Abstract
The N gene of tobacco (Nicotiana tabacum) is a typical resistance (R) gene engendering localization of tobacco mosaic virus (TMV) infection and the elicitation of a hypersensitive necrotic response. The consensus model for R gene-derived resistance is at the level of protein:protein interactions, in which proteins of the pathogen interact with already present receptor-like proteins produced by the plant's R genes. This article demonstrates, by quantitative real-time reverse transcription-PCR analysis, that in tobacco carrying the dominant allele N, a basal level of transcription indeed occurs in noninfected plants. However, accumulation of N-mRNA in infected plants indicates that transcription is stimulated by TMV infection (up to 38-fold in locally infected leaves and up to 165-fold in upper, noninoculated leaves). Potato virus Y infection did not result in accumulation of N-mRNA, indicating a specific TMV-related phenomenon. The possible uncoupling of viral restriction from necrosis is discussed.
Plants carry resistance (R) genes that often engender resistance to specific pathogens. The current consensus model is based on Flor's (1942, 1956) gene-for-gene theory, whereby a product of a pathogen's gene (avirulent gene; avr) interacts with a receptor-like protein produced from the plant's R gene (Keen, 1992). This triggers a cascade of downstream-transduced events leading to resistance. Several avr and R genes have been isolated, substantiating the gene-for-gene model (e.g. van der Ackerveken et al., 1993; Matthieu et al., 1994; Whitham et al., 1994; Bent, 1996; De Wit, 1997; Warren et al., 1998; Ellis et al., 1999; Richter and Ronald, 2000). The model suggests a protein:protein interaction for the initiation of the resistance pathway, in which the R-gene product is a cytoplasmic receptor and the pathogen gene provides the ligand. Leu-rich repeat (LRR) domains of proteins are known to be involved in protein:protein interactions (Lee and Vallee, 1990; Suzuki et al., 1990; Warren, et al., 1998).
The tobacco (Nicotiana tabacum) resistance gene N is a typical R gene, introduced into tobacco from Nicotiana glutinosa (Holmes, 1938). Native tobacco cultivars (tobacconn) are susceptible to tobacco mosaic virus (TMV) infection, allowing the systemic spread of the virus and causing mosaic symptoms. Tobacco cultivars into which the N. glutinosa N gene has been introgressed (tobaccoNN) localize TMV infection to cells adjacent to the site of viral entry and develop a hypersensitive response in the form of local necrotic lesions. The avr gene carried by TMV is the helicase domain of the viral 126-kD protein that is part of the virus replicase complex, since a mutation in this viral gene overcomes localization (Padgett and Beachy, 1993) and the expression of this fragment in transgenic plants engenders necrosis in tobacco (Abbink et al., 1998; Erickson et al., 1999). The N. glutinosa N gene has been isolated and analyzed (Whitham et al., 1994). It was found to be a typical receptor, carrying a sequence similar to the Drosophila Toll and the mammalian interleukin-1R (TIR) receptor. It also carries nucleotide-binding (NBS) and LRR domains. Several R-gene products carry similar motifs (e.g. van der Biezen et al., 2002).
Alternative splicing of the N gene's primary transcript is stimulated by TMV infection, and the two resultant proteins are required for full resistance (Dinesh-Kumar and Baker, 2000). Structural-functional analysis has indicated that the three N domains (TIR, NBS, and LRR) play a role in N-induced resistance (Dinesh-Kumar et al., 2000). The involvement of protein kinases has also been indicated in the N-derived hypersensitive response (Jin et al., 2003).
This article indicates that N-gene transcription is also up-regulated by TMV infection.
RESULTS
Expression of the N Gene and Its Homologs in Tobacco Leaves
Tobacco carries a multitude of N-related gene sequences, which are probably clustered together (Whitham et al., 1994). Therefore, any expression analysis needs to differentiate between the expression of the N gene itself and possible expression of the N-homologous sequences. To that end, we chose a section of the 3′ variable region of N to serve as a probe in northern analyses and to be amplified in real-time reverse transcription (RT)-PCR assays. The variable-region sequence should also differentiate between the N gene and other R genes sharing motifs such as NBS and LRR.
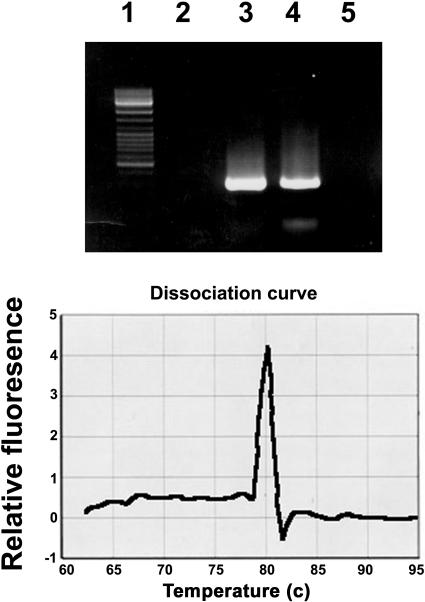
We amplified, by RT-PCR, a 140-bp segment of the variable region from a total RNA preparation extracted from noninfected and TMV-infected leaves of tobaccoNN and obtained a single, homogenous band. No such band was observed by similarly amplifying RNA extracted from TMV-infected tobacconn (Fig. 1, top, lane 1). To determine that no other sequence of similar size was amplified along with the N-specific band, we performed a temperature-dissociation analysis (Fig. 1, bottom), which indicated that the amplified product was indeed a single band. Figure 2 shows that the sequence of the amplified product was identical to the published one (GenBank accession no. U15605), indicating N-gene specificity. Furthermore, as described below, the amplified PCR product, serving as a probe in northern analyses, hybridized to N-derived RNA. Hybridization to n-derived RNA was negligible, if at all (Fig. 3). We thus concluded that the aforementioned PCR product is N specific and that this sequence is not expressed by any other N-related gene. This amplicon was therefore selected for the quantitative analyses of N mRNA.
Figure 1.
Top, Electrophoretic pattern of the product amplified by RT-PCR with primers derived from the 3′ variable region of the N gene. Primer sequences are shown in “Materials and Methods.” RNA templates served for RT-PCR were extracted from tobacco. Lane 1, Size markers. Lane 2, Noninfected tobacconn. Lane 3, Noninfected tobaccoNN. Lane 4, A noninfected systemic leaf of tobaccoNN, locally infected with TMV. Lane 5, A TMV-infected systemic leaf of tobacconn. Bottom, Temperature dissociation curve of the band shown in lane 3 of the top frame.
Figure 2.
FASTA analysis between the sequence of the PCR product of the 3′ variable region of N (lane 3 of Fig. 1) and the actual sequence of that region of the N gene.
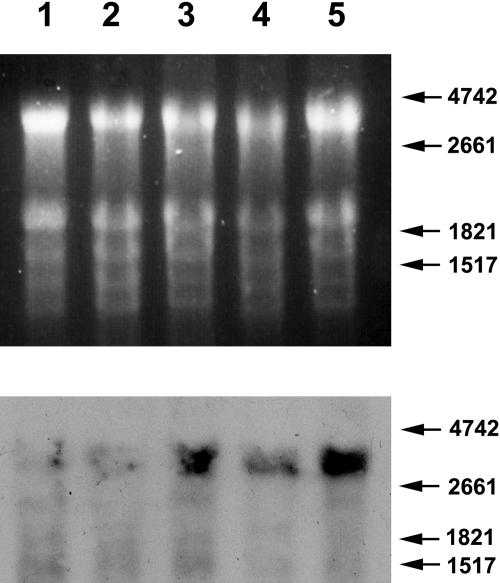
Figure 3.
Northern analysis of N-gene expression. Top, Ethidium bromide-stained gel pattern. Bottom, Pattern of fluorescent bands following hybridization of the gel shown in the top frame to the DIG-labeled PCR product shown in Figure 1. Numbers and arrows on the right indicate the positions of RNA size markers. Lanes were loaded with RNA extracted of a leaf from non-inoculated tobacconn (lane 1), a leaf from TMV-infected tobacconn (lane 2), a locally infected leaf of tobaccoNN (lane 3), a leaf from noninoculated tobaccoNN (lane 4), and a systemic (noninfected) leaf from TMV-infected tobaccoNN (lane 5).
Expression of the N Gene in TobaccoNN and Tobacconn
Northern-blot analyses were carried out with total RNA extracts from noninfected and TMV-infected tobaccoNN and tobacconn. The aforedescribed N-specific PCR product of the 3′ variable region of N served as a probe. Figure 3 indicates that tobacconn hardly transcribe any detectable N sequence, while tobaccoNN did transcribe N mRNA. The northern analysis also indicated a basal level of N expression in noninoculated leaves, which was stimulated in locally infected leaves as well as in upper, noninfected ones.
The primary transcripts of N are alternatively spliced, resulting in two types of mRNAs, both playing a role in N functions. However, the two transcripts differ in size by only 70 bases (Dinesh-Kumar and Baker, 2000) and are therefore inseparable upon northern analysis. The band reacting with the N-specific probe is approximately 3,400 bases long, agreeing with the expected size of 3,430 bases deduced from the aforementioned GenBank sequence.
Quantifying Levels of N-Gene Expression in TMV-Infected Tobacco Leaves
Real-time RT-PCR was employed to corroborate the results of the northern analysis indicating TMV-directed stimulation of N expression and to quantify the extent of that stimulation. The level of N transcripts was determined at various times following inoculation (20, 30, and 40 h in inoculated leaves and 24, 42, and 72 h and 10 d in upper leaves). One leaf of each tobaccoNN plant was inoculated with 100 μg/mL purified TMV. Real-time PCR analysis was performed with RNA extracted from inoculated leaves (localizing TMV infection) and noninoculated upper leaves of infected plants (the third leaf above the inoculated one which remained TMV-free). In inoculated leaves, N-mRNA accumulation was determined at times prior to the onset of necrosis. Necrosis did not develop in upper leaves, and samples from those leaves were collected up to 10 d postinoculation. A basal level of constitutive N-gene expression was noted in noninfected tobaccoNN (Fig. 3; Tables I and II). The level of mRNA in both locally infected and upper leaves increased with time of infection (Tables I and II). RT-PCR analysis did not result in any N-specific band when RNA from tobacconn (noninfected as well as 10 d after TMV inoculation) served as a template.
Table I.
Accumulation of N-mRNA in locally infected leaves of tobaccoNN
| Time after TMV Inoculation | Δcta | Fold Accumulation over Noninfected Leavesb |
|---|---|---|
| Noninoculated | 0.0074 | – |
| 20 h | 0.660 | × 8.9 ± 1.62 |
| 30 h | 0.283 | × 38.2 ± 1.01 |
| 40 h | 0.236 | × 31.89 ± 1.27 |
Average of triplicates.
Accumulation of RNA was calculated against internal standards of rRNA as described in “Materials and Methods.”
Table II.
Accumulation of N mRNA in upper, noninfected leaves of TMV-inoculated tobaccoNN
| Time after TMV Inoculation | Δcta | Fold Accumulation over Noninoculated Leavesb |
|---|---|---|
| Noninoculated | 0.29 | – |
| 24 h | 1.18 | × 4.06 ± 1.22 |
| 42 h | 2.44 | × 8.41 ± 1.20 |
| 72 h | 47.83 | × 164.93 ± 1.56 |
| 10 d | 21.86 | × 75.38 ± 1.16 |
Average of triplicates.
Accumulation of RNA was calculated against internal standards of rRNA as described in “Materials and Methods.”
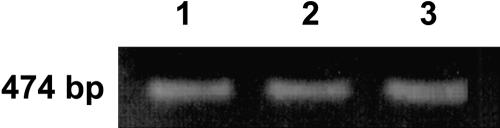
The real-time PCR results confirmed the northern analysis indications. A basal level of N expression occurs in tobaccoNN but not in tobacconn. In tobaccoNN, N-mRNA is accumulated approximately 30-fold in infected leaves and up to approximately 165-fold in upper leaves (Tables I and II), indicating stimulation of expression. In order to further ascertain that RT-PCR was carried out with the same quantity of template RNA in all samples, we checked the levels of another housekeeping gene (actin) in the same RNA samples used for the real-time PCR assays. RT-PCR was performed to only 22 cycles, so that it would not reach saturated levels for ethidium bromide detection. Figure 4 demonstrates that expression of actin was at about the same level in all samples.
Figure 4.
RT-PCR of tobaccoNN RNA with primers of actin. PCR was conducted as described in “Materials and Methods.” Lane 1, Template RNA from noninoculated leaf. Lane 2, Template RNA from a leaf locally infected with TMV, 20 h postinoculation. Lane 3, Template RNA from a systemic leaf from a plant locally infected with TMV, 72 h postinoculation.
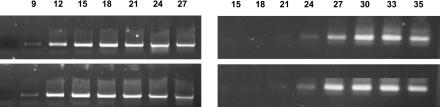
The stimulation of N expression could be attributed to a TMV-specific response or to a nonspecific disease (or stress) response. In addressing this problem, similar experiments were carried out with Potato virus Y (PVY)-infected tobaccoNN. RNA extracts from 72-h PVY-infected tobaccoNN leaves were subjected to quantitative RT-PCR assays (Fig. 5). N-mRNA bands in both noninfected and PVY-infected assays became noticeable at the same cycle, indicating similar levels of N-mRNA accumulation. Consequently, it seems that the increase in N-mRNA accumulation is specific to TMV infection and is not a general response to stress or infection.
Figure 5.
Quantitative RT-PCR of N-mRNA from noninfected and PVY (72 h postinoculation)-infected leaves. Left, PCR with 18S-rRNA-specific primers. Right, PCR with N-specific primers. In each section, the top row represents samples from noninfected tobaccoNN and the bottom row those from PVY-infected leaves. Cycle numbers are shown above each section.
DISCUSSION
Since Nicotiana spp. carry sequences homologous to that of the tobaccoNN N gene, it was necessary to establish that we were specifically detecting only N expression. To that end, we compared tobacconn, which does not carry the N gene, to its isoline (tobaccoNN) harboring that gene. Northern-blot analysis indicated that tobacconn does not express N-derived mRNA, therefore interference from other N-related genes was not likely to occur when studying N expression. However, being such a crucial point, we corroborated this by RT-PCR as well. We chose the 3′ variable region of N for amplification because it is present in both types of the alternatively spliced N-mRNA, and it is not part of any of the common R-gene domains, making interference from other R genes unlikely as well. The resultant RT-PCR product was shown to be homogenous (and by temperature-dissociation analysis also unique). The sequence of the RT-PCR product was identical to that segment of the N gene (Fig. 2).
The current consensus for R-gene activity is based on interactions between proteins. The product of the R gene is constitutively expressed, and following activation of the R protein by a product of the pathogen's avr gene, a pathway leading to resistance begins (Flor, 1942; Bent, 1996; De Wit, 1997; Warren et al., 1998; Ellis et al., 1999; Richter and Ronald, 2000). This article reports on the stimulation of one R gene's expression by a pathogen.
The N gene is a representative of R genes that encode putative receptors. The results indicate constitutive expression of the N gene, as N-mRNA was detected in noninoculated tobaccoNN leaves. Therefore, possible interaction between a pre-existing N protein and an incoming viral-directed protein activator cannot be ruled out. Upon infection, however, the rate of N-mRNA accumulation increased considerably. In leaves that were actually inoculated with TMV, maximum N-mRNA accumulation (at 30 h after inoculation) was about 38-fold that of noninoculated leaves and leveled off at 40 h. At this point, the leaves started to show necrosis, and the cessation of mRNA stimulation may be attributed to the beginning of apoptosis. The lack of N-mRNA stimulation in PVY-infected tobaccoNN indicated that accumulation of N-mRNA is probably specific to TMV infection and not a general stimulation caused by other infections or stress.
It is particularly interesting to note the accumulation of N mRNA to higher levels in upper, noninoculated leaves. N-mRNA accumulation in upper leaves of TMV-infected plants was 165-fold higher than that in noninfected plants. Accumulation increased for 72 h and remained at an elevated level for at least 10 d. These leaves were TMV-free and did not develop necrosis. Moreover, they have been shown to develop resistance toward several viruses (Ross, 1961; Loebenstein, 1963; Bozarth and Ross, 1964). It has been suggested that restriction of viral replication and movement is due to a virus-induced viral inhibitor and that antiviral substances do appear following viral infection, particularly in tobaccoNN (Sela and Appelbaum, 1962; Loebenstein and Ross, 1963; Sela et al., 1964, 1978; Antignus et al., 1975, 1977; Sela, 1986; Edelbaum et al., 1990; Gera et al., 1990). The transduction of a signal for N-mRNA accumulation from inoculated to upper leaves is coupled to the induction of systemic resistance that develops without activation by a viral component. This suggests that the transduced development of resistance is an event occurring downstream of N-protein activation. Since the induction signal for a necrotic pathway is not systemically transduced, the two downstream events known to result from N-protein activation by the virus are uncoupled. It may be postulated that activation of the N protein by a viral product leads to direct or indirect activation of a transcription factor(s), as suggested by Whitham et al. (1994). These, in turn, stimulate several different metabolic pathways. Necrosis and viral restriction may result from two different, simultaneously activated pathways, producing seemingly coupled phenotypes (Greenberg et al., 1994; Lamb, 1994). They may also interact functionally, namely, the activation of a certain cascade of events needs TMV in order to result in necrosis, and, hence, the restriction of TMV also restricts necrosis to the TMV lesions. The N-originated resistance mechanism develops in upper leaves and affects several viruses (Ross, 1961). However, in these cases, necrosis does not develop due to the lack of TMV.
MATERIALS AND METHODS
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of material. Obtaining any permission will be the responsibility of the requestor.
Plants
The following plants were used throughout the project: Nicotiana tabacum var. Samsun (referred to as tobacconn), developing systemic mosaic symptoms but not necrosis upon TMV infection, and N. tabacum var. Samsun NN (referred to as tobaccoNN), into which the N gene of Nicotiana glutinosa has been introgressed and which reacts locally upon TMV infection, restricting viral spread and causing local necrotic lesions. Tobacconn and tobaccoNN are, in fact, isolines of the tobacco var. Samsun. Information about the genetic makeup of these two tobacco variants is summarized in Sela (1981). Plants were grown in an insect-proof greenhouse at 24°C ± 2°C under a 16-h photoperiod of regular neon lighting.
Extraction of Nucleic Acids
For RNA extraction, tissue samples were collected, immediately frozen in liquid nitrogen, and then kept at −80°C. Total RNA was extracted from these tissues using the RNeasy plant minikit (Qiagen, Valencia, CA), including the QIAshredder spin columns. RNase-free DNase was then added to the column, and after 40-min incubation with the DNase, the RNA was eluted with RNase-free water. About 3 μg of RNA was obtained from 100 mg of leaf tissue.
Real-Time RT-PCR
Measured amounts of RNA (100 ng for N-gene expression analyses and 100 pg for 18S rRNA internal controls) were subjected to one-step RT-PCR using the SYBR Green PCR master mix with Taqman reverse transcriptase (Applied Biosystems, Foster City, CA). Real-time RT-PCR was conducted in GeneAmp PCR System 5700 (Applied Biosystems). Precautions were taken to ascertain reliable quantitative results: Log-linear dilution curves were performed with primers for the N gene as well as with primers for the 18S rRNA. The selected amplicon was small (140 bp). The correlation value of the N-gene curve was 0.997221 and that of the 18S rRNA curve was 0.983780; both values and the difference between them were well within the allowable parameters. Reactions performed without reverse transcriptase or without template did not result in any product. PCR cycles were as follows: 1 cycle of 30 min at 48°C and 10 min at 95°C, followed by 40 cycles each of 15 s at 95°C, 30 s at 60°C, and 45 s at 72°C.
Each N-gene peak was given an arbitrary quantitative value correlated to the 18S rRNA peak, according to the formula 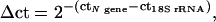 ct being the cycle threshold. Rates of stimulation of RNA expression were calculated from the Δct values at various time points. An sd for the rate of stimulation was calculated from the sds of the N-gene (s1) and 18S-rRNA (s2) cts according to the formula
ct being the cycle threshold. Rates of stimulation of RNA expression were calculated from the Δct values at various time points. An sd for the rate of stimulation was calculated from the sds of the N-gene (s1) and 18S-rRNA (s2) cts according to the formula 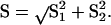
The following are the primers for all N-related RT-PCR assays, including real-time RT-PCR. Forward primer (bases 6,119–6,144 of the N gene; GenBank accession no. U15605): 5′TTCTTTGTACCTTTTGCTGGCTTAT3′. Reverse primer (bases 6,259–6,232 of the above): 5′CTCTGGTCCTTCTTTATACAACAAAC3′. Temperature-dissociation analysis (displayed by the GeneAmp 5700 system) confirmed the homogeneity of the PCR product (Fig. 1). The amplified region was from exon 4, which is not spliced out upon alternative splicing (Dinesh-Kumar and Baker, 2000). Similarly, a primer pair for tobacco rRNA was selected from the published sequence (GenBank accession no. AJ236016). The sequence of the forward primer was 5′AGGAATTGACGGAAGGGCA3′ (bases 1,142–1,161 of 18S-rRNA). The sequence of the reverse primer was 5′GTGCGGCCCAGAACATCTAAG3′ (bases 1,466–1,446 of the above). Amplification of rRNA served as an internal control for expression of a housekeeping gene. The primers were selected by the Primer Express software of GeneAmp.
RT-PCR and Quantitative RT-PCR
RT-PCR for the expression of actin was performed on 100 ng of RNA with the following primers. The forward primer was 5′GTCTGGTGATGGTGTTAGC3′ (bases 4,282–4,301 of accession no. X63603) and the reverse primer was 5′CCTATCAGCAATTCCAGGAAAC3′ (bases 4,756–4,735 of the above). For quantitative PCR, measured amounts (25 ng for N-gene expression analysis and 50 pg for 18S rRNA) of total RNA extracts of noninfected and PVY-infected tobaccoNN (72 h post inoculation) were subjected to RT-PCR with the same aforementioned primers used for real-time PCR. Samples were drawn at three-cycle intervals and analyzed by gel electrophoresis. The system was calibrated so that the appearance of the 18-rRNA-specific band in noninoculated and PVY-inoculated RNA extracts will not differ by more than a single cycle, ascertaining that similar levels of RNA were used as template in reactions from noninfected and PVY-infected leaves. The first cycle showing a visible band was indicative on the relative amount of RNA.
Northern-Blot Analysis
Total RNA was extracted from noninoculated and TMV-inoculated tobacconn leaves and from noninoculated and TMV-inoculated tobaccoNN leaves, as well as from upper, noninoculated leaves of locally infected tobaccoNN. Formaldehyde was added to the RNA to 1.8% and warmed to 65°C. The RNA, 15 μg per lane (in light of the real-time PCR results, only 1.5 μg of RNA was loaded in the case of upper leaves of inoculated plants), was electrophoresed on a 1.2% agarose gel at 70 V, 4°C with stirring. The previously described amplified N-RNA product was digoxigenin labeled and served as a probe for hybridization. Detection was performed with the DIG luminescent detection kit (Roche Diagnostics GmbH, Mannheim, Germany). RNA sizes were estimated by comparison to electrophoresed RNA Molecular Weight Markers I (Roche). Hybridization was carried out at high stringency (0.1× SSC; 65°C).
Acknowledgments
We thank the Wolfson Foundation for the use of facilities contributed to the Plant Science Institute.
This work was supported by the TEVEL consortium and by the Minerva Otto Warburg Center for Agricultural Biotechnology.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044859.
References
- Abbink TEM, Tjernberg PA, Bol JF, Linthorst HJM (1998) Tobacco mosaic virus helicase domain induces necrosis in N gene-carrying tobacco in the absence of virus replication. Mol Plant Microbe Interact 11: 1242–1246 [Google Scholar]
- Antignus Y, Sela I, Harpaz I (1975) A phosphorus-containing fraction associated with antiviral activity in Nicotiana spp. carrying the gene for localization of infection. Physiol Plant Pathol 6: 159–168 [Google Scholar]
- Antignus Y, Sela I, Harpaz Y (1977) Further studies on the biology of an antiviral factor (AVF) from virus-infected plants and its association with the N-gene of Nicotiana species. J Gen Virol 35: 107–116 [DOI] [PubMed] [Google Scholar]
- Bent AF (1996) Plant disease resistance genes: function meets structure. Plant Cell 8: 1757–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozarth RF, Ross AF (1964) Systemic resistance induced by localized virus infection: extent of change in uninfected plant parts. Virology 24: 446–455 [DOI] [PubMed] [Google Scholar]
- De Wit PJGM (1997) Pathogen avirulence and plant resistance: a key role for recognition. Trends Plant Sci 2: 452–458 [Google Scholar]
- Dinesh-Kumar SP, Baker B (2000) Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci USA 97: 1908–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Wai-Hong T, Baker BJ (2000) Structure-function analysis of tobacco mosaic virus resistance gene N. Proc Natl Acad Sci USA 97: 14789–14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelbaum E, Ilan N, Grafi G, Sher N, Stram Y, Novick D, Tal N, Sela I, Rubinstein M (1990) Purification and characterization of two antivirally active proteins from tobacco by monoclonal antibodies to human β-interferon. Proc Natl Acad Sci USA 87: 588–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Lawerence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determines difference in gene-for-gene specificity. Plant Cell 11: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson FL, Holzberg S, Calderon-Urrea A, Handly V, Axtell M, Corr C, Baker B (1999) The helicase domain of the TMV replicase proteins induces the N-mediated defense response in tobacco. Plant J 18: 67–75 [DOI] [PubMed] [Google Scholar]
- Flor HH (1942) Inheritance of pathogenicity in a cross between physiological races 22 and 24 of Melampsora lini. Phytopathology 32: 5 [Google Scholar]
- Flor HH (1956) The complementary genic systems in flax and flax rust. Adv Genet 8: 29–54 [Google Scholar]
- Gera A, Loebenstein G, Salomon R, Franck A (1990) Inhibitor of virus replication from protoplasts of a hypersensitive tobacco cultivar infected with tobacco mosaic virus is associated with a 23-K protein species. Phytopathology 80: 78–81 [Google Scholar]
- Greenberg JT, Guo A, Klessig DF, Ausubel FM (1994) Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell 77: 551–563 [DOI] [PubMed] [Google Scholar]
- Holmes FO (1938) Inheritance of resistance to tobacco mosaic disease in tobacco. Phytopathology 28: 553–561 [Google Scholar]
- Jin HL, Liu YD, Yang KY, Kim CY, Baker B, Zhang SQ (2003) Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J 33: 719–731 [DOI] [PubMed] [Google Scholar]
- Keen NT (1992) The molecular biology of disease resistance. Plant Mol Biol 19: 109–122 [DOI] [PubMed] [Google Scholar]
- Lamb CJ (1994) Plant disease resistance genes in signal perception and transduction. Cell 76: 419–422 [DOI] [PubMed] [Google Scholar]
- Lee FS, Vallee BL (1990) Modular mutagenesis of human placental ribonuclease inhibitor, a protein with leucine-rich repeats. Proc Natl Acad Sci USA 87: 1879–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebenstein G (1963) Further evidence on systemic resistance induced by localized necrotic virus infections in plants. Phytopathology 53: 306–308 [Google Scholar]
- Loebenstein G, Ross FA (1963) An extractable agent, induced in uninfected tissues by localized virus infections, that interferes with infection by tobacco mosaic virus. Virology 20: 507–517 [DOI] [PubMed] [Google Scholar]
- Matthieu HAJJ, Cozijnsen TJ, De Wit PJGM (1994) Host resistance to fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367: 384–386 [DOI] [PubMed] [Google Scholar]
- Padgett HS, Beachy RN (1993) Analysis of tobacco mosaic virus strain capable of overcoming N-gene-mediated resistance. Plant Cell 5: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter TE, Ronald PC (2000) The evolution of disease resistance. Plant Mol Biol 42: 195–204 [PubMed] [Google Scholar]
- Ross AF (1961) Systemic acquired resistance induced by localized virus infection in plants. Virology 14: 340–358 [DOI] [PubMed] [Google Scholar]
- Sela I (1981) Plant-virus interactions related to resistance and localization of viral infections. Adv Virus Res 26: 201–237 [DOI] [PubMed] [Google Scholar]
- Sela I (1986) Preparation and measurement of an antiviral protein found in tobacco cells after infection with tobacco mosaic virus. Methods Enzymol 11: 734–744 [DOI] [PubMed] [Google Scholar]
- Sela I, Appelbaum SW (1962) Occurrence of an antiviral factor in virus-infected plants. Virology 17: 543–548 [DOI] [PubMed] [Google Scholar]
- Sela I, Harpaz I, Birk Y (1964) Separation of a highly active antiviral factor from virus-infected plants. Virology 22: 446–451 [DOI] [PubMed] [Google Scholar]
- Sela I, Hauschner A, Mozes R (1978) The mechanism of stimulation of the antiviral factor (AVF) in Nicotiana leaves. Virology 89: 1–6 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Choe HR, Nishida Y, Yamawakikataoka Y, Ohnishi S, Tamaoki T, Kataoka T (1990) Leucine-rich repeats and carboxyl terminus are required for interaction of yeast adenylate cyclase with RAS proteins. Proc Natl Acad Sci USA 87: 8711–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ackerveken GFJM, Vossen P, De Wit PJGM (1993) The avr9 gene race-specific elicitor of Cladosporium fulvum is processed by endogenous and plant proteases. Plant Physiol 103: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JDG (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signaling components. Plant J 29: 439–451 [DOI] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW (1998) A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10: 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B (1994) The product of the tobacco mosaic virus resistance gene N: similarity to Toll and the Interleukin-2 receptor. Cell 78: 1101–1115 [DOI] [PubMed] [Google Scholar]