Abstract
The endogenous brassinosteroids in the dwarf mutant lk of pea (Pisum sativum) were quantified by gas chromatography-selected ion monitoring. The levels of castasterone, 6-deoxocastasterone, and 6-deoxotyphasterol in lk shoots were reduced 4-, 70-, and 6-fold, respectively, compared with those of the wild type. The fact that the application of brassinolide restored the growth of the mutant indicated that the dwarf mutant lk is brassinosteroid deficient. Gas chromatography-selected ion monitoring analysis of the endogenous sterols in lk shoots revealed that the levels of campestanol and sitostanol were reduced 160- and 10-fold, respectively, compared with those of wild-type plants. These data, along with metabolic studies, showed that the lk mutant has a defect in the conversion of campest-4-en-3-one to 5α-campestan-3-one, which is a key hydrogenation step in the synthesis of campestanol from campesterol. This defect is the same as that found in the Arabidopsis det2 mutant and the Ipomoea nil kbt mutant. The pea gene homologous to the DET2 gene, PsDET2, was cloned, and it was found that the lk mutation would result in a putative truncated PsDET2 protein. Thus it was concluded that the short stature of the lk mutant is due to a defect in the steroidal 5α-reductase gene. This defect was also observed in the callus induced from the lk mutant. Biosynthetic pathways involved in the conversion of campesterol to campestanol are discussed in detail.
Brassinosteroids (BRs) are steroidal plant growth hormones that are synthesized from plant sterols. The major biosynthetic pathway leading to brassinolide and castasterone, which are deemed biologically active BRs, has been established (Fujioka and Yokota, 2003). BRs are involved in diverse physiological processes, such as stem elongation, tracheary element differentiation, root inhibition, phytohormone synthesis and response, cold and drought stress responses, and regulation of gene expression (Clouse and Sasse, 1998; Li and Chory, 1999; Khripach et al., 2000; Bishop and Yokota, 2001; Bishop and Koncz, 2002; Clouse, 2002). The crucial role played by BRs in plant growth and development has been determined by the identification and characterization of BR mutants in Arabidopsis, pea (Pisum sativum), tomato (Lycopersicon esculentum), and rice (Oryza sativa). To date, the majority of research has been carried out on the Arabidopsis BR biosynthetic mutants, det2 (Li et al., 1996, 1997; Fujioka et al., 1997; Noguchi et al., 1999b), dwf4 (Azpiroz et al., 1998; Choe et al., 1998), cpd (Szekeres et al., 1996), dwf5 (Choe et al., 2000), dwf7 (Choe et al., 1999b), sax1 (Ephritikhine et al., 1999), dwf1/dim (Klahre et al., 1998; Choe et al., 1999a; Takahashi et al., 1995), and fackel (Jang et al., 2000), as well as the BR perception and signal transduction mutants, bri1 (Clouse et al., 1996; Li and Chory, 1997; Noguchi et al., 1999a), bak1 (Nam and Li, 2002), brs1 (Li et al., 2001), bin2 (Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002), brz1 (Wang et al., 2002), and bes1 (Yin et al., 2002). Furthermore, several mutants were also characterized from pea, tomato, and rice. The BR biosynthetic mutants include pea lkb (Nomura et al., 1997), tomato dwarf (Bishop et al., 1999) and dumpy (Koka et al., 2000), and rice brds (Hong et al., 2002; Mori et al., 2002) and d2 (Hong et al., 2003). Mutants that are insensitive to and have defects in perception of BRs include pea lka (Nomura et al., 1997, 2003), tomato cu3 and abs (Montoya et al., 2002), and rice d61 (Yamamuro et al., 2000).
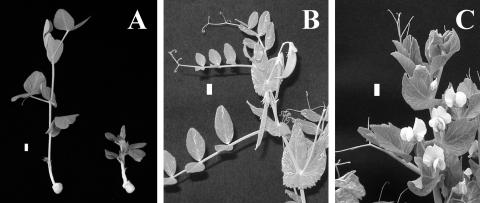
The lka and lkb mutants of pea show common dwarf phenotypes referred to as erectoides, which are not rescued by GA (Reid and Ross, 1989). The lka mutant is defective in the BR receptor kinase PsBRI1 (Nomura et al., 2003). The lkb mutant has a defect in the 24-hydrogenase that converts 24-methylenecholesterol to campesterol (Nomura et al., 1999; Schultz et al., 2001). In addition, the lkc and lk mutants of pea have been classified as erectoides. The lkc mutant shows weak dwarfism (Reid et al., 1991) and is slightly insensitive to both BR and GA (T. Yokota, unpublished data), although its genetic lesion has not been clarified. The pea mutant lk is the most severe dwarf and is characterized by short internodes, reduced yield, dark green foliage, brittle stems with increased diameter, very short peduncles and petioles, and increased apical dominance (Fig. 1; Reid, 1986). Interestingly, the gene lk appears to be largely epistatic to genes involved in the GA response (e.g. la crys slenders; Reid, 1986). In this article, we investigate the effect of BRs on the growth of the lk mutant and also analyze the endogenous BRs and sterols, resulting in the conclusion that the lk mutant is BR deficient because of impaired sterol biosynthesis. Feeding experiments using synthetic sterol substrates and molecular analyses were conducted to pinpoint the defect in the lk mutant.
Figure 1.
Morphology of the lk mutant and wild type. A, 21-d-old seedlings of LK (212+; left) and lk (212−; right). B, Adult plant of LK. C, Adult plant of lk. Bar represents 1 cm.
RESULTS
The Dwarf Phenotype of the lk Mutant Was Rescued by Treatment with Brassinolide
Treatment of the fourth internode with as little as 1 ng of brassinolide could restore its growth to that of wild-type plants (Fig. 2). The effect of brassinolide leveled off at a dosage of 10 ng. Such an effective recovery of the dwarfism, together with the earlier finding that changes in GA level or response may not be related to the lk mutation (Lawrence et al., 1992), suggests that the lk mutant is BR deficient. At a dosage of 33 ng, the fourth internode became thick and twisted, with petioles being epinastic.
Figure 2.
Effect of brassinolide on the growth of lk seedlings. Growth profiles were recorded 3 d after the fourth internodes of 8-d-old lk seedlings were treated with solvent only (A), 1 ng brassinolide (B), 3.3 ng brassinolide (C), 10 ng brassinolide (D), and 33 ng of brassinolide (E). F, Wild-type seedling treated with solvent only. Bar represents 1 cm.
The Levels of BRs Are Reduced in lk Seedlings
The endogenous levels of castasterone, 6-deoxocastasterone, and 6-deoxotyphasterol in 12-d-old shoots of lk seedlings were 4-, 70-, and 6-fold lower than those of wild-type plants, respectively (Table I), indicating that the lk mutant has a blockage in BR biosynthesis.
Table I.
Endogenous levels of BRs in 12-d-old shoots of the pea mutant lk and wild-type plants
| BR | Wild Type | lk |
|---|---|---|
| Brassinolide | nda | nd |
| Castasterone | 446 | 114 |
| Typhasterol | nd | nd |
| 3-Dehyroteasterone | trace | 42 |
| Teasterone | nd | nd |
| 6-Deoxocastasterone | 4,302 | 59 |
| 6-Deoxotyphasterol | 926 | 160 |
| 6-Deoxo-3-dehydroteasterone | nd | nd |
| 6-Deoxoteasterone | 33 | 180 |
Data (ng kg−1 fresh weight) are means of two GC-SIM runs.
nd, Not detected, or no reliable data were obtained due to impurities.
The lk Mutant Has a Defect in the Hydrogenation of Campesterol and Sitosterol
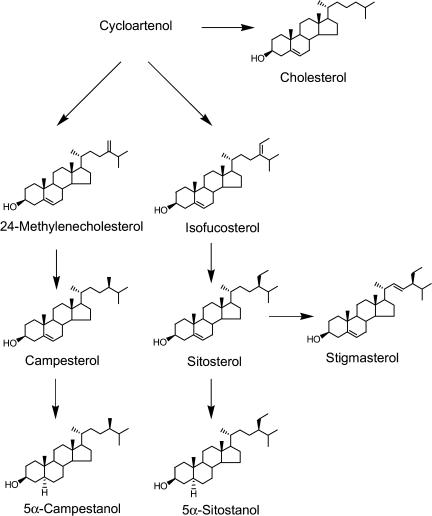
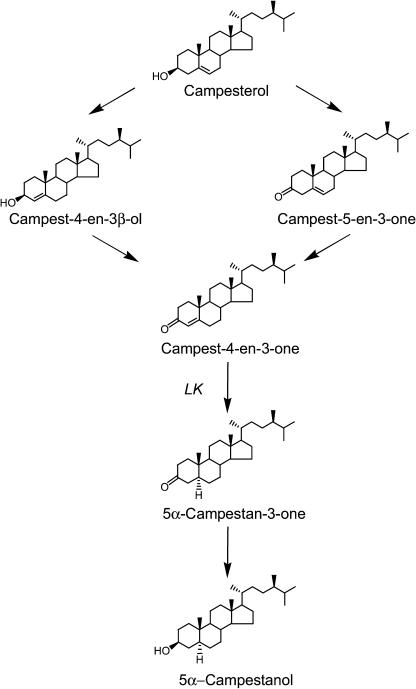
In lk shoots, campesterol, sitosterol, stigmasterol, and cholesterol, which are bulk sterols considered to be end-pathway sterols (Fig. 3), are present at comparable levels to those of wild-type plants (Table II). However, the level of campestanol, a hydrogenation product of campesterol, was two orders of magnitude lower compared with wild-type plants. Because campestanol is not separable from its 24-epimer (dihydrobrassicasterol), the quantitative data given may express the sum of campestanol and its 24-epimer. Furthermore, the level of sitostanol was also reduced, although to a lesser extent as compared with campestanol (Table II). Thus, the lk mutant seemed to have a defect in the hydrogenation of campesterol to campestanol and of sitosterol to sitostanol. It has been demonstrated that, in Arabidopsis, the synthesis of campestanol from campesterol proceeds via three intermediates, campest-4-en-3β-ol, campest-4-en-3-one, and 5α-campestan-3-one (Fig. 4). Therefore, we analyzed these steroids in the shoots of wild-type and lk seedlings, revealing that lk seedlings contained higher levels of campest-4-en-3β-ol and campest-4-en-3-one, but a lower level of 5α-campestan-3-one compared with wild-type seedlings. These findings suggest that the lk mutation results in a loss of 5α-reductase activity.
Figure 3.
Biosynthetic pathway of plant sterols.
Table II.
Endogenous levels of sterols in 17-d-old shoots and calli of the pea mutant lk and wild-type plants
| Sterol | Shoot
|
Callus
|
||
|---|---|---|---|---|
| Wild Type | lk | Wild Type | lk | |
| 24-Methylenecholesterol | ndb | nd | 2.4 | nd |
| Campesterol | 30.6 | 22.8 | 16.7 | 20.9 |
| Campest-4-en-3β-ol | nd | 0.46 | nac | na |
| Campest-4-en-3-one | 0.52 | 1.0 | na | na |
| 5α-Campestan-3-one | 0.012 | 0.004 | na | na |
| 5α-Campestanol | 1.6 | 0.01 | 0.7 | 0.04 |
| Isofucosterol | 8.9 | 5.8 | 16.7 | 7.0 |
| Sitosterol | 73.2 | 84.5 | 41.1 | 67.4 |
| Stigmasterol | 102.0 | 105.9 | 17.6 | 24.5 |
| 5α-Sitostanol | 3.3 | 0.3 | 2.0 | 0.6 |
| Cholesterol | 1.8 | 2.2 | 6.0 | 9.8 |
| Total sterols | 221.4 | 221.5 | 103.2 | 130.2 |
| End-pathway sterolsa | 207.6 | 215.4 | 81.4 | 122.6 |
Data are expressed as μg g−1 fresh weight.
End-pathway sterols, cholesterol+campesterol+sitosterol+stigmasterol.
nd, Not detected.
na, Not analyzed.
Figure 4.
Biosynthetic pathway from campesterol to campestanol.
Calli induced from shoots of lk and wild-type seedlings were also examined for the levels of the endogenous sterols (Table II). It was found that the level of campestanol and sitostanol is reduced in the lk callus, indicating that the effects of the lk mutation were also observable in callus derived from lk shoots (Table II).
Steroid 5α-Reductase Activity Is Lost in the lk Mutant
To ascertain the reaction lacking in the lk mutant, campesterol and the intermediates between campesterol and campestanol were labeled by deuteriums in the side chain (Noguchi et al., 1999b) and fed to apical tissues of lk and wild-type plants. In wild-type plants, all of these [2H6]compounds were converted to [2H6]campestanol (Table III), indicating the biosynthetic sequence of campesterol → campst-4-en-3β-ol → campest-4-en-3-one → 5α-campestan-3-one → 5α-campestanol is present in garden pea. The fact that [2H6]4-en-3β-ol and/or [2H6]3-one were not detected as intermediates in any feeding experiments (Table III) indicates that the lifetimes of 4-en-3β-ol and 3-one are very short.
Table III.
Metabolism of [2H6]-labeled sterols in 13-d-old shoots of the pea mutant lk and wild-type plants
| Substrate
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Product | [2H6]Campesterol
|
[2H6]Campest-4-en-3β-ol
|
[2H6]Campest-4-en-3-one
|
[2H6]5α-Campestan-3-one
|
||||
| Wild Type | lk | Wild Type | lk | Wild Type | lk | Wild Type | lk | |
| [2H6]Campesterol | 0.94 | 1.12 | ||||||
| [2H6]Campest-4-en-3β-ol | nda | nd | 3.68 | 2.82 | nab | na | na | na |
| [2H6]Campest-4-en-3-one | 1.27 | 3.70 | 3.89 | 2.47 | 3.78 | 13.5 | na | na |
| [2H6]5α-Campestan-3-one | nd | nd | nd | nd | nd | nd | 2.34 | 2.45 |
| [2H6]5α-Campestanol | 0.82 | nd | 1.00 | nd. | 1.98 | nd | 0.51 | 1.06 |
Each substrate (25 μg) was incubated with sliced apical portions of pea. The amounts of metabolites (μg g−1 fresh weight) are means of two GC-SIM runs.
nd, Not detected.
na, Not analyzed.
In lk explants, [2H6]campesterol and [2H6]4-en-3β-ol were metabolized to [2H6]4-en-3-one. [2H6]4-En-3-one was not further metabolized, while [2H6]3-one was converted to [2H6]campestanol (Table III). These results clearly show that the lk mutation prohibits the conversion of 4-en-3-one to 3-one. Thus the LK gene should encode a steroid 5α-reductase, suggesting that it is homologous to the Arabidopsis DET2 gene (Li et al., 1996).
The lk Mutation Is Due to the Truncation of the Pea DET2 Homolog
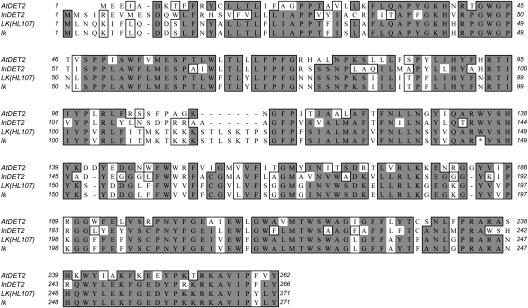
The pea steroid 5α-reductase gene homologous to the Arabidopsis DET2 gene was obtained from wild-type line JI813 and showed 58% identity and 76% similarity with DET2. Primers based on this full-length pea DET2 sequence were used to clone sequences in the wild-type lines HL107 and 212+, and in the 212− (lk) mutant. A mutation in lk was found at base 437, where guanine was changed to adenine, which corresponded to an amino acid change from Trp to a stop codon at amino acid position 146 (Fig. 5), resulting in a truncated protein. Loss of function of the LK protein ultimately leads to an early blockage in BR biosynthesis, resulting in a deficiency of the biologically active BRs, thereby explaining the extreme phenotype of the lk mutant. The sequence from JI813 differed from the HL107 (and therefore also 212+) sequence at two positions, amino acids 190 and 213. The A-G change in both cases led to a K-E amino acid change that would not be predicted to alter the protein function.
Figure 5.
Protein alignment of Arabidopsis DET2 (GenBank accession no. U53860), InDET2 (GenBank accession no. AB106360) with pea wild-type HL107 (GenBank accession no. AY573897), and lk. Conserved residues are boxed and identical residues are shown by gray shade. The first 12 amino acids of HL107 are based on the pea wild-type JI813 sequence.
DISCUSSION
This article demonstrates that the pea LK gene encodes a steroid 5α-reductase and that a lesion in the LK gene causes BR deficiency, resulting in growth suppression. The det2 mutant of Arabidopsis also has a defect in a steroid 5α-reductase (Li et al., 1996), and recently the kbt mutant of Ipomoea nil has been shown to have the same defect (Suzuki et al., 2003). The LK gene showed 60% identity and 74% similarity with DET2, and 59% identity and 77% similarity with InDET2, suggesting that the 5α-reductase gene is well conserved in the plant kingdom. The lk mutant is distinct from det2 and kbt in that its leaf blade is not as rugose and curly (Fig. 1). Furthermore, the de-etiolation characteristics of Arabidopsis and tomato mutants are not seen in lk (Symons et al., 2002). The lk mutation was found to result in a stop codon located close to the midpoint of the gene (Fig. 5). Animal steroid 5α-reductases are membrane bound and require NADPH as the sole cofactor. The human steroid 5α-reductase isozyme-2 has steroid-binding sites at both the N and C termini, as well as NADPH-binding sites in the last half of the protein (Russell and Wilson, 1994; Jin and Penning, 2001). It thus seems that the lk mutation depletes the C-terminal steroid-binding site and the NADPH-binding sites. Furthermore, the lk mutant protein also may not possess catalytic domains that have been predicted to reside near the C terminus of the rat liver steroid 5α-reductase 1 (Wang et al., 1999). Therefore, 5α-reductase activity is predicted to be seriously reduced in the lk mutant.
In both light-grown shoots and dark-grown calli of wild-type plants, the level of campestanol is approximately 5% that of campesterol (Table II), indicating that action of the LK gene is not affected by whether the tissue is differentiated or exposed to light (shoots were grown in light and calli in dark). In contrast, the relative amount of stigmasterol to sitosterol is much reduced in callus (Table II), suggesting that the dehydrogenase activity responsible for the conversion of sitosterol to stigmasterol is suppressed in callus. We recently found that the LK gene transcript levels are not significantly changed during seed growth and seed germination of pea (T. Nomura and T. Yokota, unpublished data). Expression of the DET2 gene has been shown to be ubiquitous and constitutive in Arabidopsis seedlings (Li and Chory, 1999; Bancos et al., 2002) and not to be affected by exogenous brassinolide in contrast to P450 genes involved in BR biosynthesis (Mathur et al., 1998; Goda et al., 2002; Hong et al., 2002; Montoya et al., 2002). Altogether, it seems that the 5α-reductase genes are expressed at a steady state throughout the whole plant.
Although lk plants have a serious mutation, they still contain campestanol as well as considerable levels of BRs (Tables I and II), suggesting the presence of a partially complementary gene(s). Consistent with this, there also seems to be an alternative steroid 5α-reductase in Arabidopsis (Li and Chory, 1999) and I. nil (Suzuki et al., 2003). The DET2 gene embedded in kidney 293 cells 5α-reduced testosterone, progesterone, and androstenedione (Li et al., 1997). 5α-Reduction of progesterone has also been demonstrated to occur in several plants (Stohs and El-Olemy, 1972; Lin and Heftmann, 1981; Wendroth and Seitz, 1990). So, the DET2 gene seems to 5α-reduce both C27–29 steroids and C19/21 steroids. However, steroid 5α-reductases of Solanum malacoxylon reduced campest-4-en-3-one and progesterone, but not testosterone and androstenedione (Rosati et al., 2003). Humans and rats have two steroid hormone 5α-reductases, isozymes type 1 and type 2 (Russell and Wilson, 1994; Jin and Penning, 2001). Shefer et al. (1966) demonstrated that, in rat liver, cholest-4-en-3-one was catalyzed by an enzyme different from steroid hormone 5α-reductases. Altogether, we suspect that 5α-reduction of C19/21 steroids is catalyzed by an enzyme other than DET2, and this enzyme may partially replace the action of DET2.
This article demonstrates that campest-4-en-3-one, the substrate of LK, is synthesized from campesterol (Fig. 4). Various plants have the same type of enzymatic activity that converts pregnenolone to progesterone (Bennett and Heftmann, 1965; Sauer et al., 1967; Capsi and Hornby, 1968; Stohs and El-Olemy, 1972) and sitosterol to sitost-4-en-3-one (Stohs and El-Olemy, 1971). Furthermore, we found that 4-en-3β-ol is the intermediate of this reaction in pea as observed in Arabidopsis (Noguchi et al., 1999b). However, this intermediate has been investigated only in these two plants. The pathway of 5-en-3β-ol → 5-en-3-one → 4-en-3-one has been proposed for C19/21 steroids in bacteria (Talalay and Wang, 1955), mammals (Luu-The et al., 1991), and plants (Seidel et al., 1990; Stuhlemmer and Kreis, 1996; Kreis et al., 1998; compare with Fig. 4). This two-step pathway is catalyzed by 3β-hydroxylsteroid dehydrogenase (3βHSD)/Δ5-Δ4 isomerase (Δ5-3βHSD). Mammalian Δ5-3βHSDs, members of the aldo-keto-reductase (AKR) superfamily, are membrane bound and require NAD+ (Rhéaume et al., 1991; Simard et al., 1991). In contrast, Δ5-3βHSDs of Digitalis lanata (Finsterbusch et al., 1999) and Pseudomonas testosteroni (Yin et al., 1991) are soluble enzymes belonging to the short-chain dehydrogenase/reductase family. Interestingly, Pseudomonas Δ5-3βHSD was demonstrated to have no isomerase activity (Talalay and Wang, 1955; Kawahara et al., 1962). We found that the Arabidopsis genome contains three genes homologous to D. lanata Δ5-3βHSD, with 75% similarity, and four genes similar to human Δ5-3βHSDs, with approximately 50% similarity, indicating the possible presence of 4-en-3-one as the intermediate in plants. It is intriguing to investigate whether Δ5-3βHSDs catalyze the 5-en-3-one pathway or 4-en-3β-ol pathway or both. Furthermore, it will be interesting to investigate if the substrates of Δ5-3βHSDs are C19/21 steroids or C27–29 steroids or both. In rat, hepatic C27 steroid Δ5-3βHSDs are involved in the synthesis of bile acids and cholestanol, but these enzymes were demonstrated to be different from one another and also distinct from C19/21 steroid Δ5-3βHSDs (Björkhem et al., 1972; Björkhem and Karlmar, 1974). The dwarf mutant sax1 of Arabidopsis was predicted not to convert 22-hydroxycampesterol to 22-hydroxycampest-4-en-3-one resulting in BR deficiency (Ephritikhine et al., 1999), suggesting that SAX1, which has not yet been cloned, may play a key role in clarifying the function of plant Δ5-3βHSDs. On the other hand, Streptomyces spp. and Brevibacterium are known to produce flavoproteins that catalyze the conversion of exogenous cholesterol to cholest-4-en-3-one (Horii et al., 1990; Gadda et al., 1997; Yamashita et al., 1998; Venkatramesh et al., 2003). However, a BLAST search indicated that the Arabidopsis genome does not comprise their homologous genes.
We also demonstrated that pea tissue exerts 3βHSD activity converting 5α-campestan-3-one to 5α-campestan-3β-ol (Table III; Fig. 4). Human 3βHSDs have been known to interconvert 5α-androstan-3-ones and 5α-androstan-3β-ols in vitro (Rhéaume et al., 1991). D. lanata 3βHSD was demonstrated to catalyze the interconversion of 5α-pregnane-3,20-dione and 5α-pregnane-3β-ol,20-one and of 5β-pregnane-3,20-dione and 5β-pregnane-3β-ol,20-one (Finsterbusch et al., 1999). Recently, mammalian 3α-hydroxysteroid dehydrogenase (3αHSD), a member of the AKR superfamily as is 3βHSD (Jin and Penning, 2001), was found also to have 3βHSD activity that deactivates 5α-dihydrotestosterone into 5α-androstan-3β,17β-diol (Steckelbroeck et al., 2004). Furthermore, D. purpurea AKR proteins (DpAR1 and DpAR2) have been shown to reduce C21 steroids with 3- and/or 20-carbonyl groups (Gavidia et al., 2002). These findings suggest that 3βHSDs of C28 plant steroids are also members of AKR proteins.
MATERIALS AND METHODS
Plant Material and Harvesting
The pure lines of pea (Pisum sativum) used in this study were lines 212+ (LK) and 212− (lk). The lk mutation (erectoides) originated as a spontaneous mutation in cv Cefalonia Rogue (John Innes line 885; Reid, 1986). Line 212+ is the wild type for 212−, the lk line generated by crossing the original mutant with Hobart line 107.
Unless otherwise stated, pea seedlings were grown either in a growth cabinet or greenhouse under the following conditions. In growth cabinets (Nihon Ikakikai, Tokyo), pea seeds were sown in a tray filled with moist vermiculite and grown under continuous fluorescent light (Toshiba, 40-W daylight-white tube; approximately 240 μmol m−2 s−1 at top of plant). The temperature was maintained at 25°C for the first 3 d, then lowered to 20°C for further growth. In the greenhouse, pea seedlings were grown at 26°C on Super Mix A (Sakatanotane, Yokohama, Japan) under natural light supplemented with fluorescent light, making a 13-h light/11-h dark regime.
For analysis of genes, seeds of each genotype were germinated and grown under an 18-h photoperiod in a heated greenhouse and shoots were harvested after 14 d of growth, frozen in liquid nitrogen, ground to a fine powder, and stored at −70°C.
Calli of the lk Mutant and Wild Type
Wild-type and lk seed were surface sterilized with 70% ethanol for 20 s and antiformin (×10) for 15 min, rinsed with sterilized water, and aseptically sown on 0.9% agar medium containing Murashige and Skoog inorganic salts and incubated at 25°C in a growth cabinet under continuous fluorescent light. Calli were induced in the dark at 25°C from the stems of 11-d-old wild-type and lk plants on a Murashige and Skoog agar medium containing 0.9% agar, 3% Suc, and 1 mg L−1 of 2,4-dichlorophenoxyacetic acid. These calli were subcultured three times under the same conditions before sterol analysis.
Application of Brassinolide
Brassinolide dissolved in 5 μL of ethanol containing 0.15% Tween 20 was applied directly to the fourth internode of 8-d-old seedlings grown in a growth cabinet when the third leaf was fully expanded. Control seedlings were treated just with solvent. Three days after treatment, the lengths of the fourth internodes were measured.
Gas Chromatography-Selected Ion Monitoring
A Jeol JMS-AX 505 instrument equipped with a DB-5 column (0.25 mm × 15 m; 0.25-μm film thickness; J & W Scientific, Folsom, CA) was used. The column oven temperature was set to 170°C for the first 1.5 min, elevated to 280°C at 37°C min−1, and then to 300°C at 1.5°C min−1. The carrier gas was He at the flow rate of 1 mL min−1, the injection port temperature was 260°C, and the samples were introduced by splitless injection.
Extraction and Purification of BRs
Aerial parts of 12-d-old lk seedlings grown in a greenhouse (30.7 g fresh weight) were harvested from 66 seedlings whose average height was 5.2 cm, while those of wild-type plants (71.1 g fresh weight) were harvested from 74 seedlings whose average height was 19.6 cm. The materials were extracted with methanol and [2H6]-labeled internal standards were added to the extract prior to reduction to an aqueous residue. To the extract of the wild-type shoots, the following [2H6]-labeled internal standards were added: 0.2 μg each of [2H6]brassinolide, [2H6]typhasterol, [2H6]3-dehydroteasterone, [2H6]teasterone, [2H6]3-dehydro-6-deoxoteasterone, and [2H6]6-deoxoteasterone, along with 0.5 μg each of [2H6]castasterone, [2H6]6-deoxocastasterone, and [2H6]6-deoxotyphasterol. Half the amounts of the standards were added to the lk shoot extract. The extract was partitioned twice between ethyl acetate and 0.5 m dipotassium hydrogen phosphate (pH 9). The ethyl acetate phase was evaporated to dryness and partitioned three times between n-hexane and 80% methanol. The latter phase was evaporated to dryness and purified on silica gel (Wako gel C300, 0.5 g) eluted with chloroform, and then with chloroform containing 0.5%, 7%, 10%, and 20% methanol. The 7% methanol fraction was purified on Sephadex LH-20 (column volume, 500 mL; Pharmacia, Uppsala), using methanol:chloroform (4:1, v/v) with collection of 10-mL fractions. On the basis of the rice lamina inclination bioassay, fractions 31 to 40 (wild type) and 32 to 39 (lk), respectively, were combined. These fractions were dissolved in methanol and passed through short columns of diethylaminosilica and octadecylsilica (ODS) successively. Eluates with methanol were subjected to HPLC on an ODS column (8 × 250 mm; Senshu Scientific, Tokyo) eluted with an acetonitrile-water gradient at 40°C at a flow rate of 2.5 mL min−1, fractions being collected every minute. The mobile phase was programmed as follows: 0 to 20 min, 45% acetonitrile; 20 to 40 min, 45% to 100% acetonitrile; 40 to 60 min, 100% acetonitrile. The following fractions were analyzed for BRs by gas chromatography-selected ion monitoring (GC-SIM): 15 (brassinolide), 21/22 (castasterone), 31 (teasterone), 36 (typhasterol and 3-dehydroteasterone), 39 (6-deoxocastasterone), 43 (6-deoxoteasterone), 45 (3-dehydro-6-deoxoteasterone), and 47 (6-deoxotyphasterol).
GC-SIM Quantitation of BRs
Random aliquots of extracts derived from pooled plant materials were analyzed in duplicate by GC-SIM. BRs were converted to either monomethaneboronates (MB) or bismethaneboronates (BMB), with pyridine-containing methaneboronic acid (2 mg mL−1) at 70°C for 30 min. Typhasterol, teasterone, 6-deoxotyphasterol, and 6-deoxoteasterone were further trimethylsilylated with N-methyl-N-(trimethylsilyl)trifluoroacetamide to yield methaneboronate-trimethylsilyl ethers (MB-TMSi). The contents of BRs were calculated from the peak area ratios of 2H0 and 2H6 M+ ions. The 2H0/2H6 ions monitored were m/z 528/534 (M+), 374/374, and 155/161 for brassinolide BMB; m/z 512/518 (M+), 358/358, and 155/161 for castasterone BMB; m/z 544/550 (M+), 529/535, and 515/521 for typhasterol MB-TMSi and teasterone MB-TMSi; m/z 470/476 (M+), 316/316, and 155/161 for 3-dehydroteasterone MB; m/z 498/504 (M+), 273/273, and 155/161 for 6-deoxocastasterone BMB; m/z 530/536 (M+), 440/446, and 215/215 for 6-deoxotyphasterol MB-TMSi and 6-deoxoteasterone MB-TMSi; and m/z 456/462 (M+), 231/231, and 155/161 for 3-dehydro-6-deoxoteasterone MB.
Extraction and Purification of Sterols
For shoots, the aerial parts of lk (1.2 g fresh weight; average height, 5.2 cm) and wild-type plants (1.8 g fresh weight; average height, 17.7 cm) were harvested from three 17-d-old seedlings grown in a greenhouse. For calli, 1 g fresh weight of material was used. These materials were extracted with methanol:chloroform (4:1, v/v) with homogenizing. The extract was partitioned twice between ethyl acetate and 0.5 m dipotassium hydrogen phosphate buffer, and the ethyl acetate phases were combined and evaporated to dryness. A portion (100 mg fresh-weight equivalent) of the residual solid was spiked with 1 μg of [2H6] campestanol and heated with 1 N sodium hydroxide in methanol at 80°C for 1.5 h. After evaporating the solvent, the hydrolysate was partitioned twice between chloroform and water. The chloroform phases were combined and evaporated to dryness, dissolved in chloroform, and passed through a column of silica gel (0.3 g). The eluate with chloroform was evaporated to dryness and analyzed by GC-SIM.
GC-SIM Quantitation of Sterols
Random aliquots of extracts derived from pooled plant materials were analyzed in duplicate by GC-SIM under the conditions used for BRs. Sterols were trimethylsilylated with BSTFA at room temperature. The levels of sterols were determined using calibration curves constructed from the ratios of the M+ peak area of [2H6]campestanol TMSi (m/z 480) to those of cholesterol TMSi (m/z 458), 24-methylenecholesterol TMSi (m/z 470), campesterol/24-epicampesterol TMSi (m/z 472), campestanol/24-epicampestanol TMSi (m/z 474), stigmasterol TMSi (m/z 484), sitosterol TMSi (m/z 486), sitostanol TMSi (m/z 488), and isofucosterol TMSi (m/z 484). Campesterol and 24-epicampesterol (22-dihydrobrassicasterol), as well as campestanol/24-epicampestanol, were analyzed as a mixture because they were not resolved by GC.
Feeding Experiments
[2H6]-labeled precursor sterols were prepared as described elsewhere (Noguchi et al., 1999b). Each sterol (25 μg) was dissolved in 3 mL of acetone and mixed with 10 mL of sterilized 0.1 m potassium phosphate buffer (pH 7.4) containing 0.1% Tween 20. The mixture was placed under a stream of N2 to give a sterol emulsion. Apical portions including the apex were excised from two 13-d-old seedlings, which were aseptically grown under the same conditions as used for callus production, briefly washed with sterile water, sliced by 1-mm width, and placed in a 30-mL flask that contained 5 mL of an emulsified [2H6]-labeled sterol. The mixtures were gently stirred on a rotary shaker (90 rpm) for 5 h at 25°C under the same conditions as used for growing the pea seedlings. After incubation, the explants were rinsed with sterilized water and processed under the previously described protocol for extraction and purification of sterols. Quantitation of the metabolites was carried out by GC-mass spectrometry using the absolute calibration curves prepared for the respective metabolites.
Isolation of the DET2 Homolog from Pea
Total RNA extraction was performed using the QIAquick RNeasy kit (Qiagen, Clifton Hill, Victoria, Australia). To remove contaminating DNA, the total RNA was treated using a DNA-free kit (Ambion, GeneWorks, South Australia). The full sequence of the DET2 homolog in pea was isolated by screening 200,000 plaques of a Lambda Zap II cDNA library (Stratagene, La Jolla, CA), prepared from RNA isolated from flowering shoot apices of John Innes line 813 (Hobart line 5ly). The probe used to screen the library was based on the DET2 homolog in HL107 plants, which was partially isolated using degenerate primers (Gregory, 1999).
Sequencing
Reverse transcription (RT)-PCR experiments were performed with 5 μg of total RNA with the GibcoBRL/Life Technologies SuperScript Preamplification system for first-strand cDNA synthesis (Life Technologies, Melbourne, Australia). Standard PCR reactions (Sigma Technical Bulletin; Sigma, New South Wales, Australia) using Taq polymerase were subjected to a PCR program consisting of an initial denaturation at 94°C for 1 min and then 35 cycles of 94°C for 5 s, 50°C for 30 s, and 72°C for 1 min, with a final extension step of 15 min at 72°C. PCR reactions were purified using the QIAquick PCR Purification kit (Qiagen). Primers were designed to cover the entire coding region (based on the JI813 sequence). Oligonucleotide sequences are shown 5′ to 3′ (the numbers shown correspond to positions in the cDNA sequence; F refers to the left forward primer while R refers to the right reverse primer): 1F, 5-TGAATCAAAAGATATTCTTACAAGACG-31; 1R, 851-TGCAACACACCCCAAATTAC-832; 2F, TGACACACAAACATAGTACCTCACA; 2R, 272-AGAAAGGGGGTGATGAGGAT-253; 3R, 363-TGGGGTTTTCGATAAAGTTGA-343. Sequencing reactions were subjected to a PCR program consisting of 40 cycles of 96°C for 20 s, 50°C for 20 s, and 60°C for 4 min, in a Perkin-Elmer thermal cycler using the quick start CEQ 2000 Dye Terminator Cycle Sequencing (DTCS) kit (Perkin-Elmer, Foster City, CA). Sequencings were performed on the capillary fluorescence Beckman Coulter Sequencer (Beckman Instruments, Fullerton, CA). Sequence data analysis was achieved with Sequencher software and MacVector (Accelrys, San Diego).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY573897.
Acknowledgments
We thank Kyomi Shibata and Yumiko Yamada for technical assistance.
This work was supported by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (grant no. 1146007 to T.Y.) and the Australian Research Council (to J.B.R. and L.H.J.K.), by the Human Frontier Research Program (grant no. 2000–162 to T.Y.), by a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists, and by a study visit grant of the Royal Society (to T.N.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.043786.
References
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA (1998) An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancos S, Nomura T, Sato T, Molnár G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RD, Heftmann E (1965) Progesterone: biosynthesis from pregnenolone in Holarrhena floribunda. Science 149: 652–653 [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Koncz C (2002) Brassinosteroid and plant steroid hormone signaling. Plant Cell 14: S90–S110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JD, Kamiya Y (1999) The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA 96: 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Yokota T (2001) Plant steroid hormones, brassinosteroids: current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol 42: 114–120 [DOI] [PubMed] [Google Scholar]
- Björkhem I, Einarsson K, Gustafsson JÅ (1972) 3β-Hydroxy-Δ5-C19- and C21-steroid oxidoreductase activity in rat liver. Steroids 19: 471–476 [DOI] [PubMed] [Google Scholar]
- Björkhem I, Karlmar KE (1974) Biosynthesis of cholestanol: conversion of cholesterol into 4-cholesten-3-one by rat liver microsomes. Biochim Biophys Acta 337: 129–131 [DOI] [PubMed] [Google Scholar]
- Caspi E, Hornby GM (1968) Biosynthesis of plant sterols-III. Mechanism of saturation on ring B in pregnenolone during its conversion to digitoxigenin in Digitalis lanata. Phytochemistry 7: 423–427 [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Oguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, et al (1999. a) The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol 119: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, et al (1999. b) Arabidopsis dwf7/ste is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11: 207–221 [PMC free article] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee M-O, Yoshida S, Feldmann KA, Tax FE (2002) Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semi-dominant and defective in a glycogen synthase kinase3 β-like kinase. Plant Physiol 130: 1506–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA (2000) Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J 21: 431–433 [DOI] [PubMed] [Google Scholar]
- Clouse SD (2002) Brassinosteroid signal transduction: clarifying the pathway from ligand perception to gene expression. Mol Cell 10: 973–982 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Pagant S, Fujioka S, Takatsuto S, Lapous D, Caboche M, Kendrick RE, Barbier-Brygoo H (1999) The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 18: 315–320 [DOI] [PubMed] [Google Scholar]
- Finsterbusch A, Lindemann P, Grimm R, Eckerskorn C, Luckner M (1999) Δ5-3β-hydroxysteroid dehydrogenase from Digitalis lanata Ehrh. – a multifunctional enzyme in steroid metabolism? Planta 209: 478–486 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Li J, Choi Y, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama T, Yokota T, Chory J, et al (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Ann Rev Plant Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Gadda G, Wels G, Pollegioni L, Zucchelli S, Ambrosius D, Pilone M, Ghisla S (1997) Characterization of cholesterol oxidase from Streptomyces hygroscopicus and Brevibacterium sterolicum. Eur J Biochem 250: 369–376 [DOI] [PubMed] [Google Scholar]
- Gavidia I, Pérez-Bermúdez P, Seitz HU (2002) Cloning and expression of two novel also-keto reductases from Digitalis purpurea leaves. Eur J Biochem 269: 2842–2850 [DOI] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory D (1999) The role of brassinosteroids in shoot elongation. BSc (Hons) thesis, The University of Tasmania, Australia
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves ands stem. Plant J 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii M, Ishizaki T, Paik SY, Manome T, Murooka Y (1990) An operon containing the genes for cholesterol oxidase and a cytochrome P-450-like protein from a Streptomyces sp. J Bacteriol 172: 3644–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J (2000) A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev 14: 1485–1497 [PMC free article] [PubMed] [Google Scholar]
- Jin YJ, Penning TM (2001) Steroid 5α-reductases and 3α-hydroxysteroid dehydrogenases: key enzymes in androgen metabolism. Best Pract Res Clin Endocrinol Metab 15: 79–94 [DOI] [PubMed] [Google Scholar]
- Kawahara FS, Wang SF, Talalay P (1962) The preparation and properties of crystalline Δ5-3-ketosteroid isomerase. J Biol Chem 237: 1500–1506 [PubMed] [Google Scholar]
- Khripach V, Zhabinskii V, Groot AD (2000) Twenty years of brassinosteroids: steroidal plant hormones warrant better crops for the XXI century. Ann Bot (Lond) 86: 441–447 [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Yoshida S, Chua N-H (1998) The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10: 1677–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka CV, Cerny RE, Gardner RG, Noguchi T, Fujioka S, Takatsuto S, Yoshida S, Clouse SD (2000) A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol 122: 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis W, Hensel A, Stuhlemmer U (1998) Cardenolide biosynthesis in foxglove. Planta Med 64: 491–499 [Google Scholar]
- Lawrence NL, Ross JJ, Mander LN, Reid JB (1992) Internode length in Pisum. Mutants lk, lka and lkb do not accumulate gibberellins. J Plant Growth Regul 11: 35–37 [Google Scholar]
- Li J, Biswas MG, Chao A, Russell DW, Chory J (1997) Conservation of function between mammalian and plant steroid 5α-reductases. Proc Natl Acad Sci USA 94: 3554–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1999) Brassinosteroid actions in plants. J Exp Bot 50: 275–282 [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC (2001) BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 5916–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Lin JT, Heftmann E (1981) Stereospecific reduction of progesterone by Pisum sativum. Phytochemistry 20: 1017–1022 [Google Scholar]
- Luu-The V, Takahashi M, de Launoit Y, Dumont M, Lachance Y, Labrie F (1991) Evidence for distinct dehydrogenase and isomerase sites within a single 3β-hydroxysteroid dehydrogenase/5-ene-4-ene isomerase protein. Biochemistry 30: 8861–8865 [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593–602 [DOI] [PubMed] [Google Scholar]
- Montoya T, Nomura T, Farrar K, Kaneta T, Yokota T, Bishop GJ (2002) Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 14: 3163–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, et al (2002) Isolation and characterisation of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol 130: 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999. a) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J (1999. b) Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol 120: 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Bishop GJ, Kaneta T, Reid JB, Chory J, Yokota T (2003) The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant Physiol 36: 291–300 [DOI] [PubMed] [Google Scholar]
- Nomura T, Kitasaka Y, Takatsuto S, Reid JB, Fukami M, Yokota T (1999) Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol 119: 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T (1997) Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol 113: 31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242: 161–173 [DOI] [PubMed] [Google Scholar]
- Reid JB (1986) Internode length in Pisum. Three further loci, lh, ls and lk. Ann Bot (Lond) 57: 577–592 [Google Scholar]
- Reid JB, Ross JJ (1989) Internode length in Pisum. Two further gibberellin-insensitivity genes, lka and lkb. Physiol Plant 75: 81–88 [Google Scholar]
- Reid JB, Ross JJ, Hasan O (1991) Internode length in Pisum: gene lkc. J Plant Growth Regul 10: 11–16 [Google Scholar]
- Rhéaume E, Lachance Y, Zhao HF, Breton N, Dumont M, de Launoit Y, Trudel C, Luu-The V, Simard J, Labrie F (1991) Structure and expression of a new complementary DNA encoding the almost exclusive 3β-hydroxysteroid dehydrogenase/Δ5-Δ4-isomerase in human adrenals and gonads. Mol Endocrinol 5: 1147–1157 [DOI] [PubMed] [Google Scholar]
- Rosati F, Danza G, Guarna A, Cini N, Racchi ML, Serio M (2003) New evidence of similarity between human and plant steroid metabolism: 5α-reductase activity in Solanum malacoxylon. Endocrinology 144: 220–229 [DOI] [PubMed] [Google Scholar]
- Russell DW, Wilson JD (1994) Steroid 5α-reductase: two genes/two enzymes. Annu Rev Biochem 63: 25–61 [DOI] [PubMed] [Google Scholar]
- Sauer HH, Bennett RD, Heftmann E (1967) Pregnenolone metabolism in Digitalis lanata. Phytochemistry 6: 1521–1526 [Google Scholar]
- Schultz L, Kerckhoffs LHJ, Klahre U, Yokota T, Reid JB (2001) Molecular characterization of the brassinosteroid-deficient lkb mutant in pea. Plant Mol Biol 47: 491–498 [DOI] [PubMed] [Google Scholar]
- Seidel S, Kreis W, Reinhard E (1990) Δ5-3β-hydroxysteroid dehydrogenase/Δ5-Δ4-ketosteroid isomerase (3β-HSD), a possible enzyme of cardiac glycoside biosynthesis, in cell cultures and plants of Digitalis lanata EHRH. Plant Cell Rep 8: 621–624 [DOI] [PubMed] [Google Scholar]
- Shefer S, Hauser S, Mosbach EH (1966) Studies on the biosynthesis of 5α-cholestan-3β-ol. I. Cholestenone 5α-reductase of rat liver. J Biol Chem 241: 946–952 [PubMed] [Google Scholar]
- Simard J, Melner MH, Breton N, Low KG, Zhao HF, Periman LM, Labrie F (1991) Characterization of macaque 3β-hydroxy-5-ene steroid dehydrogenase/Δ5-Δ4 isomerase: structure and expression in steroidogenic and peripheral tissues in primate. Mol Cell Endocrinol 75: 101–110 [DOI] [PubMed] [Google Scholar]
- Steckelbroeck S, Jin Y, Gopishetty S, Oyesanmi B, Penning TM (2004) Human cytosolic 3α-hydroxysteroid dehydrogenases (3α-HSDs) of the aldo-keto reductase (AKR) superfamily display significant 3β-hydroxysteroid dehydrogenase (3β-HSD) activity: implications for steroid hormone metabolism and activation. J Biol Chem 279: 10784–10795 [DOI] [PubMed] [Google Scholar]
- Stohs SJ, El-Olemy MM (1971) Δ4-β-sitosten-3-one from β-sitosterol by leaf homogenates. Phytochemistry 10: 2987–2990 [Google Scholar]
- Stohs SJ, El-Olemy MM (1972) Pregnenolone and progesterone metabolism by cardenolide producing plants. Phytochemsitry 11: 2409–2413 [Google Scholar]
- Stuhlemmer U, Kreis W (1996) Cardenolide formation and activity of pregnane-modifying enzymes in cell suspension cultures, shoot cultures and leaves of Digitalis lanata. Plant Physiol Biochem 34: 85–91 [Google Scholar]
- Suzuki Y, Saso K, Fujioka S, Yoshida S, Nitasaka E, Nagata S, Nagasawa H, Takatsuto S, Yamaguchi I (2003) A dwarf mutant strain of Pharbitis nil, Uzukobito (kobito), has defective brassinosteroid biosynthesis. Plant J 36: 401–410 [DOI] [PubMed] [Google Scholar]
- Symons GM, Schultz L, Kerckhoffs LKJ, Davies NW, Gregory D, Reid JB (2002) Uncoupling brassinosteroid levels and de-etiolation in pea. Physiol Plant 115: 311–319 [DOI] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán A, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua NH (1995) The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev 9: 97–107 [DOI] [PubMed] [Google Scholar]
- Talalay P, Wang VS (1955) Enzymic isomerization of Δ5-3-ketosteroids. Biochim Biophys Acta 18: 300–301 [DOI] [PubMed] [Google Scholar]
- Venkatramesh M, Karunanandaa B, Sun B, Gunter CA, Boddupalli S, Kishore GM (2003) Expression of a Streptomyces 3-hydroxysteroid oxidase gene in oilseeds for converting phytosterols to phytostanols. Phytochemistry 62: 39–46 [DOI] [PubMed] [Google Scholar]
- Wang M, Bhattacharyya AK, Taylor MF, Tai HH, Collins DC (1999) Site-directed mutagenesis studies of the NADPH-binding domain of rat steroid 5α-reductase (isozyme-1) I: analysis of aromatic and hydroxylated amino acid residues. Steroids 64: 356–362 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He JX, Chen M, Vafeados D, Yang YL, Fujioka S, Yoshida S, Asami T, et al (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wendroth S, Seitz U (1990) Characterization and localization of progesterone 5α-reductase from cell cultures of foxglove (Digitalis lanata EHRH). Biochem J 266: 41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Toyama M, Ono H, Fujii I, Hirayama N, Murooka Y (1998) Separation of the two reactions, oxidation and isomerization, catalyzed by Streptomyces cholesterol oxidase. Protein Eng 11: 1075–1081 [DOI] [PubMed] [Google Scholar]
- Yin SJ, Vagelopoulos N, Lundquist G, Jörnvall H (1991) Pseudomonas 3β-hydroxysteroid dehydrogenase. Primary structure and relationships to other steroid dehydrogenases. Eur J Biochem 197: 359–365 [DOI] [PubMed] [Google Scholar]
- Yin YH, Wang ZY, Mora-Garcia S, Li JM, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]