Summary
MHC restriction is a fundamental tenet of T cell biology, but the underlying mechanisms have remained controversial. The extent to which T cell receptors are biased towards MHC proteins in particular has been widely discussed. In a recent paper, Sharon et al. report direct evidence for co-evolution between TCR and MHC genes, helping explain how MHC compatibility and bias can be encoded within T cell receptors.
MHC restriction is a fundamental tenet of T cell biology, yet how a single receptor simultaneously engages both self (MHC protein) and non-self (peptide) remains a topic of considerable discussion. The parameters of this are whether MHC restriction is a result of an intrinsic bias between TCRs and MHC proteins, or whether it is a functional consequence of T cell selection or signaling mechanisms. Over the past decade, much research has been performed investigating these two extremes and possibilities in between. Structures of TCR-peptide/MHC complexes revealed that TCRs often – although not exclusively – align their germline-encoded loops alongside the MHC protein and their hypervariable loops along the peptide. This alignment, whereby the most diverse portion of the receptor tends to be paired with the most diverse portion of the ligand, could be compatible with MHC restriction emerging from an intrinsic physical bias between TCR and MHC genes. Indeed, Garcia and colleagues noted in the late 2000s that multiple structures with TCRs and mouse class II MHC proteins showed similar patterns of contacts between germline-encoded loops and MHC α helices [1], mutation of which impaired TCR binding or T cell development in mice [2].
On the other hand, the structural alignment could result from biological mechanisms that ensure “proper” binding, perhaps to ensure peptides are appropriately sampled by hypervariable loops, the TCR is able to signal, and/or co-receptor is able to engage the TCR-peptide/MHC complex [3]. Moreover, with the accumulation of more structures, it has become apparent that although patterns of TCR-MHC interactions can be conserved, there are no conserved TCR-MHC contacts at the atomic level, even with TCRs that share the same variable domains [4]. In one case, the same TCR shifts dramatically over the same MHC when the peptide is altered, altering the patterns of TCR-MHC interactions [5]. Singer and colleagues have shown that in MHC-deficient mice TCRs can bind other targets [6]. More recently, Rossjohn and colleagues identified TCRs that bound with “reverse” polarity [7, 8]. In this case, portions of an MHC molecule that are usually bound by one TCR chain interact with the other chain of the TCR.
Although some of the structural variances noted above had been previously predicted [9], what has been lacking is a concrete demonstration of actual linkage between TCRs and MHC genes. Given the highly polymorphic nature of the MHC, an intrinsic bias should result in genetic links with TCRs. The recent studies of Sharon et al. published in Nature Genetics provide evidence for this link [10]. Using expression quantitative trait loci (eQTL) analysis, the authors show linkage between multiple MHC and TCR genes. The signals are strongest for class II MHC proteins, and some of the signals map to polymorphic sites that contact the germline regions of the receptor in TCR structures with class II MHC/peptide complexes (Figure 1). The work therefore provides a significant piece of data that has until now been missing – TCR genes do indeed appear to have co-evolved with MHC proteins. The convergence with structural data strongly suggests that at least one of the underlying reasons is to ensure that the repertoire of TCR genes is compatible with polymorphic MHC proteins.
Figure 1. Molecular Interactions between MHC and TCR.
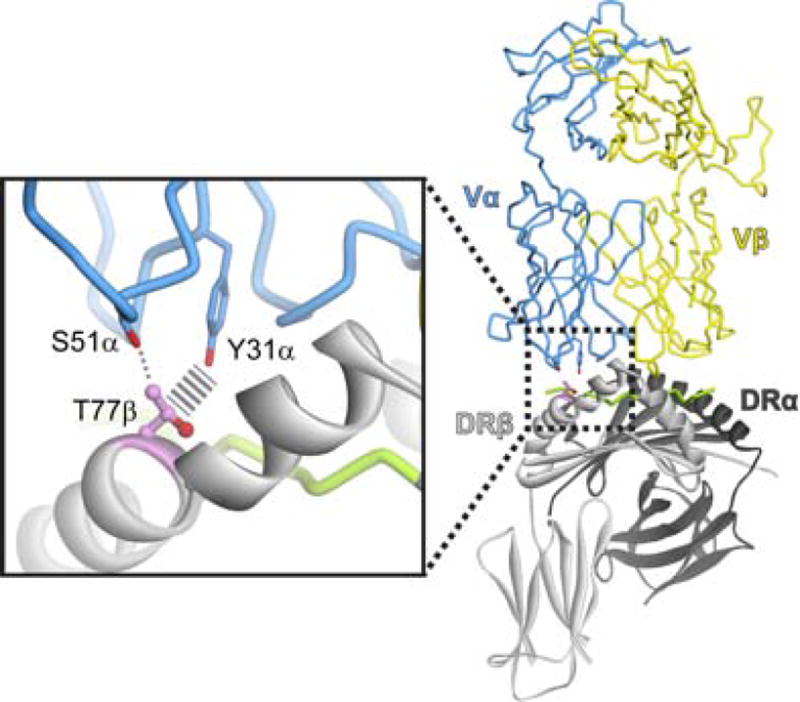
Structural overview of the complex between the class II MHC protein HLA-DR4 and the αβ TCR HA1.7 as presented in [15], highlighting the polymorphic position 77 in the DRβ chain and the interactions it forms with germline residues of the CDR1α and CDR2α germline loops in the HA1.7 TCR. The identify of position 77β co-varies with TCR genes as shown in Sharon et al. [10].
How best to reconcile this new data with the wide structural variances seen in TCR-peptide/MHC complexes and the fact that TCRs can bind targets other than MHC proteins? With >1018 theoretically possible TCRs, it is not difficult to imagine that some TCRs would bind in an unusual manner, or bind non-MHC targets. Perhaps the answer is found in the word compatible and the fact that a bias does not necessarily reflect a hard-wired necessity. Blevins and colleagues showed that TCRs binding to the common class I MHC protein HLA-A*0201 must counteract a positive charge cluster on the MHC α1 helix that is almost unique to A*0201 [11]. TCRs use negative charges in germline loops to do this, but can do so in a variety of ways. Yet Blevins et al. showed that the capacity to offset charges on A*0201 is built-in to human TCR genes – an evolved compatibility. If this were absent, TCRs would be required to offset the charges in other ways, limiting the number of TCRs that could engage A*0201 and thus the range of peptide antigens that could be recognized. The fact this does not happen leads to an intrinsic bias between TCRs and A*0201 by definition. Similar features of evolved compatibility, with ranges of compatibility and strengths of interatomic “glue” likely exist elsewhere between TCR and MHC proteins, contributing to the genetic linkage detected by Sharon et al. [10].
While the detection of a TCR-MHC genetic linkage provides a concrete step forward in understanding MHC restriction, several questions remain to be addressed. For example, how does an evolutionarily-driven compatibility influence positive and negative selection of thymocytes? Is a bias in the intact αβ TCR already manifest in the pre-TCR stage, when all thymocytes express the same pre-TCR-α chain [12]? Does a genetic bias influence initial affinity and bond lifetimes to set in motion the TCR signaling mechanisms and the inherent cross-reactivity of T cells [13, 14]? How do biases contribute to autoimmunity, and can transplantation across MHC barriers, for example after reconstitution with allogeneic hematopoietic stem cells, “break” their influence? Are co-evolved regions targets for biochemical manipulation for improving designer T cells? Lastly, how does a bias integrate with other biophysical parameters to generate not only a wide range of structural solutions but also unique effector functions associated with T cell activation? Understanding these questions and TCR-MHC recognition strategies in general will allow greater insight into immune function and influence progress in areas such as autoimmunity, graft rejection, and anti-tumor immunity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Garcia KC, et al. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott-Browne JP, et al. Germline-encoded amino acids in the [agr][bgr] T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rangarajan S, Mariuzza R. T cell receptor bias for MHC: co-evolution or co-receptors? Cell Mol Life Sci. 2014:1–10. doi: 10.1007/s00018-014-1600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadinski Brian D, et al. A Role for Differential Variable Gene Pairing in Creating T Cell Receptors Specific for Unique Major Histocompatibility Ligands. Immunity. 2011;35:694–704. doi: 10.1016/j.immuni.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borbulevych OY, et al. TCRs Used in Cancer Gene Therapy Cross-React with MART-1/Melan-A Tumor Antigens via Distinct Mechanisms. J Immunol. 2011;187:2453–2463. doi: 10.4049/jimmunol.1101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tikhonova Anastasia N, et al. αβ T Cell Receptors that Do Not Undergo Major Histocompatibility Complex-Specific Thymic Selection Possess Antibody-like Recognition Specificities. Immunity. 2012;36:79–91. doi: 10.1016/j.immuni.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beringer DX, et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat Immunol. 2015;16:1153–1161. doi: 10.1038/ni.3271. [DOI] [PubMed] [Google Scholar]
- 8.Gras S, et al. Reversed T Cell Receptor Docking on a Major Histocompatibility Class I Complex Limits Involvement in the Immune Response. Immunity. 2016;45:749–760. doi: 10.1016/j.immuni.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Marrack P, et al. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharon E, et al. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat Genet. 2016;48:995–1002. doi: 10.1038/ng.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blevins SJ, et al. How structural adaptability exists alongside HLA-A2 bias in the human αβ TCR repertoire. Proceedings of the National Academy of Sciences. 2016;113:E1276–E1285. doi: 10.1073/pnas.1522069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallis RJ, et al. Pre-TCR ligand binding impacts thymocyte development before alphabetaTCR expression. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:8373–8378. doi: 10.1073/pnas.1504971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu LC, et al. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 14.Zarnitsyna VI, et al. Estimating the diversity, completeness, and cross-reactivity of the T cell repertoire. Frontiers in immunology. 2013;4:485. doi: 10.3389/fimmu.2013.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennecke J, et al. Structure of a covalently stabilized complex of a human alphabeta T- cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. Embo J. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]


