Abstract
Meristems within the plant body differ in their structure and the patterns and identities of organs they produce. Despite these differences, it is becoming apparent that shoot and root apical and vascular meristems share significant gene expression patterns. Class III HD-Zip genes are required for the formation of a functional shoot apical meristem. In addition, Class III HD-Zip and KANADI genes function in patterning lateral organs and vascular bundles produced from the shoot apical and vascular meristems, respectively. We utilize both gain- and loss-of-function mutants and gene expression patterns to analyze the function of Class III HD-Zip and KANADI genes in Arabidopsis roots. Here we show that both Class III HD-Zip and KANADI genes play roles in the ontogeny of lateral roots and suggest that Class III HD-Zip gene activity is required for meristematic activity in the pericycle analogous to its requirement in the shoot apical meristem.
The primary plant body is composed of root and shoot systems along the apical-basal axis. Activity of the shoot and root apical meristems are responsible for producing the above-ground lateral organs and stems and the below ground primary root, respectively. Lateral organs initiate from the flanks of the shoot apical meristem and the founder cells in the root pericycle, while the vascular tissues are produced by the meristematic activity of the procambium and cambium (Steeves and Sussex, 1989). While these meristems differ significantly from each other both in their structure and in the patterns of organs they produce, they share at least some common functional genetic components. For example, SHOOT MERISTEMLESS (STM) is required for the maintenance of the shoot apical meristem and is expressed in the vascular meristems (Barton and Poethig, 1993; Long et al., 1996), and a CLAVATA signaling system acts to control meristem size in both the shoot and root apical meristems (Clark et al., 1993; Casamitjana-Martinez et al., 2003). However, the extent of functional genetic conservation among the different meristems is largely unknown.
Class III HD-Zip gene family members (PHABULOSA [PHB], PHAVOLUTA [PHV], REVOLUTA [REV], ATHB8, and ATHB15) are putative transcription factors that have been shown to be required for the establishment of the apical meristem and proper pattern formation in lateral organs and vascular tissue in the aerial portion of the plant body. Their expression in the shoot is confined to the apical meristem, the adaxial portions of lateral organs, and procambium and xylem tissue (Baima et al., 1995, 2001; McConnell and Barton, 1998; McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003; Ohashi-Ito and Fukuda, 2003; Eshed et al., 2004). Semidominant gain-of-function phb-1d mutations result in adaxialization of lateral organs (McConnell and Barton, 1998), while gain-of-function rev-10d mutations cause radialization of vascular bundles in the stem (Emery et al., 2003). Both expression and phenotypic analyses have shown ATHB8 to be involved in procambium and xylem cell differentiation (Baima et al., 1995, 2001). Triple loss-of-function phb-6 phv-5 rev-9 plants not only exhibit radialized abaxialized cotyledons but also a complete loss of a functional shoot apical meristem (Emery et al., 2003). These data demonstrate that the Class III HD-Zip genes have a fundamental role in the shoot in establishing a functional apical meristem and polarity in lateral organs and also have a role in establishing polarity in vasculature within the stem.
Acting antagonistically to the Class III HZ-Zip genes are the KANADI genes, which also encode putative transcription factors. KANADI genes are expressed in a pattern complementary to that of the Class III HD-Zip genes in the shoot; KANADI expression is in the developing phloem and abaxial regions of lateral organs early in development (Kerstetter et al., 2001; Emery et al., 2003; Eshed et al., 2004). While KANADI genes do not appear to be required for proper meristem function, they are needed for pattern formation of organs produced by the shoot apical and vascular meristems. The phenotype of kan1-2 kan2-1 kan3-1 plants is similar to that of rev-10d mutants in that the vasculature in the stem is radialized and similar to that of phb-1d mutants in that the lateral organs are adaxialized (Emery et al., 2003; Eshed et al., 2004). Expression of KANADI genes in the shoot apical meristem causes complete abolishment of meristematic activity and uniform expression of KANADI genes in developing lateral organs causes complete abaxialization of those organs (Eshed et al., 2001; Kerstetter et al., 2001; Emery et al., 2003). The complementary nature of both the phenotypes and the expression of the Class III HD-Zip and KANADI genes has led to the model that the juxtaposition of function of these two gene families is required for the proper establishment of pattern formation of all lateral organs and vasculature in the aerial portion of the plant (Eshed et al., 2001; Emery et al., 2003).
In contrast to the primary root and shoot meristems, which are established in embryogenesis, lateral roots are formed de novo from cell divisions initiated in the pericycle of the stele within the primary root. As the lateral root primordium organizes to form a root meristem, it pushes its way through the cell layers of the primary root to emerge into the rhizosphere. By this stage it has acquired all of the tissue types and most of the gene expression patterns observed in the primary root (for reviews, see Casimiro et al., 2003; Casson and Lindsey, 2003). In Arabidopsis, the initial event in lateral root formation is an asymmetric anticlinal division of 6 to 11 founder cells in the pericycle adjacent to the xylem pole to form shorter cells (stage I; stages defined by Laskowski et al., 1995; Malamy and Benfey, 1997). The central founder cells then undergo periclinal divisions, which increase the number of cell layers (stage II). Subsequently, both periclinal and anticlinal divisions increase the size and girth of the primordium (stages III through V). Just prior to emergence from the primary root, the primordium acquires morphological similarity with the root apical meristem, with layers that correspond to a root cap, epidermis, cortex, and endodermis surrounding the central stelar cells (stages VI and VII). Lateral root emergence appears to be due to expansion of existing cells rather than cell division. Cells in postemergent roots continue to expand, but active cell division at the apex indicates the presence of a functional apical meristem. It is not clear at what point the functional apical meristem first arises in the lateral root. Excised lateral roots transferred to a hormone-free environment have the potential to develop an apical meristem at a stage when the lateral root consists of only 3 to 5 cell layers (stage III; Laskowski et al., 1995). In addition, ß-glucuronidase (GUS) marker lines for the endodermis, epidermis, and cortex are visible as early as stage V, VI, and VII, respectively (Malamy and Benfey, 1997). Malamy and Benfey (1997) do conclude, however, that radial organization in the lateral root precedes active meristem formation.
Auxin is a key regulator of lateral root development (Blakely et al., 1982; Laskowski et al., 1995) that is necessary for both the initiation and development of lateral roots (Celenza et al., 1995; Reed et al., 1998; Casimiro et al., 2001; Bhalerao et al., 2002; Marchant et al., 2002; Benková et al., 2003). These studies separate lateral root development into two distinct phases: an early phase in which polar auxin transport is required to deliver auxin to the site of lateral initiation and a later phase in which the developing lateral root becomes hormone autonomous. To date, most mutations associated with lateral root development are in genes encoding components of the hormone transport and signal transduction pathways.
Because of the fundamental roles of Class III HD-Zip genes in meristem function and the complementary nature of the Class III HD-Zip and KANADI gene families in pattern formation in tissues produced by both the shoot apical and vascular meristems, we undertook an investigation of the function of both gene families in roots. Here we show that the KANADI and Class III HD-Zip gene families are expressed in complementary patterns in developing lateral roots and play functional roles in lateral root formation.
RESULTS
Class III HD-Zip and KANADI Gene Family Members Exhibit Complementary Expression Patterns in Lateral Roots
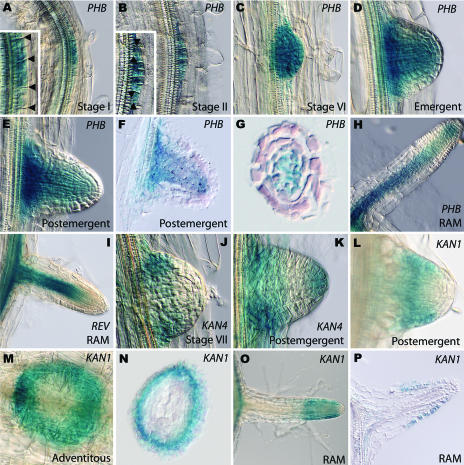
Expression patterns of several Class III HD-Zip genes were observed using GUS expression in promoter GUS fusion lines. A PHB gene trap line and a REV enhancer trap line whose expressions in the aerial portion of the plant reflect in situ hybridization data were used. As the PHB::GUS line is a gene trap, we focused our detailed analysis on this line as it likely reflects the native gene expression. Additionally, a direct promoter fusion of ATHB8 was analyzed. PHB and REV seem to have a similar expression pattern in lateral roots. However, because strong REV expression in the xylem obscures the expression pattern in initiating lateral root primordia, REV expression in stage I and II lateral roots could not be observed. PHB expression is observed as soon as lateral root primordia become morphologically distinct from the rest of the pericycle, after the initial asymmetric division in the pericycle founder cells (Stage I; Fig. 1A). PHB expression remains high throughout the lateral root during stages I to VII prior to emergence (Fig. 1, B and C). At and subsequent to emergence, PHB expression becomes limited to the central region of the lateral root (Fig. 1, D–H). REV and ATHB8 expressions parallel that of PHB during these stages. Subsequently, both PHB and REV are expressed within the differentiating vasculature, with PHB and ATHB8 expressed in the central region of the root apex but REV expression excluded from these cells (Compare Fig. 1, H and I).
Figure 1.
KANADI and Class III HD-Zip expression in Arabidopsis lateral roots. A to H, G to M, and O, Whole mounts showing GUS expression. F, G, N, P, Eight-micrometers wax sections showing GUS expression. A to H, PHB expression. A to C, PHB is detected throughout the lateral root primordia. A, At stage I, anticlinal divisions are observed (arrowheads). B, By stage II, periclinal divisions are evident (arrowheads). C, PHB is expressed throughout stage VI primordia. D, At emergence, PHB is seen to become restricted to the central region of the primordia. E and F, Postemergent primordia do not show PHB gene expression in the outer cell layers. G, This cross section near the root apex clearly illustrates PHB is expressed centrally. H, PHB gene expression in the lateral root apical meristem (RAM). The early expression patterns of both REV and ATHB8 are identical to PHB and therefore are not shown here. I, REV expression in the lateral root apex is not detected in the apical meristem, but is expressed in the vascular cambium. J, KAN4 expression first becomes apparent in Stage VII primordia. K, Postemergence, KAN4 expression is detected in the root cap and in the periphery at the base of the developing lateral root primordium. L to P, KAN1 is expressed in the periphery of lateral root apices. L, KAN1 is first detected in the apex of postemergent lateral roots. M, KAN1 expression in a top view of an adventitious root. N, Cross section near the root apex. O and P, KAN1 expression in the root apical meristem. Note that KAN2 expression is not shown as it exactly matches that of KAN1. KAN3::GUS lines show no expression in either lateral root primordia or root apices.
The expression pattern of the four KANADI genes was observed using KAN1, KAN2, and KAN3 direct promoter GUS fusions and a two component system with KAN4 driving LhG4 in a 6Op::GUS plant (Moore et al., 1998). Because we utilized GUS lines from three separate KANADI genes, all of which show peripheral expression in developing lateral roots, we are confident that this reflects native KANADI expression. All four genes are expressed at low levels in the phloem of the root (Emery et al., 2003; data not shown), with KAN1, KAN2, and KAN4 expressed in additional domains in roots. Subsequent to lateral root initiation, at about stage VII, KAN4 expression is confined to the periphery of the lateral root (Fig. 1J). KAN1 and KAN2 have similar spatial expression patterns in the periphery of lateral roots, but expression commences at the stage of emergence (Fig. 1L). The expression of KAN1 and KAN2 remains consistent in postemergent lateral roots in that expression continues in the periphery of the root apex (Fig. 1, M–P). KAN4, however, remains at the periphery and is also detected in the root cap of the elongating lateral root (Fig. 1K). In situ hybridization experiments confirm the expression of PHB in the central domain and KAN4 in the periphery of the primary root apex. The mutually exclusive expression pattern of the KANADI and the Class III HD-Zip genes in roots is reminiscent of their complementary expression patterns in the aerial organs of the plant.
Changes in KANADI and Class III HD-Zip Activity Do Not Affect Root Vascular Development
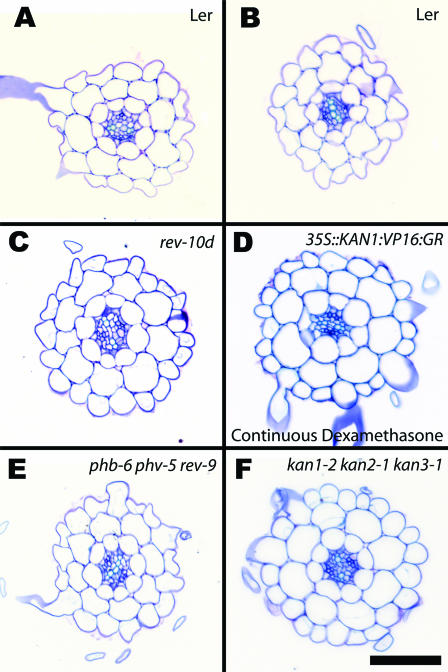
As Class III HD-Zip and KANADI genes are both expressed in developing vasculature in the roots, we inspected the vascular anatomy in mature roots in gain- and loss-of-function alleles from both gene families. There are no major aberrations in the vascular anatomy of any of the mutant genotypes analyzed (Fig. 2).
Figure 2.
Cross sections of roots of wild-type plants and genotypes with altered KANADI and Class III HD-Zip functions. All plants were sectioned 5 DAG except 35S::KAN1:VP16:GR grown continuously on dexamethasone, which was sectioned 7 DAG, as this was when the root was the same length as the other plants. All sections were made at the base of the root toward the hypocotyl to ensure that the vascular cylinder was at maturity. A and B, Landsberg erecta showing the variation seen between individual roots. A, Xylem poles, stained light blue, that do not connect across the center of the vascular cylinder. B, Xylem extending completely across the vascular cylinder. C to F, None of the genotypes with altered KANADI or Class III HD-Zip functions show any clear deviation in vascular development from wild type. Each shows the two clear xylem poles juxtaposed with two poles of smaller phloem cells. C, rev-10d. D, 35S::KAN1:VP16:GR grown continuously on dexamethasone. E, phb-6 phv-5 rev-9. F, kan1-2 kan2-1 kan3-1. Size bar represents 100 μm.
Changes in KANADI and Class III HD-Zip Activity Alter Lateral Root Development
At 10 d after germination (DAG), wild-type Landsberg erecta (Ler) plants produce a primary root averaging 2.4 cm long, from which an average 5.4 lateral roots/cm have initiated (Table I and Fig. 3A). Since both Class III HD-Zip and KANADI genes are expressed in initiating lateral roots, we examined the effects of gain- and loss-of-function alleles from both gene families on the development of lateral roots (Table I; Table II; Fig. 3).
Table I.
Analysis of lateral root production in response to auxin
| Genotype | No Treatment
|
Auxin 70 h
|
||
|---|---|---|---|---|
| Length of Primary Root | Lateral Roots per cm Primary Root | Length of Primary Root | Lateral Roots per cm Primary Root | |
| Landsberg erecta | 2.4 ± 0.6 | 5.4 ± 1.1 | 1.8 ± 0.4 | 11.0 ± 1.7 |
| stm-1 | 2.1 ± 0.7 | 5.2 ± 1.3 | 1.5 ± 0.4 | 12.0 ± 1.6 |
| phb-6 phv-5 rev-9 | 2.1 ± 0.5 | 2.1 ± 1.7 | 1.7 ± 0.3 | 7.1 ± 1.8 |
| phb-1da | 1.1 ± 0.4 | 4.5 ± 1.7 | 1.0 ± 0.3 | 9.0 ± 3.0 |
| rev-10d | 1.7 ± 0.4 | 8.0 ± 1.3 | 1.3 ± 0.4 | 13.0 ± 2.2 |
| kan1-2 kan2-1 kan3-1 | 1.9 ± 0.6 | 2.0 ± 0.9 | 1.4 ± 0.3 | 10.5 ± 1.7 |
| 35S::KAN1:GR | 2.1 ± 0.5 | 5.9 ± 1.1 | 1.8 ± 0.4 | 11.1 ± 1.2 |
| 35S::KAN1:VP16:GR | 2.4 ± 0.6 | 5.5 ± 0.7 | 1.7 ± 0.5 | 11.8 ± 1.1 |
n ≥ 20
As many phb-1d plants did not form a primary root system, the average length of primary roots and the number of laterals per centimeter of primary root were calculated from plants that did produce a primary root with laterals. ± Represents the sd of the data set.
Figure 3.
Whole root systems of wild type and genotypes with altered KANADI and Class III HD-Zip functions 10 DAG with and without auxin treatment. A, C, E, G, K, M, Untreated plants. B, D, F, H, L, Plants treated with auxin for 70 h. I, 35S::KAN1:GR treated with dexamethasone for 94 h. J, 35S::KAN1:GR treated with dexamethasone for 94 h and auxin for 70 h. N, 35S::KAN1:VP16:GR treated continuously with dexamethasone. O, 35S::KAN1:VP16:GR treated continuously with dexamethasone and auxin for 70 h. P, Closer view of the root tip in O. Size bars represent 2 mm.
Table II.
Analysis of lateral root production in genotypes with altered KANADI functions
| Genotype | Continuous Dex
|
Dex 70 h
|
Dex 94 h
|
|||
|---|---|---|---|---|---|---|
| Length of Primary Root | Lateral Roots per cm Primary Root | Length of Primary Root | Lateral Roots per cm Primary Root | Length of Primary Root | Lateral Roots per cm Primary Root | |
| Landsberg erecta | 2.4 ± 0.6 | 5.7 ± 1.0 | 2.4 ± 0.4 | 5.8 ± 1.4 | 2.1 ± 0.4 | 5.6 ± 0.9 |
| 35S::KAN1:GR | 0 | 0 | 2.3 ± 0.5 | 4.0 ± 1.0 | 2.1 ± 0.6 | 3.1 ± 0.9 |
| 35S::KAN1:VP16:GR | 1.1 ± 0.2 | 0 | 2.4 ± 0.4 | 5.5 ± 1.3 | 2.2 ± 0.4 | 5.0 ± 1.5 |
| Genotype | Continuous Dex + Auxin 70 h
|
Simultaneous Dex + Auxin 70 h
|
Dex 94 h + Auxin 70 h
|
|||
|---|---|---|---|---|---|---|
| Length of Primary Root | Lateral Roots per cm Primary Root | Length of Primary Root | Lateral Roots per cm Primary Root | Length of Primary Root | Lateral Roots per cm Primary Root | |
| Landsberg erecta | 1.5 ± 0.2 | 11.0 ± 1.6 | 1.6 ± 0.2 | 11.2 ± 1.5 | 1.7 ± 0.4 | 11.0 ± 1.9 |
| 35S::KAN1:GR | 0 | 0 | 1.8 ± 0.4 | 9.8 ± 1.3 | 1.9 ± 0.5 | 6.3 ± 1.3 |
| 35S::KAN1:VP16:GR | 1.0 ± 0.2 | 3.9 ± 1.8 | 1.7 ± 0.3 | 11.0 ± 1.2 | 1.8 ± 0.4 | 10.7 ± 1.6 |
n ≥ 20; Dex, Dexamethasone; ± represents the sd of the data set.
Loss of Class III HD-Zip activity, as exemplified by phb-6 phv-5 rev-9 triple mutants, results in the production of fewer lateral roots than in wild-type plants (2.1 lateral roots/cm, P < 0.0001), while the length of the primary root was not severely affected (average of 2.1 cm, P > 0.05; Table I; compare Fig. 3, A to E). However, since it has been reported that early initiation of lateral roots requires shoot-derived auxin (Bhalerao et al., 2002) and phb phv rev plants do not produce a shoot system, we examined the root system of shoot meristemless-1 (stm-1) plants, which also do not produce a shoot system. The number of lateral roots produced in stm-1 plants is not significantly different from that of wild-type plants (5.2 lateral roots/cm in stm-1, P > 0.05; Casimiro et al., 2001). In contrast to the reduction in lateral root formation in loss-of-function alleles, in a gain-of-function allele of REV, rev-10d, the number of lateral roots is increased as compared to Ler (average of 8.0 lateral roots/cm, P < 0.0001; compare Fig. 3, A to G). It was also observed that the length of the primary root is reduced to 1.7 cm/plant in rev-10d (P < 0.0001). Surprisingly, approximately one-third of phb-1d plants do not produce a significant primary root with the root system being composed mostly of adventitious roots initiated from the hypocotyls (Fig. 3M). Even when a primary root is present in phb-1d plants, its length is reduced to 1.1 cm/plant (P < 0.0001; Fig. 3K). In phb-1d plants that do produce a primary root system, the number of lateral roots produced is not significantly less than in wild-type plants (P > 0.05).
Surprisingly, loss of KANADI activity, as in kan1-2 kan2-1 kan3-1 triple mutants, also results in reductions in both the length of the primary root (1.9 cm/plant, P = 0.0283) and the number of lateral roots as compared to Ler (2.0 lateral roots/cm, P < 0.0001; compare Fig. 3, A to C). Because constitutive expression of KANADI activity results in seedling lethality (Eshed et al., 2001; Kerstetter et al., 2001), we constructed lines in which KANADI activity is inducible. Fusions of the glucocorticoid receptor (GR) with a protein of interest have the ability to create a steroid hormone-dependent conditional allele (Dalman et al., 1991; Lloyd et al., 1994; Sablowski and Meyerowitz, 1998). Because the cauliflower mosaic virus 35S promoter drives constitutive expression, 35S::KAN1:GR transgenic plants were constructed to allow us to constitutively activate the KAN1 protein upon application of dexamethasone (Benfey et al., 1990). While no difference from wild-type plants was observed in the absence of dexamethasone application (P > 0.05), germination of 35S::KAN1:GR plants on plates containing dexamethasone results in complete inhibition of both shoot and root meristematic activities.
Translational fusions of the strong transcriptional activation domain VP16 to transcription factors have the ability to turn the resulting fusion protein into either strong transcriptional activators or potentially reduce their inhibitory activity (Triezenberg et al., 1988; Cousens et al., 1989; Parcy et al., 1998; Ohta et al., 2001). The weaker phenotype associated with the 35S::KAN1:VP16:GR allow plants to germinate and grow on plates containing dexamethasone. While the aerial portions of these plants produce a normal number and pattern of lateral organs, lateral roots are never observed at 10 DAG (Fig. 3N). However, close inspection demonstrated that these plants occasionally produce lateral root primordia, which are all arrested at stage II. 35S::KAN1:VP16:GR plants do not produce mature lateral roots even after several weeks of growth on dexamethasone-containing media. However, if the primary root grows such that it is no longer in contact with the growth media, occasionally lateral roots are produced, presumably due to locally reduced transgene function. Controls were performed to show that there is no effect of dexamethasone on Ler plants (Table II, all values for P > 0.05).
Effects of Auxin Application
A 70-h application of 1 μm of the auxin indole-3-acetic acid (IAA) to Ler plants (immediately prior to observation at 10 DAG) results in an increased production of lateral roots by 5.6 lateral roots/cm (11.0 lateral roots/cm as compared to 5.4 lateral roots/cm in untreated seedlings; Table I; Fig. 3, A and B). This auxin response in Ler plants is not affected by dexamethasone application (Table II, all values for P > 0.05). Most mutants have a similar response to auxin as does Ler, with an increase of about 5.6 lateral roots/cm upon auxin application (values for P > 0.05). However, four genotypes differ from the wild type in their response to exogenous auxin. Of the Class III HD-Zip and KANADI genotypes, the only mutant line with an increased responsiveness to auxin was the kan1-2 kan2-1 kan3-1 triple mutant, which produced 8.5 lateral roots/cm more than untreated plants (P < 0.0001; Fig. 3, C and D). The mutant lines that had a decrease in the responsiveness to auxin are the KANADI gain-of-function genotypes. 35S::KAN1:VP16:GR plants, which do not produce a single lateral root after 10 d when grown from germination on plates containing dexamethasone, still have a reduced capacity to respond to auxin, producing 3.9 lateral roots/cm (P = 0.0166; compare Fig. 3, N to O). When 35S::KAN1:GR is pretreated with dexamethasone for 24 h, it has a significantly reduced response to auxin of only 3.2 lateral roots/cm (P = 0.0003; Fig. 3, I and J). The presence of the KAN1:GR protein alone does not cause a cessation in growth of lateral roots because simultaneous application of both auxin and dexamethasone for 70 h to 35S::KAN1:GR plants produced an additional 5.8 lateral roots/cm, about as many lateral roots as Ler plants produced in response to auxin (P > 0.05), suggesting that perhaps the action of auxin is faster than that of activating the KAN1:GR protein. The response of 35S::KAN1:VP16:GR plants pretreated with dexamethasone to auxin is comparable to that of Ler plants (P > 0.05). Thus, loss of KANADI activity results in a greater sensitivity to exogenous auxin, whereas increased KANADI activity results in a reduced sensitivity. It is of interest to note that while the phb-6 phv-5 rev-9 triple mutants did not have a significantly different response to auxin (5.0 lateral roots/cm, P > 0.05), the stm-1 plants did have a greater response to auxin as compared to wild-type plants (6.8 lateral roots/cm, P = 0.0403).
DISCUSSION
Members of the Class III HD-Zip and KANADI gene families act in an antagonistic manner to pattern several tissues in the shoot, including meristems, vascular tissues, and lateral organs. In organs produced from the shoot meristem and vascular bundles in the stem, Class III HD-Zip and KANADI genes have complementary expression patterns and complementary loss- and gain-of-function phenotypes. Here we demonstrate that this complementarity is conserved with respect to expression patterns and, to some extent, mutant phenotypes in lateral root ontogeny.
Complementary Class III HD-Zip and KANADI Gene Expression Patterns
Class III HD-Zip expression initiates throughout the lateral root primordium as anticlinal cell divisions within the pericycle define the future position of lateral roots, suggesting that these genes play a role early in lateral root development. Class III HD-Zip gene expression becomes restricted to the central region of lateral roots at emergence at the same time KANADI gene expression is first detected in the periphery. Complementary Class III HD-Zip and KANADI gene expression continues throughout lateral root development. Their complementary patterns suggest that these genes may antagonistically regulate each other's expression in the lateral root. In contrast to the shoot system in which Class III HD-Zip and KANADI genes are required for proper radial pattern formation within the stem, that the radial pattern in lateral roots is already established by the time Class III HD-Zip gene expression becomes restricted to the central region and KANADI gene expression is detected in the periphery suggests that these gene families are not involved in establishing radial patterns in lateral roots. Additionally, that we don't see any significant change in vascular anatomy in any of the Class III HD-Zip or KANADI mutants analyzed suggests that these genes are also not required for radial pattern formation in the primary root.
Complementary Class III HD-Zip and KANADI Loss- and Gain-of-Function Phenotypes
Phenotypic analysis of Class III HD-Zip and KANADI mutants has uncovered a role for these genes in lateral root development. That phb-6 phv-5 rev-9 plants produce fewer lateral roots and rev-10d plants produce more lateral roots than the wild type implicates a role of Class III HD-Zip genes in promoting lateral root formation. That the triple loss-of-function still produces lateral roots, minimally affects the primary root, and shows no obvious change in vascular development may be due to functional redundancy with ATHB8 and ATHB15. While phb-1d mutants often do not have a significant primary root system, they exhibit little change in the pattern of lateral root formation. In contrast, rev-10d mutations do not alter primary root growth as severely but significantly increase the production of lateral roots. This resembles the situation in the shoot where phb-1d primarily affects tissues formed from the apical meristem, while rev-10d primarily affects tissues formed from the vascular cambium. Since lateral roots arise from vascular tissue, it is not unreasonable to expect that the rev-10d gain-of-function allele would have a greater effect than the phb-1d gain-of-function allele.
Continuous ectopic activity of KAN1:VP16:GR from germination completely eliminates the formation of mature lateral roots, while continuous ectopic expression of KAN1::GR completely inhibits primary root growth. While we cannot predict the exact functional outcome of the fusion of the VP16 domain to the KAN1 protein, the difference between these transgenes illustrates that KANADI function in the primary root can be separated from that in lateral root formation. Lateral root primordia that are produced in the 35S::KAN1:VP16:GR plants when they are grown on continuous dexamethasone are arrested at or before stage II, postinitiation, but prior to organization of the lateral root meristem. These phenotypes could be due to the ability of KANADI genes to down-regulate Class III HD-Zip expression or function (Eshed et al., 2001). However, in contradiction to this simple hypothesis, the kan1-2 kan2-1 kan3-1 triple mutant both reduces the length of the primary root and produces fewer lateral roots. This is particularly striking because the kan1-2 kan2-1 kan3-1 phenotype closely mirrors the phenotypes of both phb-1d and rev-10d in the shoot (McConnell and Barton, 1998; Emery et al., 2003). In the root, however, the kanadi triple mutant has a shorter primary root like the Class III HD-Zip gain of function mutants but produces fewer lateral roots in direct opposition to the rev-10d phenotype. These results show that KANADI function can both promote as well as reduce lateral root formation.
Relationship of Class III HD-Zip and KANADI Genes with Auxin in Lateral Root Formation
Auxin has been shown to be a primary regulator of lateral root development, and it has been shown previously that ATHB8 gene expression can be up-regulated in the shoot vasculature by auxin application (Baima et al., 1995), suggesting that in at least some developmental processes, Class III HD-Zip genes function downstream of auxin. rev-10d was the only genotype examined that has increased numbers of lateral roots in the absence of exogenous auxin; however, rev-10d plants did not have an increase in their response to exogenous auxin application. These data suggest that REV has the ability to promote lateral root formation in the absence of exogenous auxin, but this increase is not due to an increase in sensitivity to auxin. Perhaps the REV gain-of-function allele is activating a genetic program that stimulates pericycle cells to attain lateral root identity in a process independent of auxin, a scenario consistent with the hypothesis that Class III HD-Zip genes may act downstream of auxin action.
Most KANADI and Class III HD-Zip mutants examined did not have an altered response to auxin as compared to wild-type plants. The exceptions to this generalization are lines in which KANADI activity is either increased or decreased. Loss of KANADI activity (kan1-2 kan2-1 kan3-1) resulted in an increased responsiveness to auxin, and, conversely, gain of KANADI activity (35S::KAN1:GR and 35S::KAN1:VP16:GR) resulted in a reduced response to auxin. One hypothesis is that alterations in KANADI activity result in changes in the effects of auxin on the activation of the Class III HD-Zip genes, placing KANADI genes downstream of auxin action in the production of lateral organs in the root. That 35S::KAN1:VP16:GR plants separate KANADI function in lateral root initiation and subsequent development may reflect a competition between repression of Class III HD-Zip activity by KANADI and promotion of Class III HD-Zip activity by auxin. The fact that auxin application rescues the ability of 35S::KAN1:VP16:GR plants to produce mature lateral roots supports this hypothesis.
It is interesting that stm-1 plants have a statistically significant increase in their responsiveness to auxin. Though this result just made the cutoff for significance with a P = 0.0403 and the increase is not large, an additional 1.2 lateral roots/cm, the results in Casimiro et al. (2001) show an increase of about 2 lateral roots/cm when plants are treated with 0.1 μm of the auxin 1-naphthylacetic acid for 10 d. The fact that phb-6 phv-5 rev-9 plants, which also do not produce an apical meristem, do not also have an increased response to auxin suggests an additional role for the STM gene in the auxin response in lateral root formation.
Relationship of Class III HD-Zip and KANADI Genes in the Root Apical Meristem
With respect to the primary root, both gain-of-function KAN1:GR and gain-of-function phb-1d genotypes have a deleterious effects on the function of the root apical meristem, with KAN1:GR completely eliminating root apical meristem function when grown from germination on dexamethasone and phb-1d occasionally eliminating function. KAN1:GR likely eliminates Class III HD-Zip activity, which in turn compromises the function of root meristems, both primary and lateral. In contrast, phd-1d likely affects the development of the root apex during embryogenesis, as it has a minimal effect on lateral root meristems. In addition, overall root length is reduced in both KANADI gain-of-function KAN1:VP16:GR and loss-of-function kan1-2 kan2-1 kan3-1 genotypes and in Class III HD-Zip gain-of-function (phb-1d and rev-10d) genotypes. While Class III HD-Zip loss-of-function (phb-6 phv-5 rev-9) does not appear to affect overall root length, it will be interesting to observe if removing additional family members will have an effect on the root apical meristem analogous to their loss-of-function effects on the pericycle, shoot, and vascular meristems.
CONCLUSIONS
Based on their complementary expression patterns, and in most cases, complementary phenotypes, we propose that Class III HD-Zip and KANADI genes have functions in lateral root ontogeny analogous to their functions in the shoot. In this scenario, Class III HD-Zip activity would promote meristematic activity in the pericycle possibly in response to auxin maxima. Postinitiation, Class III HD-Zip expression becomes limited to the root meristems as they become organized via KANADI activity repressing Class III HD-Zip expression in the periphery of the emerging lateral roots. Reduction in formation of lateral roots in loss-of-function Class III HD-Zip genotypes and increased formation in gain-of-function Class III HD-Zip genotypes are consistent with this hypothesis. The surprising reduction in lateral root numbers in plants with reduced KANADI activity could be due to a requirement for juxtaposition of Class III HD-Zip and KANADI functions in the stele to allow for outgrowth of the lateral root primordia from the pericycle, analogous to the situation for lamina growth in developing leaves. As we do not have complete loss-of-function of either Class III HD-Zip or KANADI genes in the root, we cannot determine if either of these genetic pathways is absolutely required for lateral root formation. However, in analogy with their function in the shoot where Class III HD-Zip activity is required for shoot apical meristem formation, we believe that Class III HD-Zip activity is a prerequisite for de novo meristem formation in the pericycle, suggesting the possibility that Class III HD-Zip activity may be required for all meristems in angiosperms.
MATERIALS AND METHODS
Genetics and Molecular Biology
Loss-of-function genotypes have been described previously: kan1-2 kan2-1 kan3-1 and phb-6 phv-5 rev-9 (Emery et al., 2003), phb-1d (McConnell and Barton, 1998), and stm-1 (Barton and Poethig, 1993).
Isolation of the KAN1 cDNA is described in Eshed et al. (2001). The GR domain was obtained from Doris Wagner and the VP16 domain was obtained from Detlef Weigel. The vector pART7 contains the 35S promoter and the 3′OCS terminator sequence (Gleave, 1992). The GR domain was cloned into pART7 using a BamHI/XbaI restriction digest. The translational fusion of the GR domain to the KANADI cDNA was accomplished by adding a BamHI site to the 3′ end of the cDNA using the primer 5′-TATAGGATCCCCTTTCTCGTGCCAATCTGGTCTGCCTAATGT-3′. This fragment was then cloned into pART7 GR with a XhoI/BamHI restriction digest. The translational fusion of the VP16 domain to the GR domain was constructed by adding a 5′ KpnI site using the primer 5′-TATATGGTACCATGCATGCCCCCCCGACCGATGTCAGCCT-3′, and a 3′ BamHI site was added using the primer 5′-TATAGGATCCCCCTGCAGCCCACCGTACTCGTCAATTCCAA-3′. This PCR product was cloned into pART7 GR using a KpnI/BamHI restriction digest. The translational fusion of the KAN1 cDNA to the VP16:GR domains was constructed by adding a 3′ KpnI site using the primer 5′-ATATAGGTACCTTTCTCGTGCCAATCTGGTCTGCCTAATGT-3′. This PCR product was then cloned into pART7 VP16:GR using a XhoI/KpnI restriction digest. All translational fusions were sequenced to verify the accuracy of the PCR reaction.
REV and PHB expression analyses utilized rev-9, a T-DNA enhancer trap insertion in the 5′ untranslated region 364 bases upstream of the translation start site (Eshed et al., 1999; Emery et al., 2003), and phb-6 (SGT4606), a Ds gene trap allele in the first exon of PHB (Parinov et al., 1999). ATHB 8::GUS is a 7.8-kb fragment of Columbia genomic DNA 5′ of the ATG of ATHB8 (amplified using the primers 5′-AAGTCGACCATCAAGGCAAAACAGAGAAGG-3′ and 5′-CGCTGCAGCTTTGATCCTCTCCGATCTCTC-3′) subcloned into pRITAI with a SalI/PstI restriction digest. KAN1/2/3::GUS lines are described in Eshed et al. (2001) and Emery et al. (2003). For KAN4≫GUS, a 7.1-kb fragment 5′ of the ATG of KAN4 was amplified adding a 5′ SalI site using the primer 5′-TTGTCGACCTTATCACTATGGCGTACAATGTG-3′ and a 3′ ClaI site using the primer 5′-TTATCGATCAAAGAATGTGTGGAAAGAGAACG-3′. This fragment was amplified from Colombia genomic DNA and subcloned in front of LhG4 with a SalI/ClaI restriction digest (Moore et al., 1998).
The NotI fragments of these plasmids were introduced into the binary vector pMLBART. All plasmids were introduced into Agrobacterium strain ASE by electroporation and transformed into Ler wild-type plants except KAN4::LhG4, which was transformed into a plant containing 6Op::GUS. Transgenic plants were selected by resistance to the herbicide Basta.
Plant Growth and Conditions
Seeds were sterilized by soaking in 95% ethanol for 15 min and 2% sodium hypochlorite and 0.1% Tween 20 for 15 min and washing twice with sterile water, 5 min each wash. Seeds were placed on plates containing 0.7% agar, Murashige and Skoog salts, and 1% Suc. Plants were grown on plates for 7 DAG and then transferred to plates supplemented with 1 μm IAA and/or 30 μm dexamethasone. For dexamethasone pretreatment experiments, plants were grown on Murashige and Skoog plates for 6 DAG, transferred to 30 μm dexamethasone-containing plates for 24 h, and then transferred to plates containing dexamethasone and 1 μm IAA for 70 h. Controls were transferred directly to 1 μm IAA at 7 DAG. Plants were grown in a Percival Intellus Environmental Controller (Perry, Iowa) at 22°C with 16 h days.
Histology and Histochemistry
Histological staining of GUS activity was performed according to McConnell and Barton (1998). The FeCN concentration was changed as follows: for rev-9, we used 10 mm; for KAN1::GUS and KAN2::GUS, we used 5 mm; and for KAN4≫GUS, phb-6, and ATHB8::GUS, we used 2.5 mm. Roots were visualized as whole mounts under a Zeiss (Jena, Germany) Stemi SV11 dissecting microscope and a Zeiss Axioskop 2 compound microscope equipped with a Zeiss Axiocam digital camera using both bright field and differential interference contrast optics. Stained roots were fixed overnight in 4% paraformaldehyde, dehydrated in ethanol, infiltrated into d-Limonene, embedded into Paraplast-Xtra (Fisher, Loughborough, Leicestershire, UK), and subsequently sectioned at 8 μm.
Roots to be analyzed for aberrations in vascular pattern were fixed overnight in 1.5% glutaraldehyde, 1% paraformaldehyde, and 4% acrolein. They were then dehydrated in ethanol, embedded into JB-4 Plastic (Polysciences, Warrington, PA) as per the manufacturer's instructions, and sectioned at 5 μm. Prior to visualization, sections were stained in 0.1% toluidine blue for 5 min and rinsed in deionized water for 5 min.
In situ hybridization experiments were performed on primary roots of seedlings 5 DAG. Antisense digoxigenin labeled RNA was produced to full length PHB and KAN4 cDNAs.
Statistical Analysis
Lateral roots were quantified by counting the total number of roots along the primary root and dividing by the total length of the primary root. At least 20 plants per treatment per genotype were counted. One-way ANOVA was used to evaluate the difference in means in root length and number of lateral roots per centimeter. Welch ANOVA was used when appropriate to adjust for unequal variances between means. Tukey's honestly significant difference mean-separation test was used to compare the means of all Ler controls. Comparison of the auxin response of mutants to Ler was evaluated using a two-way ANOVA with a full factorial design to consider the interaction between treatment and genotype.
Acknowledgments
We thank Chuck Gasser and Anat Izhaki for helpful discussion and comments on this manuscript; John Emery, Yuval Eshed, V. Sundaresan, Klaus Palme, Doris Wagner, Detlef Weigel, and Kim Richardson for providing materials used in this project; Sandy Floyd for technical assistance in plastic sectioning and microscopy; and Kathren Murrell Stevenson for assistance with statistical analysis.
This work was supported by the National Science Foundation (grant nos. IBN 9986054 and IBN 0234347 to J.L.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.040196.
References
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G (1995) The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182 [DOI] [PubMed] [Google Scholar]
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G (2001) The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK, Poethig RS (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Benfey PN, Ren L, Chua NH (1990) Combinatorial and synergistic properties of CaMV 35S enhancer subdomains. EMBO J 9: 1685–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G (2002) Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Blakely LM, Durham M, Evans TA, Blakely RM (1982) Experimental studies on lateral root formation in radish seedling roots: I. General methods, developmental stages, and spontaneous formation of laterals. Bot Gaz 143: 341–352 [Google Scholar]
- Casamitjana-Martinez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Lindsey K (2003) Genes and signalling in root development. New Phytol 158: 11–38 [Google Scholar]
- Celenza JL Jr, Grisafi PL, Fink GR (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM (1993) CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418 [DOI] [PubMed] [Google Scholar]
- Cousens DJ, Greaves R, Goding CR, O'Hare PO (1989) The C-terminal 79 amino acids of the herpes simplex virus regulatory protein, Vmw65, efficiently activate transcription yeast and mammalian cells in chimeric DNA-binding proteins. EMBO J 8: 2337–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalman FC, Scherrer LC, Taylor LP, Akil H, Pratt WB (1991) Localization of the 90-kDa heat shock protein-binding site within the hormone-binding domain of the glucocorticoid receptor by peptide competition. J Biol Chem 266: 3482–3490 [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of Arabidopsis. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Emery JF, Bowman JL (2004) Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 6: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollma K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM (1995) Formation of lateral root meristems is a two-stage process. Development 121: 3303–3310 [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science 266: 436–439 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Barton MK (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery JF, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Moore I, Galweiler L, Grosskopf D, Schell J, Klaus P (1998) A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci USA 95: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H (2003) HD-Zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol 44: 1350–1358 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395: 561–566 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK (1998) Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM (1989) Patterns in Plant Development, Ed 2. Cambridge University Press, Cambridge, UK
- Triezenberg SJ, Kingsbury RC, McKnight SL (1988) Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev 2: 718–729 [DOI] [PubMed] [Google Scholar]