RDD site; all 11 alignments that mismatch the genome are shown. B. Read alignments around a positive control RDD site. Plotted are the positions of read alignments to the genome surrounding the known A→I editing site in AZIN1 [12] (on the forward strand this site appears as T→C). The format is the same as in A. For plotting, we randomly sampled 15 alignments that match the genome at the RDD site, and 15 alignments that do not match the genome at the site. C. Position biases in alignments around RDD sites. For each RDD site with at least five reads mismatching the genome, we calculated the fraction of reads with the mismatch (or the match) at each position in the alignment of the RNA-seq read to the genome (on the + DNA strand). Plotted is the average of this fraction across all sites, separately for the alignments which match and mismatch the genome. D. Histogram of p-values for the position bias test. For each RDD site with at least five reads mismatching the genome, we calculated a p-value for the position bias test (Supplementary Information). Plotted is the histogram of these p-values. If these sites were not consistently biased, the distribution of p-values would be uniform; this is indicated with the dashed grey line.
We asked whether the patterns seen in Figure 1A are typical among RDD sites. Indeed, mismatches to the genome at RDD sites are dramatically enriched at the ends of RNA sequencing reads; this contrasts with reads that match the genome at these sites (Figure 1C). This pattern is evidence that many of the RDD sites are false positives due to mapping or sequencing errors.
To quantify what fraction of RDD sites may be false positives, we used metrics developed in for calling single nucleotide polymorphisms (SNPs) from Illumina sequencing data. In this context, it is known that a search for mismatches between aligned reads and a genome will result in large numbers of false positive SNPs, many of which can be filtered out based on various criteria [2; 4; 8; 9]. We used two criteria based on comparing, at each RDD site, the alignments of RNA sequencing reads that match the genome with the alignments of reads that mismatch the genome–a test for position bias and a test for strand bias (Supplementary Information). These tests provide quantitative measures for the intuition that there should be no systematic differences in strand or start position between alignments of reads covering the two alternative genotypes at a site, and are similar to tests implemented in SNP-calling packages [4; 9].
In Figure 1D, we show the histogram of p-values for the position bias test for the 7,812 RDD sites with at least five reads supporting both bases. There is a clear skew towards low p-values, indicating pervasive technical artifacts. At a p-value threshold of 0.01, 87% of these RDD sites fail either the strand bias test or the position bias test (at a p-value threshold of 0.05, the corresponding number is 93%). To test the specificity of these filters, we compared the reported RDD sites to a database of known A→I RNA editing sites [10]. There are 23 sites in common between the two data sets; of these, 21 (91%) pass both of the filters. This indicates that we are largely only removing false positives.
Genetic variation is another source of false positives; an additional 1% of the putative RDD sites appear instead to be known genetic variants in these individuals (Supplementary Material). In total, we estimate that at least 88% (at a p-value threshold of 0.01) to 94% (at a p-value threshold of 0.05) of the RDD sites are likely false positives. This is probably an underestimate of the true false positive rate, since some false positive sites will pass the bias tests by chance and there are additional, unannotated SNPs in the genome.
Given the above results, we re-examined the validation experiments done by Li et al.[1]. These experiments are of two types. First, at 11 sites, the authors confirmed that the RDD event was absent from genomic DNA but present in cDNA by Sanger sequencing. At six of these 11 sites, the event is of the type A→G, and four of these six are present in a database of known A→I RNA editing sites [10]; these are likely true positives. Of the remaining five sites, three fall in a single gene–HLA-DQB2–that is copy number variable in these individuals [11], and one–in the gene DPP7–overlaps a known SNP (at which the reported RDD type matches the known alleles) [2]. We suggest that the authors have detected genetic variation rather than RNA-DNA differences at these sites. In sum, these experiments identify two previously unknown sites of A→I RNA editing, and provide evidence for a single G→A event.
The second validation experiment involved identifying peptides corresponding to RDD events. In their Table 3, Li et al.[1] provide 17 examples where both the “DNA form” (the unaltered version) and the “RNA form” (the modified version) of peptides were detected via mass spectrometry. All but one of these sites fail the bias tests described above. We propose that the “RNA forms” of these peptides are in most cases normal forms produced by paralogous genes. Indeed, examination of the “RNA forms” revealed that seven match both the reported protein and additional proteins equally well, and four of the remaining 10 match other proteins (in addition to the reported protein) with a single additional mismatch (Table 1; Supplementary Material). It cannot be ruled out that the “RNA forms” of these proteins are instead normal forms caused by genetic variation in their paralogs. An additional possibility is that some “RNA forms” result from sequencing errors in the peptides.
Table 1. Characteristics of RDD sites reported in peptides.
We re-evaluated the peptides presented in Table 3 of Li et al. (2011). Repeated from that table are the gene names, positions and types of RDD sites, and “RNA forms” of protein sequences. We additionally show the numbers of aligned reads that mismatch the genome at each site, and the p-values from the tests for position bias and strand bias at each site. P-values in red are less than 0.01. We used BLAST to search the human genome for matches to the peptides; given are the names of additional genes (apart from the one reported by Li et al. [1]) that match the peptide equally well (since these are the “RNA forms” of the peptides, the best matches have a single mismatching amino acid), and the number of genes with one additional mismatch (for a total of two mismatches) to the peptide. Mismatches are defined as either a substitution or an insertion/deletion of a single amino acid.
| Protein | Position (hgl8) | RDD type | # RDD reads | “RNA form” peptide sequence | P-values (dist., strand) | Equally good matches | # additional close matches |
|---|---|---|---|---|---|---|---|
| AP2A2 | chr11:976858 | T→G | 3 | DLALESMCTLASSEFSHEAVK | 0.01, 0.59 | AP2A1 | 0 |
| DFNA5 | chr7:24705225 | T→A | 23 | VFPQLLCITLNGLCALGR | 8 × 10–21, 2 × 10–7 | - | 0 |
| ENO1 | chr1:8848125 | T→C | 336 | EGPELLK | 9 × 10–65, 8 × 10–13 | C7orf25, ABCF1 | >20 |
| ENO3 | chr17:4800624 | T→G | 8 | LAQSNGWGGMVSHR | 0.76, 0.0005 | - | 2 |
| FABP3 | chr1:31618424 | T→A | 3 | MVDAFLGTR | 0.007, 0.07 | - | 1 |
| FH | chr1:239747217 | T→A | 37 | KEYDTFGELK | 1 × 10–43, 2 × 10–20 | - | 0 |
| HMGB1 | chr13:29935772 | T→A | 10 | MSSNAFFVQTCR | 1 × 10–9, 1 × 10–8 | HMGB2 | 2 |
| NACA | chr12:55392932 | G→A | 16 | DIELVMSQANVSR | 3 × 10–8, 0.80 | - | 1 |
| NSF | chr17:42161411 | T→C | 13 | LLDYVPIGPR | 2 × 10–9, 0.07 | - | 0 |
| POL2RB | chr4:57567852 | T→A | 17 | IISDGQK | 4 × 10–10, 0.0007 | MLKN1, CUL4B | >20 |
| RAD50 | chr5:131979610 | T→G | 9 | WRQDNLTLR | 1 × 10–6, 0.01 | - | 0 |
| RPL12 | chr9:129250509 | A→G | 518 | HSGDITFDEIVNIAR | 1 × 10–187, 7 × 10–12 | - | 0 |
| RPL32 | chr3:12852658 | G→T | 356 | SAQLAIR | 6 × 10–95, 8 × 10–12 | RBM46 | >20 |
| RPS3AP47* | chr4:152243651 | C→A | 81 | EVQKNDLK | 1 × 10–62, 1 × 10–12 | - | 3 |
| SLC25A17 | chr22:39520485 | A→G | 3 | TTHMVLLGIIK | 0.002, 0.06 | - | 0 |
| TUBA1* | chr2:219823379 | A→G | 33 | EDMAALGK | 4 × 10–6, 6 × 10–13 | CCDC85B, TUBA1B, TUBA1C | 9 |
| TUBB2C | chr9:139257297 | G→A | 9 | LHFFMPDFAPLTSR | 0.007, 0.31 | TUBB8, TUBB4Q, TUBB6, TUBB2B, TUBB2A, TUBB, TUBB4 | 1 |
The RefSeq name for TUBA1 is TUBA4A, and the RefSeq name for RPS3AP47 is RPS3A.
In summary, we estimate that a minimum of 88-94% of the RDD sites identified by Li et al.[1] are false positives due to mapping errors, sequencing errors, and genetic variation. It is possible that the remainder of RDD sites contain examples of novel mechanisms of gene regulation.
Supplementary Material
References
- 1.Li M, et al. Science. 2011 [Google Scholar]
- 2.1000 Genomes Project Consortium et al. Nature. 2010;467:1061. [Google Scholar]
- 3.Bass BL. Annu Rev Biochem. 2002;71:817. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Depristo MA, et al. Nat Genet. 2011;43:491. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura K, et al. Nucleic Acids Res. 2011 [Google Scholar]
- 6.Erlich Y, Mitra PP, de la Bastide M, McCombie WR, Hannon GJ. Nat Methods. 2008;5:679. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meacham F, et al. Nature Precedings. 2011 [Google Scholar]
- 8.Li H, Ruan J, Durbin R. Genome Res. 2008;18:1851. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, et al. Bioinformatics. 2009;25:2078. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiran A, Baranov PV. Bioinformatics. 2010;26:1772. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 11.Conrad DF, et al. Nature. 2010;464:704. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JB, et al. Science. 2009;324:1210. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.