Abstract
Phenotypic, genotypic, and transcript level (microarray) data from an interspecific backcross population of Eucalyptus grandis and Eucalyptus globulus were integrated to dissect the genetic and metabolic network underlying growth variation. Transcript abundance, measured for 2,608 genes in the differentiating xylem of a 91 (E. grandis × E. globulus) × E. grandis backcross progeny was correlated with diameter variation, revealing coordinated down-regulation of genes encoding enzymes of the lignin biosynthesis and associated methylation pathways in fast growing individuals. Lignin analysis of wood samples confirmed the content and quality predicted by the transcript levels measured on the microarrays. Quantitative trait locus (QTL) analysis of transcript levels of lignin-related genes showed that their mRNA abundance is regulated by two genetic loci, demonstrating coordinated genetic control over lignin biosynthesis. These two loci colocalize with QTLs for growth, suggesting that the same genomic regions are regulating growth, and lignin content and composition in the progeny. Genetic mapping of the lignin genes revealed that most of the key biosynthetic genes do not colocalize with growth and transcript level QTLs, with the exception of the locus encoding the enzyme S-adenosylmethionine synthase. This study illustrates the power of integrating quantitative analysis of gene expression data and genetic map information to discover genetic and metabolic networks regulating complex biological traits.
Wood is composed of secondary xylem, a highly specialized conductive and structural support tissue produced by lateral growth and differentiation of the meristematic vascular cambium (Fukuda, 1996). Genes expressed during the developmentally regulated process of xylogenesis determine the physical and chemical properties of wood. The product of xylogenesis represents one of the world's most important natural resources, yet relatively little is known about the genetic regulation of this process. Wood serves as a renewable source of energy, and it is a sink for atmospheric carbon. Wood is also the raw material for the global pulp and paper, and timber industries. The growth and development of trees and other woody plants are fundamental determinants of the dynamics and composition of forest ecosystems.
At the plant cell level, growth is determined by cell division and expansion. Expansion is driven primarily by internal osmotic pressure generated by water uptake. Expansion is constrained by the cell wall and depends on cell wall composition and the degree of association between its different components (Buchanan et al., 2000). Growth of secondary xylem results from a sequential developmental process that begins with cambial cell division, followed by cell expansion, secondary wall formation, lignification, and programmed cell death (Fukuda, 1996). Wood growth has been associated with the number and rate of cell divisions at the meristematic cambium (Gregory and Wilson, 1968; Wilson and Howard, 1968; Zobel and van Buijtenen, 1989). Indole-3-acetic acid has been shown to function as a positional signal for xylem differentiation (Uggla et al., 1998; Mellerowicz et al., 2001). Specific transcription factors, such as members of the R2R3-MYB gene family, are also likely to control lignification during xylogenesis (Patzlaff et al., 2003). Despite the progress in defining the molecular and cellular processes involved, the mechanisms that determine the rate of xylogenesis (wood growth) and variation in wood properties remain largely unknown.
Variation in growth rate of woody plants has been studied using quantitative trait locus (QTL) mapping (Bradshaw and Stettler, 1995; Grattapaglia et al., 1996; Plomion et al., 1996). Most studies have identified one to three major QTLs for growth traits, explaining in combination from 13% (Grattapaglia et al., 1996) to more than 27% (Wu, 1998) of the phenotypic variation in eucalypt, pine, or poplar. Identification of genes underlying growth QTLs could provide important clues about the biological processes that control trait variation. While much progress has been achieved in QTL analysis methods, only a few studies have identified specific genes that underlie QTLs (Flint and Mott, 2001; Glazier et al., 2002; Korstanje and Paigen, 2002; Morgante and Salamini, 2003).
Our understanding of the cellular and genetic mechanisms that regulate growth in forest trees can be expanded by large-scale analysis of gene expression, such as microarray analysis (Schena et al., 1995). Microarrays have identified clusters of coexpressed genes (Eisen et al., 1998) and allowed inferences about biological processes implicated in plant development and environmental responses (Wullschleger and Difazio, 2003). Recently, the value of microarrays to elucidate the genetic control of gene expression variation was demonstrated in mice and Drosophila melanogaster (Karp et al., 2000; Eaves et al., 2002; Wayne and McIntyre, 2002). Microarrays have been used to determine gene expression levels in segregating populations and identify genomic regions (gene expression QTLs, or eQTLs) explaining transcript variation in coregulated genes (Brem et al., 2002; Schadt et al., 2003; Yvert et al., 2003). When correlated with phenotypic data from a quantitative character, this approach has successfully identified positional candidate genes by colocalizing gene expression QTLs and trait QTLs (Schadt et al., 2003).
Genotypic, phenotypic, and transcript level data were integrated to identify genes controlling growth variation in the forest tree species Eucalyptus. We studied the association between phenotypic variation in diameter growth and the transcript levels of 2,608 cDNAs in the progeny of an elite hybrid of Eucalyptus grandis and Eucalyptus globulus. E. grandis is known for rapid growth and E. globulus for superior wood quality. The backcross progeny of the E. grandis × E. globulus hybrid is particularly suited for the dissection of the molecular mechanisms involved in growth variation because of the genetic diversity and wide segregation that is observed in this population. We have previously genetically characterized this E. grandis backcross progeny (Myburg et al., 2003), and QTLs for diameter growth were identified in the genetic linkage maps of the F1 hybrid backcross parent (Myburg, 2001). In the work reported here, the signal intensity measured on microarrays was used as a quantitative indicator of transcript level variation (Brem et al., 2002; Schadt et al., 2003) and was correlated with quantitative variation of tree growth in this progeny. This analysis revealed coordinated reduction of transcript levels for genes encoding enzymes involved in lignin biosynthesis in the progeny that displayed superior growth (relative to slow growing progeny). This down-regulation of lignin gene transcripts was correlated with direct measurements of lignin content. Quantitative genetic analysis of gene expression indicated that eQTLs for expression of lignin-related genes colocalize with growth QTLs, indicating common regulation.
RESULTS
QTL Analysis of Diameter Growth
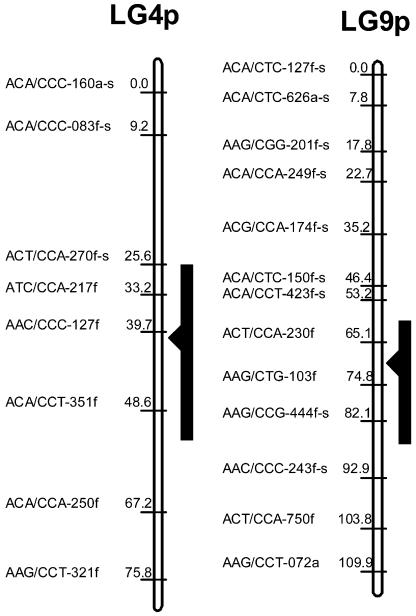
A progeny of each of 186 individuals from an F1 hybrid (E. grandis × E. globulus), backcrossed to an unrelated E. grandis, was previously genotyped with AFLP markers, and genetic maps were generated for the two progeny parents (Myburg et al., 2003). QTL analysis of diameter growth at breast height was carried out for a subset of 91 individuals from this mapping population. These individuals were cloned as rooted cuttings and represent the same range of variation in growth observed in the larger mapping population. QTL analysis by composite interval mapping (Zeng, 1993, 1994) identified two highly significant QTLs (experimentwise α = 0.01) on linkage group 4 (39.7 cM) and linkage group 9 (71.1 cM) of the F1 hybrid (paternal) map (Fig. 1), with likelihood ratios of 26.8 and 18.0, respectively. The two QTLs were of opposite effect, with the QTL on linkage group 4 having a positive additive effect of 1.5 cm, and the QTL on linkage group 9 a negative additive effect of 1.25 cm. The two QTLs jointly explain approximately 30% of the phenotypic variation in growth observed in this family. A marginally significant QTL (likelihood ratio of 12) with negative effect was detected on linkage group 11 (12 cM).
Figure 1.
Growth QTLs on linkage groups 4 and 9 of the paternal map of the E. grandis × E. globulus F1 hybrid tree. AFLP markers are displayed with respective location (cM), and black bars indicate the likelihood ratio interval (experimentwise α = 0.1) of the diameter growth QTLs.
Transcript Profiles of the E. grandis Backcross Progeny
To identify genes whose expression patterns are associated with growth variation in the segregating backcross progeny, transcript levels were estimated for 2,608 cDNAs on a microarray, for each of the 91 individuals of the E. grandis backcross family. These cDNAs represent putative homologs of genes known to be, or are potentially, involved in wood formation. Differentiating xylem tissue was collected during the peak of the growing season to maximize the probability of identifying genes associated with growth variation in this cross. The microarray data were analyzed using two sequential mixed linear models (Jin et al., 2001; Wolfinger et al., 2001) to normalize the data and identify genotypic effects on gene expression. Relative transcript levels were estimated for each individual and each cDNA using least square means. The microarray raw intensity data are deposited in the Gene Expression Omnibus database.
Transcript Level and Growth Correlation
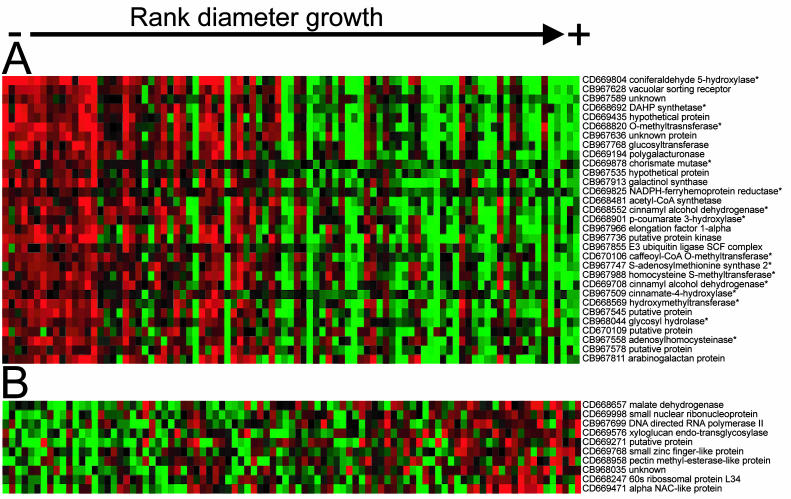
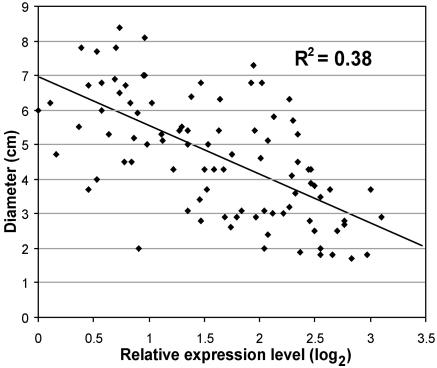
Each cDNA was tested for association between transcript level and diameter growth variation in the backcross progeny using correlation analysis (Neter et al., 1996). The expression patterns of a total of 26 genes were significantly correlated with growth, after a Bonferroni correction for 2,608 tests (individual test significance threshold of 0.000019, corresponding to an experimentwise α = 0.05). A slightly less stringent criteria was adopted (individual test significance threshold of 0.0001) to include a larger sample for further analysis. Lowering the stringency added 11 genes to the set of 26 that were initially identified. The transcript levels estimated for these genes were all negatively correlated with growth (Fig. 2). The most significant correlation was observed for a cDNA representing a putative coniferaldehyde 5-hydroxylase (Cald5H; expressed sequence tag [EST] CD66980; Meyer et al., 1996), also called ferulate 5-hydroxylase, an enzyme at the branch of the phenylpropanoid pathway toward syringyl monolignol biosynthesis (Franke et al., 2000; Li et al., 2000). Transcript variation of Cald5H explains 38% of the diameter growth variation in this E. grandis backcross family (Fig. 3). The majority of the other significantly correlated genes are homologs of enzymes involved in lignin biosynthesis and the phenylpropanoid pathway, including cinnamate-4-hydroxylase (C4H; EST CB967509), 4-coumarate-3-hydroxylase (C3H; EST CD668901), caffeoyl-CoA O-methyltransferase (CCoAOMT; EST CD670106), and O-methyltransferase (OMT; ESTs CD668820 and CD670000; Fig. 4). Negative correlation was also observed for two ESTs representing the enzyme cinnamyl alcohol dehydrogenase (CAD; ESTs CD668552 and CD669708). Lack of additional sequences for Eucalyptus does not allow us to determine whether they represent the same gene or if those are independent genes from the same gene family.
Figure 2.
Association between gene expression and diameter variation. The E. grandis backcross progeny is ranked according to diameter (x axis) and negative (A) or positive (B) correlation between relative transcript level and diameter variation (y axis). Least square means estimates of transcript levels were normalized relative to the mean of each gene across the population. Red represents higher and green represents lower expression relative to the other individuals of the population, for each specific gene. Black indicates no change in mRNA levels. GenBank accession numbers and putative functions are displayed on the right. Genes represented by multiple cDNAs are represented by the most highly correlated, and those related to lignin biosynthesis are indicated by asterisks.
Figure 3.
Transcript level (log2) of the Eucalyptus homolog of Cald5H (EST CD66980), estimated by least square means, and diameter growth variation (cm) in the 91 individuals from the E. grandis backcross population.
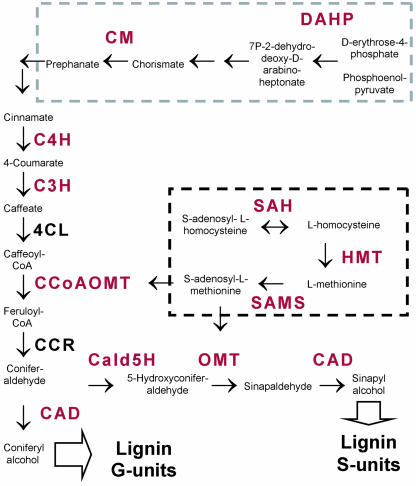
Figure 4.
Biochemical pathways involved in lignin biosynthesis. Simplified representation of the monolignol biosynthesis pathway and partial view of the Met (gray box) and shikimate (black box) pathways. Genes down-regulated in fast-growing trees are in red. Multiple arrows indicate multiple metabolic steps.
This set of coordinately regulated genes included all but two enzymes of the phenylpropanoid pathway, 4-coumarate:CoA ligase (4CL) and cinnamoyl-CoA reductase (CCR), which were represented in the microarray. CCR was represented by one cDNA (EST CB967622), which generated a product of poor quality when amplified by PCR, and, therefore, its correlation with growth remains inconclusive. Four cDNA clones representing 4CL (ESTs CD668307, CD669076, CD668571, and CD669589) were included in the array and produced nonsignificant correlation with growth, suggesting that it may not be subject to the coordinated regulation of the other enzymes of the pathway. 4CL may be subject to different regulation, or there could be no variation for its transcript levels in this cross.
cDNAs that were negatively correlated with growth included those representing genes encoding two enzymes of the shikimate pathway, phospho-2-dehydro-3-deoxyheptonate aldolase synthase (DAHP; EST CD668692) and chorismate mutase (CM; ESTs CD669878 and CB967683), and three enzymes involved in S-adenosylmethionine biosynthesis (Met metabolism), S-adenosylmethionine synthase (SAMS; EC:2.5.1.6, EST CB967747), homo-Cys S-methyltransferase (HMT; EC:2.1.1.14, ESTs CD669142, CD669275, and CD967988), and adenosylhomocysteinase (SAH; EC:3.3.1.1, EST CB967558). The shikimate and Met pathways are both involved in the biosynthesis of substrates for the phenylpropanoid pathway, l-Phe (shikimate) and S-adenosylmethionine (Met; Fig. 4). Additional genes negatively associated with growth included the putative Eucalyptus homologs of a vacuolar-sorting receptor homolog (EST CB967628), genes involved in carbohydrate metabolism (β-[1-3]-glucosyltransferase, polygalacturonase, acetyl-CoA synthetase), and several hypothetical and putative proteins.
Genes with transcript levels positively correlated with growth were not significant at the individual test significance threshold of 0.0001. Ten genes were correlated with diameter growth at a lower stringency (individual test significance threshold of 0.001; Fig. 2). This included a putative xyloglucan endo-transglycosylase (EST CD669576), which is a member of a gene family involved in cell expansion (Darley et al., 2001; Bourquin et al., 2002), and other cell wall associated genes, such as a pectin methyl-esterase (EST CD668958).
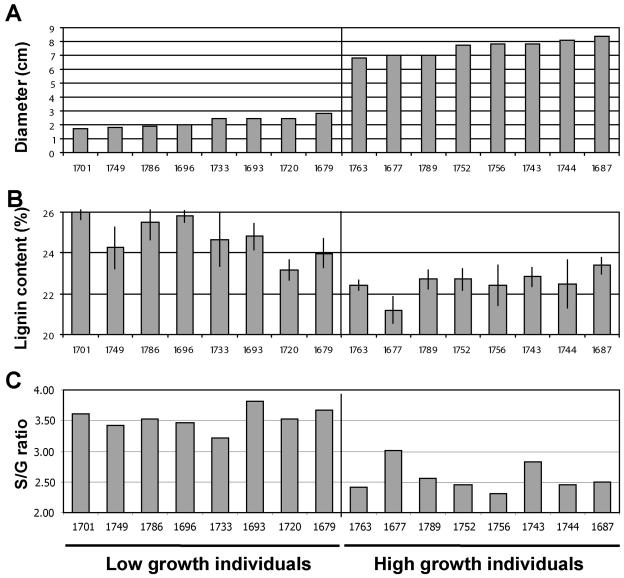
Transcript and Growth Variation Is Associated with Changes in Lignin Content and Quality
To confirm that the variation in expression of the lignin-related genes correlated with diameter growth translated into actual changes of lignin content and composition, eight individuals from the E. grandis backcross progeny were selected for lignin analysis from each of the extremes of the gene expression and growth distributions. Angiosperm lignin is composed of guaiacyl and syringyl monolignol units, which determine lignin's reactivity during hydrolysis (Sarkanen et al., 1967). Lignin composition is measured based on its constituent ratio of syringyl and guaiacyl monomers (S to G ratio). Lignin content and S to G ratios confirmed the expectation from the microarray analysis (Fig. 5). The lignin fraction (acid soluble and insoluble lignin) of the wood averaged 22.5% (0.6% se) in the fast-growing trees and 24.8% (1% se) in the slow-growing trees (Fig. 5B). Considering only the lignin fraction of the wood, lignin content was 10% lower in fast-growing individuals. Substantial differences were also observed in S to G ratios. S units were 38% more abundant in slow-growing trees (Fig. 5C), correlating well with the expected role of Cald5H in the synthesis of S units (Franke et al., 2000; Li et al., 2000), the higher Cald5H transcript levels detected in slow growers, and the colocalization of Cald5H expression QTLs with growth QTLs (see below).
Figure 5.
Growth and lignin properties of the E. grandis backcross population. Diameter growth (A), lignin content (B), and S to G ratios (C) were measured in 16 trees displaying high and low growth. Individual tree identifiers are in the x axis.
Correlated Expression of Lignin-Related Genes
Next, it was tested whether the correlation between transcript levels of the lignin-related genes and growth also translated into a strong correlation among the genes themselves. Lack of correlation among genes of the pathway would suggest independent regulation of gene expression, while the opposite would support the hypothesis that they are under a higher level of genetic control by a limited number of genetic loci. Lack of correlation may also be due to the absence of genes in an existing regulatory network, on the microarray. An analysis of correlation revealed a highly significant (P value <0.0001) association among the expression levels of the genes encoding enzymes of the phenylpropanoid (Cald5H, C4H, C3H, CCoAOMT, OMT, and CAD), shikimate (DAHP and CM), and Met (SAMS, HMT, and SAH) pathways (Table I). The strongest correlation (R2 = 0.82) was detected between two adjacent enzymes in the pathway, Cald5H and OMT, while C4H displayed comparatively weak correlations with all the other genes.
Table I.
Correlation of transcript levels estimated for genes coding for enzymes of the phenylpropanoid, shikimate, and Met pathways
| C4H | C3H | CCoAOMT | Cald5H | OMT | CADa | CADb | DAHP | CM | SAH | HMT | SAMS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C4H | 0.11** | 0.31 | 0.26 | 0.30 | 0.09* | 0.12* | 0.15* | 0.14* | 0.13* | 0.14* | 0.07** | |
| C3H | 0.54 | 0.68 | 0.57 | 0.65 | 0.51 | 0.66 | 0.52 | 0.32 | 0.50 | 0.59 | ||
| CCoAOMT | 0.58 | 0.70 | 0.48 | 0.50 | 0.54 | 0.36 | 0.44 | 0.52 | 0.51 | |||
| Cald5H | 0.82 | 0.50 | 0.55 | 0.77 | 0.60 | 0.52 | 0.63 | 0.57 | ||||
| OMT | 0.49 | 0.59 | 0.69 | 0.57 | 0.53 | 0.58 | 0.52 | |||||
| CADa | 0.66 | 0.51 | 0.28 | 0.23 | 0.34 | 0.49 | ||||||
| CADb | 0.42 | 0.42 | 0.49 | 0.57 | 0.59 | |||||||
| DAHP | 0.56 | 0.38 | 0.48 | 0.48 | ||||||||
| CM | 0.35 | 0.51 | 0.41 | |||||||||
| SAH | 0.65 | 0.50 | ||||||||||
| HMT | 0.68 |
Correlation significance was typically below 0.0001. Exceptions are indicated by * (P < 0.01) and ** (P < 0.05).
EST CD668552.
EST CD669708.
Gene Expression QTLs and Colocalization with Growth
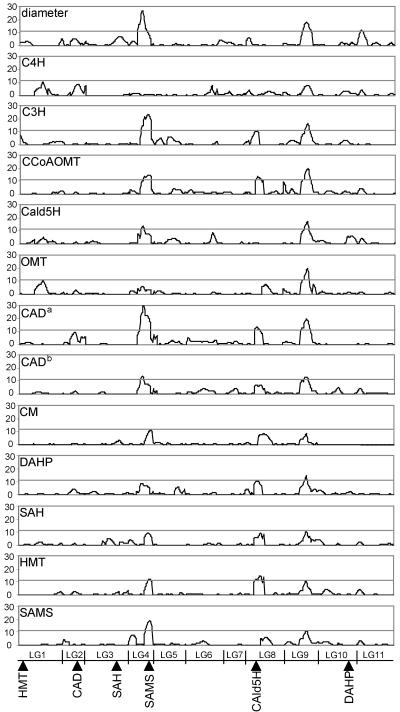
Identification of the gene or genes responsible for coordinated regulation of lignin biosynthesis and growth is difficult due to the lack of genomic information for Eucalyptus. However, mapping of QTLs for gene expression levels (eQTLs) can provide information about regulation by common trans-acting elements, where such elements are genetically located, and how many major loci are involved. The least square means estimates obtained for the cDNAs of the genes encoding enzymes of the phenylpropanoid, shikimate, and Met pathway were combined with the F1 hybrid paternal framework marker data for genome-wide QTL detection scans, using composite interval mapping. With the exception of C4H, eQTLs were detected (at likelihood ratios >11) for all the genes encoding enzymes involved in lignin biosynthesis that were previously identified as highly correlated to growth (Fig. 6). All these genes share a common eQTL, which overlaps with the QTL for growth identified in linkage group 9, with the exception of CM (likelihood ratio at linkage group 9 = 8.1). The majority of these genes also have an eQTL on linkage group 4, which in most cases colocalizes with the growth QTL identified in this linkage group. The presence of pair-wise epistatic interactions between eQTLs was evaluated by multiple interval mapping analysis (Kao et al., 1999), but no significant interactions were detected for the lignin-related genes (data not shown).
Figure 6.
Likelihood ratio profiles generated by composite interval-mapping analysis of diameter growth and expression levels of lignin-related genes. The likelihood ratio scale is indicated on the y axis. The x axis represents the eleven linkage groups of the F1 hybrid paternal map arranged end-to-end. The gray line (likelihood ratio of 11.0) represent the experimentwise α = 0.10. Growth and lignin-related gene expression QTLs colocalize on linkage group 4 (LG4p) and 9 (LG9p). Genetic location of several lignin-related genes are indicated in the lower panel. Footnotes: a, EST CD668552; b, EST CD669708.
Gene Mapping
These results suggest that transcription of these genes is either regulated by trans-acting transcriptional regulators, or that these genes colocalize to the same genomic regions. To evaluate these hypotheses, we mapped some of the genes onto the genetic map of the F1 hybrid (Fig. 6). The mapping results indicate that they are located in various linkage groups, generally not in the same location as their own eQTLs, therefore supporting the hypothesis that they are regulated by trans-acting factors. Genetic location of none of these genes overlaps the growth or gene expression QTLs, with the exception of the SAMS gene, which colocalizes with the growth and gene expression QTLs identified on linkage group 4. SAMS is involved in providing methyl groups for lignin biosynthesis and could represent a regulatory or rate-limiting step. Other regulatory genes involved in the control of transcript levels of genes of the phenylpropanoid, shikimate, and Met-pathway genes may not have been represented in the cDNA microarray or could be regulated other than at the transcript level.
DISCUSSION
We have characterized segregating transcript profiles in an interspecific backcross to E. grandis using microarrays containing 2,608 cDNAs and integrated the phenotypic, genotypic, and transcript data to identify metabolic and regulatory networks implicated in growth variation. Analysis of the association of mRNA abundance patterns in this backcross pedigree revealed that the expression of genes involved in lignin biosynthesis and associated methylation pathways is negatively correlated with diameter growth and is predictive of lignin content and quality in these trees. eQTL analysis of these genes revealed common regulatory loci and colocalization of lignin eQTLs with growth QTLs.
Lignin is the second most abundant component of wood, to which it confers strength, impermeability, and protection against pathogens. Lignin also represents a major obstacle to the efficient use of plant cell wall carbohydrates and plant fibers in food, forage, biomass energy conversion, and wood processing for pulp and paper production. High lignin levels, as one of the main sinks for carbon in the xylem, could limit availability of carbon for cell division and growth. Therefore, higher carbohydrate consumption for more lignin biosynthesis may have a negative effect on growth rate. Alternatively, the lower lignin content detected in fast-growing trees could be a secondary effect resulting from different genetic factors. However, evidence for a primary effect of lignin on growth comes from studies of transgenics where down-regulation of specific genes of the lignin biosynthetic pathway in aspen (Populus tremuloides Michx.) resulted in a significant increase in growth (Hu et al., 1999; Li et al., 2003). In loblolly pine (Pinus taeda), a 50% reduction of CAD enzyme activity was associated with a growth increase of 14% (Wu et al., 1999). Although complex biological traits, such as diameter growth, may be affected by many different physiological processes, we propose that a higher rate of lignin biosynthesis is directly related to reduced growth in this hybrid population. The negative effect of lignin on growth could be due to competition for carbon flow or other mechanisms.
Quantitative analysis of transcript variation measured in widely segregating E. grandis × E. globulus backcross progeny demonstrated the power of microarray technology to dissect complex traits and investigate metabolic networks. Microarray analysis identified coordinate transcription of genes encoding most enzymes of the monolignol biosynthesis branch of the phenylpropanoid pathway, as previously described in Arabidopsis (Harmer et al., 2000) and loblolly pine (Anterola et al., 2002). Our results suggest that this orchestrated transcription extends to other genes in associated metabolic pathways. This includes DAHP and CM from the shikimate pathway, which generates the primary substrate of the phenylpropanoid pathway, l-Phe. These two enzymes are typically inhibited in microbes by a feedback mechanism triggered by excess of l-Phe (Ogino et al., 1982). In Arabidopsis, two of the three existing isoforms of CM are inhibited by l-Phe (Mobley et al., 1999). Other coexpressed genes included SAMS, HMT, and SAH, which encode three enzymes of the Met pathway and provide methyl groups for monolignol biosynthesis. SAMS, HMT, and SAH are some of the most abundant ESTs found in cDNA libraries made from immature xylem tissue of loblolly pine undergoing compression wood formation (M. Kirst, unpublished data), a tissue with high lignin content relative to normal wood. The underexpression of SAMS has been shown to result in a decrease of lignin content in maize (Zea mays; Shen et al., 2002). The coordination of transcript profiles is consistent with the importance of the shikimate and Met pathways in lignin biosynthesis and suggests that transcription of many of the genes in these pathways are under a higher level of coordinated control.
The correlation of gene expression in a segregating progeny can also extend our knowledge about other genes in these pathways. cDNAs representing previously uncharacterized or hypothetical genes, which are strongly correlated with lignin-related genes (such as ESTs CB967589, CD669435, and CB967636) are likely to be involved in this biological process. Similarly, new functions can be tentatively assigned to previously characterized genes that had not been described in the context of lignin biosynthesis, such as a spot 3 protein and a vacuolar sorting receptor homolog (ESTs CB967628 and CD668320).
The QTLs for gene expression and growth colocalized to a high extent, suggesting that a relatively small number of major-effect genes affect growth and lignin biosynthesis in this cross. Several genes encoding enzymes in the general phenylpropanoid pathway and in the synthesis of lignin precursors contain common motifs that are recognized by specific transcription factors, such as MYBs (Borevitz et al., 2000; Patzlaff et al., 2003). The QTLs we identified here could represent such transcription factors. Alternatively, individual genes or clusters of genes could be directly involved through mechanisms such as feedback control, which is known to affect the shikimate and phenylpropanoid pathways (Blount et al., 2000; Guillet et al., 2000). Genetic mapping of candidate genes identified in this study revealed genetic colocalization of SAMS with growth and lignin gene expression QTLs. SAMS' role as the last enzyme in the synthesis of S-adenosylmethionine and its negative effect on the levels of lignin in transgenic maize plants (Shen et al., 2002) suggest that it is an important candidate for further analysis. SAMS may represent a rate-limiting step in lignin biosynthesis. Many other candidate genes that have not been mapped could be located in the same QTL interval. Therefore, a larger collection of genes might identify other candidate genes involved in the regulation of expression of lignin genes.
Efforts to genetically dissect growth traits in forest species have typically identified three to five QTLs (Bradshaw and Stettler, 1995; Grattapaglia et al., 1996; Plomion et al., 1996; Byrne et al., 1997; Wu, 1998; Kaya et al., 1999), which, in combination, accounted for from 13% to 27% of the phenotypic variation. QTLs identified in forest species are typically unstable from age to age (Verhaegen et al., 1997; Emebiri et al., 1998), implying that different genes regulate growth during different stages of growth and development. QTLs identified by phenotypic measurements carried out after several years are likely to represent the accumulated effect of QTLs over the tree lifetime (Weng et al., 2002). Therefore, expression variation assessed at maturity may not reflect differences in expression in the initial years. Growth rate measured on forest species typically has low heritability, indicating that the phenotypic variation is highly dependent on the environment. Possibly only a fraction of the composite number of loci affecting growth play a role in the variation of this hybrid cross, making it easier to dissect genetically. Additional gene expression studies of the progeny, carried out over several years, will tell us more about the stability of the correlation of lignin biosynthesis and growth.
Finally, this work follows from a previous study, in which variation in transcript abundance of the Eucalyptus homolog of the Arabidopsis RCI2 gene explained one-fourth of the phenotypic variation in wood density, twice the amount explained by a QTL identified for the trait (11%; Kirst, 2004). Here, we identified a large set of genes with correlated response relative to transcript abundance and growth, and which explained up to 38% (Cald5H) of the variation in the growth of the progeny. This accounts for more of the growth variation than that explained by each of the two major QTLs individually (18% and 12%) or jointly (approximately 30%). Previous estimates generated from QTL analysis of diameter growth on the entire mapping population indicated a lower proportion (approximately 20%) of the phenotypic variation explained. This result suggests that transcript variation at a gene underlying a QTL is a better predictor of phenotype because transcript level may represent genetic, environmental, and developmental sources of variation that are unaccounted for by the QTL analysis of growth.
MATERIALS AND METHODS
Eucalyptus grandis Backcross Pedigree, Genetic Maps, and QTL Analysis
An F1 hybrid from E. grandis (tree G50) and Eucalyptus globulus (unknown parent in a pollen mix) was backcrossed to an unrelated E. grandis individual (tree 678.2.1) to generate the E. grandis backcross population (Myburg et al., 2003). A subset of 91 individuals from the original E. grandis backcross population was clonally propagated as rooted cuttings and planted on a similar site as the original mapping population near Paysandú (Uruguay). Identification of QTLs for growth, measured as diameter at breast height at 20 months, was performed using composite interval mapping (window size of 10 cM; Zeng, 1993, 1994) in a subset of framework markers with approximately 10 cM spacing, using Windows QTL Cartographer (Department of Statistics, North Carolina State University, Raleigh; Basten et al., 2000). Likelihood ratio thresholds were determined by permutation testing (Churchill and Doerge, 1994; Doerge and Churchill, 1996).
Microarray Design
cDNAs included in the microarray were selected from a unigene set derived from approximately 14,000 ESTs sequenced from five cDNA libraries from E. grandis (differentiating xylem, juvenile and adult leaf, petiole and root) and two libraries from Eucalyptus tereticornis (flower). EST sequences were annotated based on similarity (BLASTX E value < 1E−5) to the latest version of Arabidopsis predicted protein sequences (ftp://ftpmips.gsf.de/cress/arabiprot/) and were functionally classified according to the Gene Ontology Consortium (Ashburner et al., 2000). The microarray comprised 2,608 cDNAs that included the unigene set derived from the differentiating xylem cDNA library (555 cDNAs) and ESTs annotated in the following functional categories: cell wall organization and biogenesis (GO:0007047, 117 cDNAs), cytoskeleton organization and biogenesis (GO:0007010, 91 cDNAs), secondary metabolism (GO:0009699, 187 cDNAs), protein targeting (GO:0006605, 764 cDNAs), cell communication and signal transduction (GO:0007154, GO:0007165; 514 cDNAs), stress response and defense response (GO:0006950, GO:0006952, 441 cDNAs), amino acid metabolism (GO:0006520, 166 cDNAs), nitrogen and sulfur metabolism (GO:0006807, GO:0006790, 69 cDNAs), nucleotide metabolism (GO:0009117, 113 cDNAs), phosphate metabolism (GO:0006796, 134 cDNAs), c-compound and carbohydrate metabolism (GO:0005975, GO:0006730, 387 cDNAs), cell growth, division, and DNA synthesis (GO:0007049, GO:0006259, 262 cDNAs), mRNA transcription (GO:0009299, 333 cDNAs), protein biosynthesis (GO:0006412, 182 cDNAs), transport (GO:0006810, 231 cDNAs), and energy pathways (GO:0006091, 335 cDNAs). Gene Ontology functional categories overlap and therefore many genes are classified into more than one category. Considering the annotation of the Arabidopsis predicted protein sequences, the minimum number of unique features in the cDNA microarray is estimated to be 2,000. cDNA sequences are deposited in GenBank. cDNA clones were amplified by PCR, purified in Multiscreen 96-well filtration plates (Millipore, Bedford, MA), screened for quality in agarose gels, and printed in duplicate on aminosilane-coated glass slides (Corning, Corning, NY) using an Affymetrix 417 Spotter (Santa Clara, CA).
Tissue Collection, RNA Preparation, and Gene Expression Profiling
Differentiating xylem was collected from 20-month-old trees growing in a field plot in Paysandú, Uruguay, over a period of two consecutive days during the peak of the growing season. Tissue was scraped from the entire surface of the first 2 m of the stem. To avoid RNA degradation and gene expression responses to wounding, the bark was removed progressively as the scraping proceeded from top to bottom. The entire procedure consumed less than 5 min per tree. Immediately upon each scrape, the collected tissue was submerged and stored in RNAlater solution (Ambion, Austin, TX) maintained at 10°C to 15°C and transferred to a −20°C freezer within 8 h. The tissue samples were transported to Raleigh (North Carolina) frozen in RNAlater. Upon arrival, samples were transferred to a −80°C freezer, where they remained for 2 to 4 weeks until RNA extraction was carried out. RNA extraction (Chang et al., 1993) was followed by purification in RNAeasy plant mini kit (Qiagen USA, Valencia, CA) columns. RNA integrity was evaluated on agarose gels. Absorbance 260/280 ratios between 1.8 and 2.2 were typically obtained. Total RNA was reverse transcribed, labeled, and hybridized following the aminoallyl labeling method (Hegde et al., 2000). Slides were scanned and the images processed using a ScanArray 4000 Microarray Analysis System scanner and the QuantArray software (Packard Bioscience, Meriden, CT). Signal intensity measurements were deposited in the Gene Expression Omnibus database under the accession numbers GPL348 (Platform), GSM7637 to GSM7727 (Sample), and GSE502 (Series).
Microarray Experimental Design and Statistical Analysis
The experiment followed a loop design (Churchill, 2002) to maximize the number of sampled individuals, while biologically replicating each genotype twice (Cy3 and Cy5). Raw signal intensity values were transformed (log2) and analyzed using two interconnected ANOVA models (Jin et al. 2001; Wolfinger et al., 2001), using PROC MIXED in SAS (SAS Institute, Cary, NC). The normalization ANOVA model yijk = μ + Ai + Dj + Pk + (A × D)ij + (A × P)ik + (D × P)jk + (A × D × P)ijk + ɛijk was used to account for systematic (experiment-wide) sources of variation associated with array (Ai, degrees of freedom (df) = 90, random effect), dye (Dj, df = 1, fixed effect) and pin effects (Pk, df = 3, fixed effect), and interactions. The residuals were treated as normalized values and analyzed in the following ANOVA model (gene model), where the effect of the tree genotypes was evaluated for each gene individually: rilm = μ + Ai + N(A)l(i) + Tm + ɛilm. Tm (df = 90, fixed effect) represents effect of the individual tree, or genotype, on the expression of every gene, from which least square means estimates were calculated. Array (Ai, df = 90, random effect) was included in the model to control for spot effect (Jin et al., 2001; Wolfinger et al., 2001). Each spot printed on one array, for one specific gene, may contain features that are unique (DNA concentration, for instance) relative to the same spot for that same gene, printed on a different slide. Because two different sample mRNAs (one labeled with Cy3 and the other with Cy5) are hybridized to the same slide, the array effect is included to account for covariation between the two samples because they are hybridized to the same spots. Finally, N(A)l(i) (df = 91, random effect) accounts for the spot replication within slides. Failure to account for spot replication results in an artificial inflation of the significances because they do not represent true biological replicates (not independently labeled and hybridized RNA samples; Jin et al., 2001; Churchill, 2002). Interactions between biological (e.g. tree genotype) and technical effects (e.g. array) were evaluated in a complete model and were not found to be significant. Residuals were visually inspected using JMP (SAS Institute) to confirm that consistency of error variances and normality of error terms were obtained. We have previously confirmed the value of quantitative estimates of gene expression for the same dataset (Kirst, 2004). Analyses of correlations were carried out using the Multivariate and Fit model functions in JPM release 5.0 (SAS Institute).
Lignin Content and Monolignol Composition
Cell walls were saponified with 1 m NaOH for 16 h at room temperature, and lignin was extracted and quantified following a microscale Klason method (Kaar and Brink, 1991). Absolute amounts of the guaiacyl (G units) and syringyl (S units) lignin monomers were quantified by derivatization followed by reductive cleavage and solubilization with acetyl bromide (DFRC method of Lu and Ralph, 1997). Reductive cleavage using zinc dust in acetic acid and acetylation was carried out in dichloromethane containing acetic anhydride and pyridine. Samples were dried and stored in the dark for subsequent gas chromatography analysis. The derivatization followed by reductive cleavage process generates 4-acetoxycinnamyl acetate monomers of guaiacyl and syringyl units, which were quantified by gas chromatography using standards provided by John Ralph (Dairy Forage Research Center, U.S. Department of Agriculture) and selective ion monitoring.
Gene Expression QTLs
Least square means estimates of transcript levels were used for QTL analysis using composite interval mapping (Zeng, 1993, 1994) implemented in Windows QTL Cartographer (window size of 10 cM; Basten et al., 2000). Empirical thresholds adopted were determined as described previously (Kirst, 2004).
Gene Mapping
Primers used to PCR-amplify Cald5H, DAHP, SAMS, HMT, and SAH were designed based on EST sequences derived from E. grandis xylem ESTs and are described in Table II. CAD was mapped previously in this population by Myburg (2001). Genes were mapped by genotyping 96 to 160 individuals from the original mapping population using single strand conformation polymorphism (Suzuki et al., 1990; Maruya et al., 1996) and silver staining (Bassam et al., 1991).
Table II.
Primer sequences (5′→3′)
| Gene | Primer Sequence | Annealing Temperature |
|---|---|---|
| Cald5H | F ATCTGCTGGCCTTCTACAGC | 53 |
| R GTTTTAGGCCCACGACACC | ||
| DAHP | F TCACCCAGTGGAATTTGGAT | 54 |
| R AAGCATATGGGCAGAGCAGT | ||
| SAMS | F CTCACTGAGGTCAGGAAGAA | 55 |
| R CCTTAGGAGACCTGCACAAT | ||
| HMT | F GTCCGAGCAGGCATTCTACT | 54 |
| R CCGACTTGAAGCTGCTCACT | ||
| SAH | F CAGATCTCTGTCCGAAGAAG | 54 |
| R CACCACCAGTACTCCTGAAG |
F, Forward; R, reverse.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers CB967505 to CB968059, CD667988 to CD670002, CD670004, CD670097, CD670101 to CD670112 and CD670114 to CD670137.
Acknowledgments
We thank S. Tingey and J. Vogel (DuPont) for kindly providing database information and biological materials for the microarrays; G. Gibson and R. Wolfinger for assistance in establishing the appropriate ANOVA model for the analysis of the microarray data and for comments about the manuscript; B. Sosinski and L. He (Genome Research Laboratory, North Carolina State University) for establishing the microarray facility; and C. Stasolla, L. van Zyl, D. Craig, G. Passador-Gurgel, D. Newmann, L. Solomon, and S. Fekybelu for technical assistance.
This work was supported in part by the National Science Foundation (grant no. DBI 9975806), the North Carolina State University Forest Biotechnology Industry Consortium, and by the North Carolina State University Genomics Program (fellowship to M.K.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037960.
References
- Anterola AM, Jeon JH, Davin LB, Lewis NG (2002) Transcriptional control of monolignol biosynthesis in Pinus taeda: factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J Biol Chem 277: 18272–18280 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196: 80–83 [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B (2000) QTL Cartographer, Version 1.15. A Reference Manual and Tutorial for QTL Mapping. Department of Statistics, North Carolina State University, Raleigh, NC
- Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA (2000) Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol 122: 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14: 3073–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw HD, Stettler RF (1995) Molecular genetics of growth and development in Populus. 4. Mapping QTLs with large effects on growth, form, and phenology traits in a forest tree. Genetics 139: 963–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L (2002) Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL, editors (2000) Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD
- Byrne M, Murrell JC, Owen JV, Kriedemann P, Williams ER, Moran GF (1997) Identification and mode of action of quantitative trait loci affecting seedling height and leaf area in Eucalyptus nitens. Theor Appl Genet 94: 674–681 [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 117–121 [Google Scholar]
- Churchill GA (2002) Fundamentals of experimental design for cDNA microarrays. Nat Genet (Suppl) 32: 490–495 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley CP, Forrester AM, McQueen-Mason SJ (2001) The molecular basis of plant cell wall extension. Plant Mol Biol 47: 179–195 [PubMed] [Google Scholar]
- Doerge RW, Churchill GA (1996) Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves IA, Wicker LS, Ghandour G, Lyons PA, Peterson LB, Todd JA, Glynne RJ (2002) Combining mouse congenic strains and microarray gene expression analyses to study a complex trait: the NOD model of type 1 diabetes. Genome Res 12: 232–243 [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emebiri LC, Devey ME, Matheson AC, Slee MU (1998) Interval mapping of quantitative trait loci affecting NESTUR, a stem growth efficiency index of radiata pine seedlings. Theor Appl Genet 97: 1062–1068 [Google Scholar]
- Flint J, Mott R (2001) Finding the molecular basis of quantitative traits: successes and pitfalls. Nat Rev Genet 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Franke R, McMichael CM, Meyer K, Shirley AM, Cusumano JC, Chapple C (2000) Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J 22: 223–234 [DOI] [PubMed] [Google Scholar]
- Fukuda H (1996) Xylogenesis: initiation, progression, and cell death. Annu Rev Plant Physiol Plant Mol Biol 47: 299–325 [DOI] [PubMed] [Google Scholar]
- Glazier AM, Nadeau JH, Aitman TJ (2002) Finding genes that underlie complex traits. Science 298: 2345–2349 [DOI] [PubMed] [Google Scholar]
- Grattapaglia D, Bertolucci FLG, Penchel R, Sederoff RR (1996) Genetic mapping of quantitative trait loci controlling growth and wood quality traits in Eucalyptus grandis using a maternal half-sib family and RAPD markers. Genetics 144: 1205–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RA, Wilson BF (1968) A comparison of cambial activity of white spruce in Alaska and New England. Can J Bot 46: 733–734 [Google Scholar]
- Guillet G, Poupart J, Basurco J, De Luca V (2000) Expression of tryptophan decarboxylase and tyrosine decarboxylase genes in tobacco results in altered biochemical and physiological phenotypes. Plant Physiol 122: 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J (2000) A concise guide to cDNA microarray analysis. Biotechniques 29: 548–550 [DOI] [PubMed] [Google Scholar]
- Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai CJ, Chiang VL (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17: 808–812 [DOI] [PubMed] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G (2001) The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet 29: 389–395 [DOI] [PubMed] [Google Scholar]
- Kaar WE, Brink DL (1991) Simplified analysis of acid-soluble lignin. J Wood Chem Technol 11: 465–477 [Google Scholar]
- Kao CH, Zeng ZB, Teasdale RD (1999) Multiple interval mapping for quantitative trait loci. Genetics 152: 1203–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, Cuomo PJ, Kohl J, Wahl L, Kuperman D, Germer S, et al (2000) Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol 1: 221–226 [DOI] [PubMed] [Google Scholar]
- Kaya Z, Sewell MM, Neale DB (1999) Identification of quantitative trait loci influencing annual height- and diameter-increment growth in loblolly pine (Pinus taeda L.). Theor Appl Genet 98: 586–592 [Google Scholar]
- Kirst M (2004) Transcription Regulation and Plant Diversity. PhD thesis. North Carolina State University, Raleigh, NC (http://www.lib.ncsu.edu/theses/available/etd-01042004-175350/)
- Korstanje R, Paigen B (2002) From QTL to gene: the harvest begins. Nat Genet 31: 235–236 [DOI] [PubMed] [Google Scholar]
- Li L, Zhou YH, Cheng XF, Sun JY, Marita JM, Ralph J, Chiang VL (2003) Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Natl Acad Sci USA 100: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LG, Popko JL, Umezawa T, Chiang VL (2000) 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J Biol Chem 275: 6537–6545 [DOI] [PubMed] [Google Scholar]
- Lu FC, Ralph J (1997) Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: protocol for analysis of DFRC monomers. J Agric Food Chem 45: 2590–2592 [Google Scholar]
- Maruya E, Saji H, Yokoyama S (1996) PCR-LIS-SSCP (low ionic strength single-stranded conformation polymorphism): a simple method for high-resolution allele typing of HLA-DRB1, -DQB1, and -DPB1. Genome Res 6: 51–57 [DOI] [PubMed] [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W (2001) Unravelling cell wall formation in the woody dicot stem. Plant Mol Biol 47: 239–274 [PubMed] [Google Scholar]
- Meyer K, Cusumano JC, Somerville C, Chapple CCS (1996) Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA 93: 6869–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley EM, Kunkel BN, Keith B (1999) Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis thaliana. Gene 240: 115–123 [DOI] [PubMed] [Google Scholar]
- Morgante M, Salamini F (2003) From plant genomics to breeding practice. Curr Opin Biotechnol 14: 214–219 [DOI] [PubMed] [Google Scholar]
- Myburg AA (2001) Genetic Architecture of Hybrid Fitness and Wood Quality Traits in a Wide Interspecific Cross of Eucalyptus Tree Species. PhD thesis. North Carolina State University, Raleigh, NC (http://www.lib.ncsu.edu/theses/available/etd-20010723-175234/)
- Myburg AA, Griffin AR, Sederoff RR, Whetten RW (2003) Comparative genetic linkage maps of Eucalyptus grandis, Eucalyptus globulus and their F1 hybrid based on a double pseudo-backcross mapping approach. Theor Appl Genet 107: 1028–1042 [DOI] [PubMed] [Google Scholar]
- Neter J, Kunter MH, Nachtshein CJ, Wasserman W (1996) Applied Linear Statistical Models, Ed 4. WCB/McGraw-Hill, Chicago
- Ogino T, Garner C, Markley JL, Herrmann KM (1982) Biosynthesis of aromatic compounds - C13 NMR spectroscopy of whole Escherichia coli cells. Proc Natl Acad Sci USA 79: 5828–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, et al (2003) Characterisation of a pine MYB that regulates lignification. Plant J 36: 743–754 [DOI] [PubMed] [Google Scholar]
- Plomion C, Durel CE, O'Malley DM (1996) Genetic dissection of height in maritime pine seedlings raised under accelerated growth conditions. Theor Appl Genet 93: 849–858 [DOI] [PubMed] [Google Scholar]
- Sarkanen KV, Chang H-M, Allan GG (1967) Species variation in lignins. 3. Hardwood lignins. Tappi 50: 587–590 [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302 [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470 [DOI] [PubMed] [Google Scholar]
- Shen B, Li CJ, Tarczynski MC (2002) High free methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-L-methionine synthetase 3 gene. Plant J 29: 371–380 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Orita M, Shiraishi M, Hayashi K, Sekiya T (1990) Detection of Ras gene mutations in human lung cancers by single-strand conformation polymorphism analysis of polymerase chain reaction products. Oncogene 5: 1037–1043 [PubMed] [Google Scholar]
- Uggla C, Mellerowicz EJ, Sundberg B (1998) Indole-3-acetic acid controls cambial growth in Scots pine by positional signaling. Plant Physiol 117: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaegen D, Plomion C, Gion JM, Poitel M, Costa P, Kremer A (1997) Quantitative trait dissection analysis in Eucalyptus using RAPD markers. 1. Detection of QTL in interspecific hybrid progeny, stability of QTL expression across different ages. Theor Appl Genet 95: 597–608 [Google Scholar]
- Wayne ML, McIntyre LM (2002) Combining mapping and arraying: an approach to candidate gene identification. Proc Natl Acad Sci USA 99: 14903–14906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng C, Kubisiak TL, Nelson CD, Stine M (2002) Mapping quantitative trait loci controlling early growth in a (longleaf pine x slash pine) x slash pine BC1 family. Theor Appl Genet 104: 852–859 [DOI] [PubMed] [Google Scholar]
- Wilson BF, Howard RA (1968) A computer model for cambial activity. Forest Sci 14: 77–90 [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8: 625–637 [DOI] [PubMed] [Google Scholar]
- Wu RL (1998) Genetic mapping of QTLs affecting tree growth and architecture in Populus: implication for ideotype breeding. Theor Appl Genet 96: 447–457 [DOI] [PubMed] [Google Scholar]
- Wu RL, Remington DL, MacKay JJ, McKeand SE, O'Malley DM (1999) Average effect of a mutation in lignin biosynthesis in loblolly pine. Theor Appl Genet 99: 705–710 [DOI] [PubMed] [Google Scholar]
- Wullschleger SD, Difazio SP (2003) Emerging use of gene expression microarrays in plant physiology. Comp Funct Genom 4: 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L (2003) Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet 35: 57–64 [DOI] [PubMed] [Google Scholar]
- Zeng ZB (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA 90: 10972–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel BJ, van Buijtenen JP (1989) Wood variation. Springer-Verlag, Heidelberg