Abstract
The P19 protein of tomato bushy stunt virus (TBSV) is a multifunctional pathogenicity determinant involved in suppression of posttranscriptional gene silencing, virus movement, and symptom induction. Here, we report that P19 interacts with the conserved RNA-binding domain of an as yet uncharacterized family of plant ALY proteins that, in animals, are involved in export of RNAs from the nucleus and transcriptional coactivation. We show that the four ALY proteins encoded by the Arabidopsis genome and two ALY proteins from Nicotiana benthamiana are localized to the nucleus. Moreover, and in contrast to animal ALY, all but one of the proteins are also in the nucleolus, with distinct subnuclear localizations. Infection of plants by TBSV or expression of P19 from Agrobacterium results in relocation of three of the six ALY proteins from the nucleus to the cytoplasm demonstrating specific targeting of the ALY proteins by P19. The differential effects on subcellular localization indicate that, in plants, the various ALY proteins may have different functions. Interaction with and relocalization of ALY is prevented by mutation of P19 at residues previously shown to be important for P19 function in plants. Down-regulation of expression of two N. benthamiana ALY genes by virus-induced gene silencing did not interfere with posttranscriptional gene silencing. Targeting of ALY proteins during TBSV infection may therefore be related to functions of P19 in addition to its silencing suppression activity.
The majority of plant viruses have a very small genome size (12 kb or less) and encode very few (less than 10) viral proteins. To be successful, virus infection relies on the interaction with and subversion of normal host cellular processes by specific viral proteins, many of which, it is now becoming clear, are multifunctional and target more than one host pathway. A good example is provided by the potyvirus HC-Pro protein, which was identified initially as an insect transmission factor (Govier and Kassanis, 1974). HC-Pro also functions as a protease involved in processing of the viral polyprotein in genome amplification in single cells, in systemic movement of the virus in plants, and as the determinant of synergism, in which mixed infections of a potyvirus together with a second, different virus result in symptoms that are much more severe than those produced by either virus alone (Carrington et al., 1989; Kasschau et al., 1997; Shi et al., 1997).
Recent studies have shown that the synergistic and movement functions of HC-Pro result from the interaction of this protein with a host defensive system, referred to as posttranscriptional gene silencing (PTGS), that is deployed by the plant in response to challenge by viruses (Anandalakshmi et al., 2000; Kasschau and Carrington, 2001). The PTGS system responds to double-stranded (ds) RNAs that are formed as part of the normal replication cycle of single-stranded RNA viruses. The dsRNAs are cleaved into 21 to 26 nucleotide RNAs, known as short interfering (si) RNAs (Hamilton and Baulcombe, 1999; Hammond et al., 2000). The siRNAs are incorporated into an RNA-induced silencing complex that targets full-length viral RNA by base pairing to complementary sequences, leading to their degradation. In plants, siRNAs of two size classes, 21 to 22 nucleotides (nt) and 24 to 26 nt, have been observed, and the longer class is suggested to be involved in the process of systemic spread of the silencing signal through the vascular system to initiate PTGS in upper leaves (Hamilton et al., 2002). Similar systems operate in other organisms, including the nematode Caenorhabditis elegans, the fruit fly Drosophila, and humans, although the precise details of these processes, referred to collectively as RNA silencing, may differ from one another (Cerutti, 2003; Denli and Hannon, 2003).
Many plant viruses have been shown to encode proteins, such as potyvirus HC-Pro, that target the silencing process, weakening the host defense and enhancing virus infection. There is no amino acid sequence homology apparent between the 10 or more of these so-called silencing suppressor proteins that have been identified so far from different viruses, and it is possible that each targets a different part of the PTGS machinery (Moissiard and Voinnet, 2004).
A well-studied silencing suppressor protein is the P19 protein encoded by Tomato bushy stunt virus (TBSV). In an infiltration assay, P19 was found to be one of the most effective suppressors of silencing and completely prevented the production/accumulation in plants of both short and long forms of siRNA (Hamilton et al., 2002; Havelda et al., 2003). The P19 protein of Cymbidium ringspot virus (CymRSV), a virus similar to TBSV, was shown to bind in vitro specifically to ds siRNAs generated by PTGS but not to bind to single-stranded (ss) siRNAs, long ss-, or dsRNA (Silhavy et al., 2002). More recently, the structure of P19 has been determined, revealing that the siRNA duplex is bound by a P19 homodimer and that conserved Trp residues cap either end of the ds siRNAs, ensuring the binding of only 19 to 21 nt RNAs (Vargason et al., 2003; Ye et al., 2003). Thus, silencing suppression by P19 involves sequestration of siRNAs by direct binding.
In addition, P19 is also known to be involved in several other aspects of viral infection. TBSV that does not express P19 is unable to move systemically in spinach, does move systemically in Nicotiana benthamiana but no longer induces systemic necrosis, does not induce local necrosis in Nicotiana tabacum, and has impaired local, cell-to-cell movement in pepper (Scholthof et al., 1995a, 1995b; Turina et al., 2003). Whether any of these phenotypes is related to loss of P19 silencing suppression activity or whether they represent different functions of P19 is not known.
To further understand how TBSV influences the host plant, we carried out a yeast two-hybrid analysis to identify plant proteins that interact with P19. From these studies, we have identified a family of plant ALY proteins that are targeted by the P19 silencing suppressor protein of TBSV in vivo. In plants, infection by TBSV or expression of the P19 protein alone causes intracellular redistribution of a subset of the ALY proteins from the nucleus to the cytoplasm, suggesting that P19 may affect aspects of RNA export from the nucleus. In addition, experiments were carried out to examine whether the P19-ALY interaction affects RNA silencing in the plant.
RESULTS
TBSV P19 Interacts with ALY RNA Export Factors in Yeast Two-Hybrid Screens
The TBSV P19 protein has been reported to suppress RNA silencing in Arabidopsis (Voinnet et al., 2003). To identify host proteins that might be targeted by P19, a yeast two-hybrid screen of an Arabidopsis cDNA library was performed. cDNA clones were isolated from 64 independent yeast colonies and analyzed by DNA sequencing. Fifty-five (86%) were found to contain partial coding sequences of members of the family of four Arabidopsis homologs of ALY, a gene encoding a highly conserved protein involved in transcription and RNA transport in eukaryotes. Five clones were obtained of the Arabidopsis gene At5g59950 (which we refer to as ALY1), 41 clones of At5g02530 (ALY2), 7 clones of At1g66260 (ALY3), and 2 clones of At5g37720 (ALY4).
Using information from the Arabidopsis genome sequencing project to design gene-specific 5′ and 3′ PCR primers, the complete coding region of each of the four Arabidopsis genes was amplified and cloned.
Isolation of ALY Genes from N. benthamiana and Phylogenetic Analysis
Because Arabidopsis is not a susceptible host of TBSV, we proceeded to isolate ALY genes from N. benthamiana, a highly susceptible host for TBSV and many other viruses. Although no full-length sequences of ALY from Nicotiana species were identified by database searches, it was possible to assemble full-length sequences using tomato and potato expressed sequence tag (EST) sequences and design primers for amplification of N. benthamiana ALY genes. Two full-length cDNAs (NbAly615 and NbAly617) were amplified and cloned from N. benthamiana.
There is variation in ALY gene number in animals with a single gene in humans, and two genes in mouse and Drosophila. By contrast, Arabidopsis contains four ALY genes, and from our analyses there are at least three clear ALY homologs in Solanaceous species and four or more ALY homologs in many monocots (Supplemental Fig. 1, available at www.plantphysiol.org). These findings indicate that there has been an expansion of the ALY gene family in plants, which may have provided the opportunity for diversification of expression patterns, location, and function.
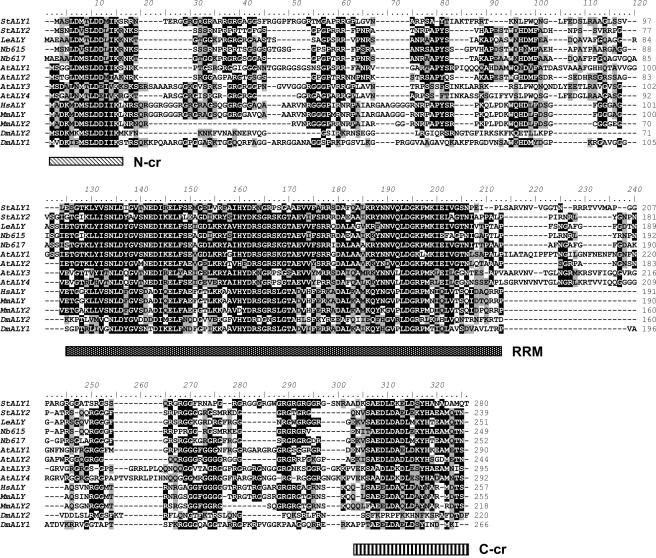
Phylogenetic analysis of the four Arabidopsis (At)ALY proteins revealed an overall sequence identity of 53% between AtALY1 and AtALY2 and 71% between AtALY3 and AtALY4 (Fig. 1). However, AtALY1 and AtALY2 have only between 35% to 37% sequence identity with AtALY3 and AtALY4. NbALY617 and NbALY615 most closely resembled AtALY1 and AtALY2 (60% and 57%, and 52% and 60% sequence identity, respectively). Nevertheless, it was not possible, using amino acid sequence alignment, to identify NbALY615 or NbALY617 as specific orthologs of any of the individual Arabidopsis ALY proteins.
Figure 1.
Alignment of ALY homologs. Mouse (Mm), human (Hs), Drosophila (Dm), Arabidopsis (At), potato (St), tomato (Le), and N. benthamiana (Nb). Residues are shaded by degree of homology. Hatched boxes under the sequences indicate conserved N-terminal (N-cr) and C-terminal (C-cr) regions and RNA-binding domain (RRM).
Interaction with TBSV P19 Involves the RRM Domain of ALY
Using the yeast two-hybrid system, the interaction between TBSV P19 and each of the six proteins encoded by the full-length Arabidopsis and N. benthamiana ALY genes was reconfirmed, both when the bait protein was P19 and the prey protein was ALY, and in the reverse arrangement. In addition, P19 was also shown to interact in this system with ALY(REF) proteins from Drosophila and humans (Fig. 2A). Using the endogenous MEL1 gene as a second reporter, relative interaction strength was measured by a semiquantitative α-galactosidase assay. In this test, NbALY617 and AtALY2 interacted the most strongly with P19 (Fig. 2B).
Figure 2.
Yeast two-hybrid analysis of P19:ALY interactions. A, Yeast growth on double (SD-LW) and triple drop-out media (SD-LWH, supplemented with 20 mm 3-AT). AD is activation domain fusion, BD is DNA-binding domain fusion. The empty activation domain plasmid (pACT2) was used as a negative control. B, Semiquantitative α-galactosidase assay to measure interaction strength.
ALY proteins are 30-kD proteins that have a highly conserved domain structure. This consists of short N-terminal and C-terminal motifs flanking two variable regions containing varying numbers of Arg-Gly-Gly (RGG) repeats embedded in nonconserved amino acid sequence. The central domain of the protein contains an RNA-binding domain (also referred to as an RNA-recognition motif [RRM]) that contains two more highly conserved subdomains, RNP1 and RNP2. The 41 clones of AtALY2 isolated from the library screen carried N-terminal deletions of different sizes. The most extensive deletion, which still could interact with P19, extended for 68 amino acids, and lacked the entire N-terminal conserved domain and two-thirds of the N-terminal variable domain. To further investigate which regions of ALY are responsible for the interaction with TBSV P19, a number of deletion mutants of the AtALY2 protein were constructed and tested in the yeast two-hybrid assay. These mutants lacked the C-terminal variable domain (mutant RSΔ), the central RRM domain (mutant HSΔ), both the central and C-terminal domains (mutant RHΔ), or carried the RRM domain alone. In these tests, mutants HSΔ and RHΔ were unable to interact with the TBSV P19, but mutant RSΔ and the RRM domain-only construct were able to interact with P19 (Fig. 3). These results demonstrate that P19 interacts with the central RRM domain of AtALY2, which is the most highly conserved region of the ALY proteins.
Figure 3.
P19 interacts in yeast with the RRM domain of AtALY2. Domain names as in Figure 1. N-vr and C-vr domains contain variable repeats of an RGG motif. Deleted regions indicated by hatched lines. l.s. is the most extensive N-terminal deletion obtained during the Arabidopsis cDNA library screen. HSΔ, RHΔ, RSΔ, and RRM of AtALY2 are described in the text. Interaction with P19 was scored by assessment of yeast growth on minimal medium and by semiquantitative α-galactosidase assay.
In Vitro Binding of the Arabidopsis ALY2 Protein to TBSV P19
The interaction between AtALY and TBSV P19 was also demonstrated using a gel overlay (far western) assay. We were unable efficiently to express full-length glutathione S-transferase (GST)-tagged ALY2, thus GST-tagged ALY2 RSΔ, lacking the C-terminal domain but retaining P19 binding activity in yeast, was expressed in Escherichia coli and transferred to polyvinylidene difluoride (PVDF) membrane. TBSV P19 labeled with 35S-Met bound predominantly to the GST-ALY2 RSΔ protein but did not bind to GST or other E. coli proteins (Fig. 4). Thus, AtALY2 consisting only of the N-terminal and RRM domains was able to interact with TBSV P19 in vitro, consistent with the interaction results obtained in yeast.
Figure 4.
Binding of P19 to ALY in vitro. Total proteins of E. coli not expressing recombinant protein (BL21), expressing GST fused with AtALY2 RSΔ (GST-ALY) or GST only (GST). A, Stained with Coomassie. B, Probed with in vitro translated P19 labeled with 35S-Met. Arrows indicate positions of full-length GST-ALY and GST proteins.
Different Subcellular Localization of ALY Proteins
ALY in other higher eukaryotes is known to localize to the nucleus. In a previous study, AtALY3 (DIP1) and AtALY4 (DIP2) were isolated in a yeast two-hybrid screen as interacting with the DNA-binding domain of poly(ADP-ribose) polymerase, and were also shown to be nuclear proteins (Storozhenko et al., 2001). To examine the subcellular localization of each of the ALY proteins, all four Arabidopsis ALY proteins and the two N. benthamiana proteins were fused to the N terminus of the green fluorescent protein (GFP) and expressed in N. benthamiana leaves either from the tobacco rattle virus (TRV) vector (MacFarlane and Popovich, 2000) or from a binary plasmid in Agrobacterium tumefaciens. Confocal fluorescence microscopy revealed that in all epidermal cells, GFP fluorescence was confined to the nucleus.
Unexpectedly, and in contrast to the situation in animals, five of the six ALY proteins labeled the nucleolus in addition to the nucleoplasm (Fig. 5, A–G). Careful comparison of the localization pattern of the four different Arabidopsis ALY proteins revealed distinct differences in labeling. ALY2 labeled the nucleoplasm strongly with bright foci embedded in a speckled network but showed very little or no labeling of the nucleolus (Fig. 5B). ALY1, ALY3, and ALY4 labeled the nucleolus strongly (Fig. 5, A, D, and E). The nucleoplasm of ALY1 contained a small number of intensely staining bodies, whereas ALY4 exhibited less intense speckling. Nucleoplasmic labeling with ALY3 was less intense than with the other ALY proteins but consistently labeled a peripheral region of the nucleolus. Differences were also observed in the intranuclear labeling patterns of the two N. benthamiana ALY proteins (Fig. 5, F and G), although each was present in the nucleolus.
Figure 5.
Nuclear localization of ALY proteins and delocalization by TBSV P19. Distribution of fluorescence in epidermal cell infiltrated with Agrobacterium expressing ALY proteins tagged with GFP is confined to the nucleus (A–G: AtALY1, AtALY2, AtALY2 HSΔ, AtALY3, AtALY4, NbALY615, and NbALY617, respectively). A, The nucleus is labeled as Nu and the nucleolus as No. Expression of TBSV P19 tagged with GFP results in fluorescence throughout the cell (H). Expression of AtALY2-GFP in the presence of TBSV, which expresses the P19 protein, resulted in relocalization of fluorescence out of the nuclei into the cytoplasm (close-up view I and field view K, compare with J in which AtALY2-GFP is expressed alone [field view]). By contrast, fluorescence remained nuclear when AtALY2 HSΔ was expressed in the presence of TBSV (L). As with AtALY2-GFP and AtALY4-GFP, NbALY617-GFP also was relocalized from the nucleus to the cytoplasm when coexpressed with P19 from Agrobacterium binary plasmids (M). However, in the presence of P19 mutants R72G and R85G, fluorescence from NbALY617-GFP remained nuclear (N and O, respectively). Bars = 10 μm, except J to L, bars = 100 μm.
The GFP fusion of AtALY2 HSΔ, lacking the central RRM domain necessary for interaction with TBSV P19 in yeast, was also localized to the nucleus rather than the nucleolus; however, the protein accumulated in large, irregular clumps inside the nucleus rather than being distributed throughout the nucleoplasm (Fig. 5C). The specific nuclear and nucleolar localizations of the ALY-GFP fusions were maintained regardless of which expression system was used (data not shown) and have also been demonstrated following expression in Arabidopsis cell cultures (J. Brown and P. Shaw, personal communication).
The TBSV P19 was also expressed as a GFP fusion either using TRV or Agrobacterium (Fig. 5H). This protein had the same localization pattern as free GFP (data not shown) and was distributed throughout the cell.
Relocalization of AtALY2 by TBSV P19
In initial experiments, to corroborate that the interaction between ALY and P19 is biologically significant in plants, N. benthamiana plants were inoculated with TBSV, and systemically infected leaves were further infiltrated with Agrobacterium expressing AtALY2-GFP, which is the ALY protein that was isolated most frequently in the yeast two-hybrid screen. After 3 d, the infiltrated leaves were examined by confocal fluorescence microscopy. In these plants, some areas of epidermal cells contained the AtALY2-GFP protein that was restricted to nuclei. However, frequently, these areas of cells were flanked by other patches of epidermal cells in which the AtALY2-GFP protein was mostly absent from nuclei and was localized to the cytoplasm (Fig. 5, K [field view], I [close up]). In control experiments, the GFP fusion of the AtALY2 HSΔ protein, which does not interact with P19 in yeast, was not relocalized from the nucleus to the cytoplasm following infection of the plants by TBSV (Fig. 5L). Thus, infection by TBSV resulted in the relocalization of AtALY2-GFP from the nucleus to the cytoplasm. The patchy distribution of areas containing relocalized AtALY2-GFP possibly represents the uneven pattern of TBSV infection in these leaves.
Furthermore, we used Agrobacterium to express the TBSV P19 protein and AtALY2-GFP protein together in N. benthamiana leaves by infiltration of mixed bacterial cultures. In this experiment also, AtALY2-GFP was relocalized from the nucleus to the cytoplasm (data not shown), demonstrating that the TBSV P19 protein is able to relocalize AtALY2-GFP in the absence of any other viral proteins.
To examine whether virus infection in general, or infection specifically with TBSV was responsible for relocalization of ALY-GFP, the AtALY2-GFP protein was expressed from Agrobacterium in plants that were infected with a different virus, potato virus Y. In these plants, AtALY2-GFP remained in the nucleus, showing that redistribution of ALY is a specific effect of TBSV infection. Similarly, expression of the potato virus Y HC-Pro and cucumber mosaic virus 2b silencing suppressor proteins using Agrobacterium did not cause relocalization of AtALY2-GFP out of the nucleus (data not shown), showing that relocalization of ALY is not a general phenomenon related to interaction with silencing suppressor proteins and is specific to TBSV P19.
TBSV P19 Relocalizes Only a Subset of ALY Proteins
To examine whether the P19-ALY interaction caused relocalization of all of the ALY proteins, each of the five other cloned ALY proteins was fused to GFP and expressed with TBSV P19 in plants by agroinfiltration. Only two of the Arabidopsis ALY proteins (AtALY2-GFP and AtALY4-GFP) and one of the N. benthamiana ALY proteins (NbALY617-GFP) were relocalized by P19 from the nucleus into the cytoplasm by P19 (Fig. 5, I, K, and M; AtALY4-GFP data not shown). By contrast, AtALY1-GFP, AtALY3-GFP, and NbALY615-GFP were not detectably relocated into the cytoplasm by P19. Thus, in planta TBSV P19 targets only a subset of the ALY proteins for relocalization.
Single-Site Mutations of P19 Abolish Interaction with and Relocalization of ALY
Previous analyzes of P19 showed that mutation of particular residues differentially affects P19 function in systemic infection, symptom production, and silencing suppression (Chu et al., 2000; Qiu et al., 2002; Turina et al., 2003). To examine whether the P19/ALY interaction could play a role in these processes, we investigated the interaction characteristics of P19 carrying the R72G (Arg at residue 72 replaced by Gly) and R85G (Arg at residue 85 replaced by Gly) mutations that previously were shown to affect all of the known in vivo properties of P19. Specifically, TBSV carrying the P19 mutation R72G did not move systemically in spinach, was not able to cause systemic lethal necrosis, and had little or no silencing suppression activity in N. benthamiana. TBSV P19 R85G had no silencing suppression activity in newly emerged leaves and had slightly impaired systemic movement in spinach but did cause systemic lethal necrosis in N. benthamiana.
The earlier experiments showed that wild-type P19 interacted strongly in yeast with one of the ALY proteins from N. benthamiana (NbALY617) and relocalized NbALY617-GFP from the nucleus (Fig. 5M). However, when we expressed the P19 mutant R72G in plants from Agrobacterium, we did not observe relocalization of NbALY617-GFP from the nucleus (Fig. 5N). Moreover, in the yeast two-hybrid system, P19 R72G showed a drastically weakened interaction with NbALY617 (Fig. 2B). Likewise, P19 R85G did not relocalize NbALY617-GFP in planta (Fig. 5O) and showed reduced interaction strength with NbALY617 in yeast.
To rule out potential instability of the two P19 mutant proteins as the reason for their inability to relocalize NbALY617, western-blot analysis of agroinfiltrated leaves was carried out. This experiment confirmed that the wild-type and P19 mutant proteins were all expressed to easily detectable levels in infiltrated tissue, although there was perhaps a 4-fold increase in the accumulation of wild-type P19 protein as compared to the two mutant proteins (Fig. 6).
Figure 6.
Expression of P19 in infiltrated tissue. Western blot of extracts of leaves infiltrated with Agrobacterium, probed with antibodies to the TBSV P19 protein. Mock is from a plant infiltrated with Agrobacterium carrying an empty binary plasmid. P19 wt are two separate plants infiltrated with Agrobacterium expressing wild-type P19 protein. P19 R72G and P19 R85G are plants infiltrated with Agrobacterium expressing either of the two mutant P19 proteins. Positions of molecular mass size markers appear at left. The location of the P19 protein is indicated at the right of the figure. ** indicates probable P19 dimers. * indicates a cross-reacting plant or Agrobacterium protein. Load indicates protein loading levels revealed by Ponceau Red staining of the blotted filter.
Silencing of Two ALY Genes in N. benthamiana by VIGS Does Not Affect Subsequent Transgene Silencing or Virus Cross-Protection
The disruption of the P19-ALY interaction by mutations in P19 that drastically affect many of its functions suggests that this interaction is important for some aspect of viral infection. To begin to examine whether the interaction with ALY affects the silencing suppression activity of P19, we carried out experiments to silence ALY expression in N. benthamiana. Transgenic plants expressing the GFP protein were inoculated with TRV carrying fragments of both NbALY615 and NbALY617 (TRV-615/617), producing a systemic infection and virus-induced gene silencing (VIGS) of the endogenous NbALY615 and NbALY617 genes. Reverse transcription (RT)-PCR was used to show that in these plants after 3 weeks, NbALY615 and NbALY617 mRNAs were either absent or very significantly reduced compared to a control mRNA (ubiquitin; Fig. 7A). Control plants were either mock inoculated or inoculated with TRV carrying part of the Arabidopsis PDS gene (TRV-PDS; Yu et al., 2003). Upper leaves of all of these plants were then infiltrated with Agrobacterium carrying a binary plasmid to induce silencing of the GFP transgene, which was monitored by UV illumination of the plants and northern-blot analysis of the GFP mRNA and siRNAs (Fig. 7B; data not shown). In these experiments, silencing of NbALY615 and NbALY617 by VIGS did not affect subsequent silencing of the GFP transgene in the infiltrated patches.
Figure 7.
Effect of inhibition of ALY expression on the local silencing of a transgene. A, VIGS of the endogenous NbALY615 and NbALY617 genes in N. benthamiana plants transgenic for GFP (line 16c) was achieved by inoculating them with TRV carrying fragments of both NbALY615 and NbALY617 (TRV-615/617). RT-PCR was used to show that after 3 weeks, NbALY615 and NbALY617 mRNAs were either absent or very significantly reduced in systemic tissue compared to control plants (last two lanes versus other lanes). Control plants were either nontransgenic, or 16c plants noninoculated (lanes Nt and Ni, respectively), or 16c plants mock-inoculated (lane M) or inoculated with TRV carrying part of the Arabidopsis PDS gene (lane TRV-PDS). Each lane represents a different individual plant. RT-PCR of ubiquitin mRNA was also performed for each plant as a control. B, Upper leaves of the same plants used in A were infiltrated with Agrobacterium carrying a 35SP/GFP/NosT T-DNA to induce silencing of the GFP transgene, which was monitored by northern-blot analysis of the GFP mRNA. Silencing of the GFP transgene (lanes M, TRV-PDS, and TRV 615/617) occurred irrespective of any preestablished silencing of NbALY615 and NbALY617 by VIGS. The normal level of GFP mRNA accumulation is indicated in lane Ni, a 16c plant that was not virus-inoculated and also not infiltrated with Agrobacterium. The ethidium bromide-stained rRNAs are shown below each lane, as loading controls.
One manifestation of RNA silencing is the phenomenon of cross-protection, where infection of a plant by a virus leads to the induction of PTGS and protection from further reinfection by a virus having the same or very similar RNA sequence (Ratcliff et al., 1999). To test whether cross-protection was affected by reduced levels of ALY, nontransgenic N. benthamiana plants were either mock-inoculated or inoculated with TRV-PDS or with TRV-615/617. The plants were maintained for 3 weeks to allow for systemic VIGS of ALY to be established. Two upper leaves of each plant were reinoculated with TRV expressing GFP (TRV-GFP), and 3 d later the plants were examined under UV light for GFP fluorescence. All four plants that were initially mock-inoculated displayed large, strongly fluorescent green lesions on all leaves that were secondarily inoculated with TRV-GFP, showing that these plants were fully susceptible to TRV infection. However, all 8 plants initially inoculated with the control virus TRV-PDS and all 12 plants initially inoculated with TRV-615/617 were completely lacking TRV-GFP-induced fluorescent lesions (data not shown). Thus, silencing of NbALY615 and NbALY617 by VIGS did not prevent the establishment of cross-protection in N. benthamiana.
Finally, we examined whether expression of ALY would affect the silencing or suppression of silencing of a transiently expressed mRNA. Thus, an Agrobacterium culture containing a binary plasmid expressing the β-glucuronidase (GUS) gene was infiltrated into N. benthamiana plants either alone or mixed with other cultures expressing ALY2 and/or P19. Expression of ALY2 did not interfere with the silencing of GUS or the suppression of silencing by P19 (data not shown). As we do not have an antibody to ALY, we were not able to confirm that ALY2 was expressed in these experiments. However, in parallel experiments using fluorescently tagged ALY and P19 proteins, whose expression was confirmed by confocal microscopy, silencing of GUS mRNA was not affected by the presence of ALY1-GFP or ALY2-GFP. In addition, coexpression with ALY2-GFP did not affect the suppression of GUS silencing induced by P19 tagged with monomeric red fluorescent protein (P19-mRFP; Supplemental Fig. 2A), despite the delocalization to the cytoplasm of ALY2-GFP induced by P19-mRFP (Supplemental Fig. 2B).
DISCUSSION
The identification of host proteins that physically interact with virus proteins is an important step in understanding how viruses make use of host components to carry out their life cycle and how the host attempts to fight against virus infection. Here, we show that a family of plant proteins, homologous to ALY/REF RNA export factors from Drosophila and mammals, is specifically targeted by the TBSV P19 pathogenicity protein. Interaction with ALY proteins from Arabidopsis and N. benthamiana was demonstrated both in vivo and in vitro, the interaction domain was mapped to a central domain conserved in plants and animals, and P19 was shown to alter subcellular localization of a subset of ALY proteins in planta. Interaction in yeast between TBSV P19 and a partial clone of a tobacco ALY homolog, referred to as Hin19, was also described in a recent article (Park et al., 2004).
Differential Localization of Plant ALY Proteins in the Nucleus
In Arabidopsis, the family of ALY proteins consists of four members. AtALY3 and AtALY4 previously were shown to bind to the DNA-binding domain of poly(ADP-Rib) polymerase and were denoted as DIP1 and DIP2, respectively (Storozhenko et al., 2001). The function of ALY has been most studied in Drosophila and mammals, which encode only one or two homologs. ALY, which is also referred to as REF, is a multifunctional protein that is involved in RNA transport from the nucleus and also in transcriptional activation. It is a constituent of the exon-junction-complex that forms during the splicing of pre-mRNAs and is located around 20 nt upstream of the exon/exon boundary (Le Hir et al., 2000a, 2000b; Stutz et al., 2000; Zhou et al., 2000). After splicing, ALY interacts with the export factor TAP to initiate transport of the mRNA through the nuclear pore complex and into the cytoplasm for translation. In addition, ALY is also known to be a transcriptional coactivator, interacting with a number of different proteins that are part of complexes that bind to enhancer sequences and activate transcription (Bruhn et al., 1997; Virbasius et al., 1999). It is proposed that in these contexts ALY functions as a chaperone to facilitate the homo- and heterodimerization of other proteins.
The function of ALY in plants is not known. GFP tagging of the Arabidopsis and N. benthamiana ALY proteins showed that they are all localized to the nucleus but that there are distinct patterns of subnuclear distribution. AtALY2 localizes to granular bodies in the nucleoplasm but is absent from the nucleolus. This pattern corresponds to that seen with human ALY, which was located in speckles in the nucleoplasm, where it colocalized with splicing factors (Zhou et al., 2000). A similar nucleoplasmic localization was found for mouse ALY-GFP; however, a splice variant lacking the N-terminal variable domain was distributed both in the nucleus, but not in speckles, and in the cytoplasm (Rodrigues et al., 2001). We have shown that the central RRM domain of AtALY2 is dispensable for nuclear import. However, this domain, which is proposed to have RNA- and protein-binding capabilities, is important for the proper subnuclear localization of ALY2.
In addition to a nucleoplasmic localization, AtALY1, AtALY3, AtALY4, NbALY615, and NbALY617 were found to accumulate in the nucleolus. This is different from the distribution seen for ALY in animals and might reflect an involvement in plant-specific processes. Also, the precise intranuclear distribution differed for each of the four Arabidopsis ALY proteins, suggesting that the different ALY family members may have specific functions. Nucleoli of plants and animals are known to differ in their structural organization, and a vectorial model has been proposed for plant nucleoli in which successive biochemical steps in rRNA maturation and ribosome biogenesis occur in concentric layers enveloping the transcription sites (Brown and Shaw, 1998). The outer region of the nucleolus, where the majority of the plant ALY proteins localize, corresponds to the granular component where ribosomal subunits are assembled and exported. This might indicate that the plant ALY proteins have different or additional functions to those of animal ALY, possibly related to the modulation of RNA-protein and RNA-RNA complex formation in the nucleolus or facilitating transport/export of different RNA species.
P19 Relocalizes a Subset of the ALY Homologs from the Nucleus to the Cytoplasm
In animals, ALY is a nuclear shuttling protein that moves out of the nucleus during RNA transport but is reimported rapidly back into the nucleus, so that at steady state it is detectable only in the nucleus. Fractionation experiments determined that P19 accumulates mostly in the cell cytoplasm (Scholthof et al., 1995a). Our results using GFP-tagged P19 also suggest a cytoplasmic location for P19, although in neither set of studies can the presence of a proportion of P19 in the nucleus be ruled out.
Interestingly, infection of plants by TBSV, as well as expression of TBSV P19 protein alone from Agrobacterium, specifically altered the subcellular localization of a subset of the ALY proteins. AtALY2-GFP, AtALY4-GFP, and NbALY617-GFP were relocalized out of the nucleus into the cytoplasm. A truncated version of AtALY2-GFP lacking the central RRM domain, which is necessary for interaction with P19 in yeast, was retained in the nucleus and was not relocalized to the cytoplasm following coexpression in plants with P19. These results provide evidence for a dynamic localization of AtALY2, AtALY4, and NbALY617 and support the view that targeting by the P19 protein interferes with their shuttling between the nucleus and cytoplasm. We suggest that the interaction between P19 and these ALY proteins most likely takes place in the cytoplasm and prevents reimport of ALY into the nucleus. However, it is possible that some P19 could enter the nucleus, bind to ALY in the nucleus, and be exported to the cytoplasm as a protein complex. In yeast, all six plant ALY homologs interacted with P19; however, only the three strongest interactors were relocalized by P19 in planta. The correlation between interaction strength and the relocalization of ALY proteins in vivo might point to a competition for binding sites between P19 and endogenous factors. This suggests that P19 interaction with ALY inhibits the normal cellular function of ALY. Alternatively, relocalization of ALY by P19 could be indicative of the recruitment of ALY activity to the region of the cell where P19 functions. Differential relocalization of ALY is further evidence that the various ALY family members probably have different functions in the cell, only some of which are directly relevant to the TBSV infection process.
Mutated P19 with Impaired Pathogenicity Function Do Not Relocalize ALY
It was shown previously that substitution in P19 of the Arg at position 72 (R72) with Gly (R72G) affects many properties of the virus, including silencing suppression activity, cell-to-cell and long distance movement, and development of necrotic symptoms (Chu et al., 2000; Qiu et al., 2002; Turina et al., 2003). A second mutant P19 (R85G) has no silencing suppression activity in plants but, in contrast to the R72G mutation, exhibits host-specific differences in systemic movement and induction of lethal necrosis (Qiu et al., 2002). Our results show that both substitutions significantly decrease interaction strength with ALY in yeast and abolish relocalization of ALY in vivo. Both of the mutant P19 proteins were readily detected by western blotting in infiltrated leaves, suggesting that the lack of interaction of these proteins with ALY is not caused by instability and failure to accumulate.
Recently, examination of the structure of P19 showed that both Arg residues form salt bridges with Glu residues in the N terminus of the protein, near to the conserved Trp cap residues. The R72G mutation particularly was predicted to bury a charged residue within the protein structures and to affect the structural integrity of the protein (Vargason et al., 2003). This could explain why this mutant protein was unable to interact with ALY in yeast and did not relocalize AtALY2-GFP, AtALY4-GFP, or NbALY617-GFP from the nucleus.
The R85G mutation might be structurally less affected, allowing P19(R85G) to retain some of its activities, although it was unable to relocalize NbALY617-GFP.
Silencing of NbALY615 and NbALY617 Does Not Inhibit PTGS in N. benthamiana
The mechanism for silencing suppression is not fully understood, although during TBSV infection, P19 is known to significantly reduce free siRNAs in the cell, preventing their incorporation into the RNA-induced silencing complex (Lakatos et al., 2004). Combined with results from in vitro experiments, P19 was proposed to sequester ds-siRNAs by direct, noncatalytic binding without the intervention of other proteins. Nevertheless, one hypothesis for the functional role of the P19-ALY interaction could be that in planta one or more of the ALY proteins are involved in the RNA silencing pathway, perhaps exporting some siRNAs from the nucleus, and that during silencing suppression, the P19 protein could obtain these siRNAs by binding to ALY. We used VIGS to silence expression of two N. benthamiana ALY proteins, NbALY615 and NbALY617, and showed that both silencing of a GFP transgene and homology-driven virus cross-protection were unaffected in these plants. In addition, overexpression of AtALY2 in plants did not affect silencing or silencing suppression of a transiently expressed GUS gene. Consequently, these results argue against a role for ALY in RNA silencing, although it should be remembered that we expect there to be additional ALY proteins in N. benthamiana that were not silenced in these experiments. One or more of these additional proteins might be the primary target of P19 or might have overlapping functions that can compensate for the loss of NbALY615 and NbALY617.
Alternative Roles for ALY in Virus Infection
Our experiments have shown that the interaction between P19 and ALY in the highly susceptible host N. benthamiana may not be connected with the known roles of the P19 protein in this plant: silencing suppression and systemic necrosis. However, the possible role of ALY in TBSV movement in this or other plant species remains to be investigated.
In animals, ALY or ALY-associated proteins are specifically targeted by viral proteins. For example, the nucleoprotein of Influenza A virus binds to UAP56, a DEAD-box helicase that itself interacts with ALY, to stimulate virus-encoded RNA polymerase activity (Momose et al., 2001). In addition, the ICP27 protein of herpes simplex virus type 1 and EB2 of Epstein-Barr virus interact with ALY (Chen et al., 2002; Hiriart et al., 2003). These animal viruses replicate in the nucleus and require strategies to facilitate export of unspliced viral RNAs into the cytoplasm. However, replication/transcription of most RNA plant viruses, including TBSV, is thought to take place in the cytoplasm and therefore would not require ALY-mediated export of viral RNAs from the nucleus. Alternatively, P19 synthesized during TBSV infection may influence ALY-mediated export of host mRNAs, either to up-regulate or down-regulate the expression of specific host proteins. This may be related to the phenomenon of host transcriptional shutoff by viruses, which is well studied in animals (Weidman et al., 2003) but is less well understood in plants (Maule et al., 2002).
CONCLUSION
Although some of the more obvious functions of plant viral proteins have been identified, it is clear that many of these proteins are multifunctional and interact with the host cellular machinery in subtle and sophisticated ways. This is particularly the case for viral pathogenicity proteins, many of which recently have been shown to act as suppressors of RNA silencing, although how this activity relates to the effects these proteins often have on virus movement and symptom development is not understood. Our discovery that the TBSV P19 pathogenicity protein relocalizes some but not all of the plant ALY proteins from the nucleus to the cytoplasm offers us a way to understand in better detail how this virus co-opts plant processes to its own aims.
MATERIALS AND METHODS
Cloning of TBSV P19
The 19 K gene was cloned from an uncharacterized T-46 isolate of TBSV from the Scottish Crop Research Institute virus collection using PCR primers homologous to the BS3 isolate of TBSV (Luis-Areteaga et al., 1996). The 19 K gene was inserted into pENTR4 (Invitrogen, Carlsbad, CA) from which it was moved by recombinational cloning into yeast vectors (see below) and into pDEST14 (Invitrogen) for in vitro coupled transcription/translation.
Yeast Two-Hybrid Screens
The TBSV P19 gene was inserted into pAS2.1 (CLONTECH, Palo Alto, CA) previously modified by the insertion of an attR-cassette (Invitrogen) to allow recombinational cloning using the Gateway system (Invitrogen). This plasmid was transformed into yeast strain PJ69-4A and used as a bait for yeast two-hybrid screenings, employing an improved interaction mating protocol described previously (Soellick and Uhrig, 2001). Mating was carried out between the bait strain and yeast strain Y187 that had been transformed with a GAL4 activation domain-fused cDNA library from Arabidopsis, and interacting clones were selected by growth of colonies at 30°C on media lacking Leu, Trp, and His (SD-LWH) and supplemented with 3 to 20 mm 3-aminotriazol (3-AT). As a second reporter, the product of the MEL1 gene was quantitated according to Lazo et al. (1978).
Cloning of Full-Length Arabidopsis ALY and N. benthamiana ALY Genes
The Arabidopsis ALY genes were amplified by RT-PCR from poly(A+) RNA isolated from leaves of young plants. The ALY1 gene was amplified with primers 579 (AAAACATGTCGACTGGATTAGATATGTCTC) and 580 (GGGAATTCTTAGTTTGTCTCCATATCTCCAGAATG), ALY2 using primers 577 (AAAACATGTCAGGTGCTTAGATATGTC) and 578 (GGGAATTCTAACTTGTTTCCATTGCCTCTTTGTGG), ALY3 with primers 573 (AAAACATGTCAGACGCTTTGAATATGACTC) and 574 (GGGAATTCCTTAAGAGATGTTCATAGCTTCAGCATG), and ALY4 with primers 575 (AAAACATGTCTGGAGCATTGAATATGACTC) and 576 (GGGAATTCTTAAGAGGTGTTCATGGCATCAGCGTG). These primers added an NcoI-compatible BspLU11I restriction site at the translation start codon of each gene, and an EcoRI site downstream of the translation termination codon. The full-length genes were cloned into pGemT-easy (Promega, Madison, WI) for sequencing and then recloned into pENTR4 for further manipulation.
Database searches did not identify any ALY sequences from Nicotiana benthamiana or other Nicotiana species, thus a search was carried out of potato and tomato EST databases using a combination of the original mouse ALY gene and the four corresponding Arabidopsis ALY homologs (At5g59950 [ALY1], At5g02530 [ALY2], At1g66260 [ALY3], and At5g37720 [ALY4]), which were used in a BLAST search against a CAP3 (Huang and Madan, 1999) EST assembly made from both potato and tomato EST sequences in dbEST (http://www.ncbi.nlm.nih.gov/dbEST/) as of October 2002. From this search, contig consensus sequences were identified that correspond to the current Institute for Genomic Research (Rockville, MD) gene indices (http://www.tigr.org/tdb/tgi/plant.shtml) TC numbers TC53565 and TC 41221 (potato) and TC 101766 (tomato). PCR amplification of cDNA synthesized from total RNA isolated from leaves of N. benthamiana plants with primer pairs 615 (CCACATGTCAAATCTKGATGTATCTCTCG) and 616 (TTTTAGTTTGTCTGCATGGCTTCTGC), and 617 (CCCATGGCTGAGGCAGCTTTGGATATG) and 618 (TTTCAGTTCGTTTGCATTGCTTCTGTATG) added BspLU11I (615) or NcoI (617) restriction sites at the translation start codon of each gene and gave products of the expected size. The full-length genes were sequenced and cloned into pENTR4. Alignment of plant, mouse, human, and Drosophila ALY protein homologs was undertaken using ClustalW with additional manual refinement. BLAST analyzes used resources at Mouse Genome Informatics (http://www.informatics.jax.org/), Ensemble (http://www.ensemble.org), and Flybase (http://flybase.bio.indiana.edu/). Arabidopsis sequences are located at http://www.tair.org/.
Deletion Mutation of AtALY2
The RRM domain of the AtALY2 gene is flanked by a HindIII restriction site (nt 325) and a SapI restriction site (nt 580). Deleting the sequence between these sites created mutant HSΔ, which encodes both N-proximal and in-frame C-proximal RGG domains but lacks the central RRM domain. Mutant RHΔ is deleted between the HindIII site and an EcoRI site in pENTR4 immediately downstream of the AtALY2 gene and encodes only the N-proximal RGG domain of the ALY2 protein fused to eight vector plasmid-encoded residues at the C terminus. Mutant RSΔ is deleted between the SapI and EcoRI sites and encodes both the N-proximal RGG domain and central RRM domain fused to eight vector-specified C-terminal residues. The RRM domain of the AtALY2 gene was amplified using primers 743 (AAAAAGCAGGCTCTAAGCTTTACATTTCAAAC) and 744 (AGAAAGCTGGGTCTAAAGAGCTGGAGCAGAAAG), to add flanking sequences for recombinational cloning into Gateway vectors (Invitrogen). The deletion mutants and isolated RRM domain were recombined into pACT2-attR and assayed for their ability to interact with the TBSV P19.
Gel Overlay Assay
The full-length Arabidopsis ALY2 gene and the HSΔ, RSΔ, and RHΔ domain deletion mutants were cloned by recombination into pGEX-attR, a modified version of pGEX-3X (Promega), for expression as C-terminal fusions to the GST peptide. The plasmids were transformed into Escherichia coli BL21, and recombinant protein expression was induced at 30°C by the addition of 0.5 mm isopropylthio-β-galactoside to the growth medium; however, only unfused GST and GST:ALY2 RSΔ were expressed to significant levels (data not shown). To examine binding in vitro of the TBSV P19 to ALY, total proteins from induced E. coli cells were fractionated by SDS-PAGE and transferred to PVDF membrane. The blotted proteins were denatured in 7 m guanidinium hydrochloride and renatured in the presence of 140 mm NaCl/2 mm dithiothreitol (DTT) according to the method of Chen et al. (2000). TBSV P19 was labeled with S35-Met by coupled in vitro transcription/translation using a commercial kit (Novagen, Madison, WI), followed by filtration through Sephadex G25 (Pharmacia, Piscataway, NJ). The blotted membrane was treated at room temperature with blocking solution (10% glycerol, 0.5 mm EDTA, 100 mm NaCl, 20 mm Tris-Cl, pH 7.5, 0.1% Tween 20, 2% fat-free milk powder), then incubated at 4°C for 4 h in 3 mL blocking solution containing 1 mm DTT and 35S-labeled TBSV P19. The blot was washed several times in blocking solution, air dried, and exposed to film.
Expression and Analysis of GFP-Tagged Proteins
The four Arabidopsis and two N. benthamiana ALY genes were PCR amplified using primers to add a NcoI site at the translation initiation codon and a XbaI site in place of the termination codon. The GFP gene was PCR amplified using primers to add a XbaI site immediately upstream of the translation initiation codon and a KpnI site immediately after the termination codon. The PCR fragments were cloned in a three-part ligation into the TRV expression vector, TRV-GFPc (MacFarlane and Popovich, 2000), that had previously been digested with NcoI and KpnI to remove the GFP gene to produce constructs TRV-AtAly1-GFP, TRV-AtAly2-GFP, TRV-AtAly2 HSΔ-GFP, TRV-AtAly3-GFP, TRV-AtAly4-GFP, TRV-NbAly615-GFP, and TRV-NbAly617-GFP. A C-terminal GFP-fusion of the TBSV P19 protein (TRV-P19-GFP) was made using the same strategy. The GFP-tagged proteins were expressed by mixing transcripts of the various TRV constructs with total RNA from plants infected with TRV RNA1. The RNAs were inoculated to N. benthamiana plants and examined 2 to 4 d later by confocal fluorescence microscopy.
For expression by agroinfiltration, the AtALY2-GFP, AtALY2 HSΔ-GFP, unfused P19, and P19-GFP fusions were PCR amplified from the TRV plasmids with primers to add a 5′ BamHI site and a 3′ KpnI site and cloned into the same sites of the binary vector pROK2. The remaining ALY-GFP gene fusions were excised as a HpaI-KpnI fragment from the TRV vector plasmids and cloned into SmaI-KpnI digested pROK2. To obtain pROK2 expressing P19 fused to mRFP1 (Campbell et al., 2002), the P19 gene was amplified by PCR with primers adding a 5′ BamHI and 3′ XbaI sites, while the mRFP gene was amplified by PCR with primers adding a 5′ XbaI and 3′ KpnI site. The resulting fragments were ligated into pROK2 linearized with BamHI and KpnI in a three-piece ligation. The constructs were transformed into Agrobacterium tumefaciens strains LBA4404 and C58C1 for delivery into N. benthamiana plants by agroinfiltration (Voinnet et al., 2000). GFP- and mRFP-derived fluorescence in individual cells was detected and recorded using a Leica (Wetzlar, Germany) TCS SP spectral confocal laser scanning microscope (Gillespie et al., 2002).
Western-Blot Analysis of P19 Expression
For analysis of P19 expression, three discs (collected using the cap of an Eppendorf tube, approximately 60 mg tissue) were excised from leaves of N. benthamiana plants 3 d after infiltration with agrobacteria carrying binary plasmids containing wild-type or mutant P19 genes. The leaf discs were frozen in liquid nitrogen, homogenized with a micropestle, and extracted with 120 μL of buffer (7 m guanidine-HCl, 1 mm EDTA, 5 mm DTT, 20 mm Tris-Cl, pH 7.5). After extraction, the samples were equilibrated in 8 m urea/20 mm Tris-Cl, pH 7.2, and boiled for 5 min, and 20-μL aliquots were separated by electrophoresis through a 10% polyacrylamide/SDS gel. The proteins were transferred to PVDF membrane and probed with a mouse monoclonal antibody (1:20,000 dilution) specific for the TBSV P19 protein. Secondary probing used a hydrogen peroxidase-conjugated goat anti-mouse antibody (catalog no. 172-1011, 1:1,000 dilution; Bio-Rad, Cambridge, MA), followed by chemiluminescent detection using the ECL system (Amersham Bioscience, Buckinghamshire, UK). Equivalent loading of total proteins in each sample was examined by staining of the blotted membrane using Ponceau Red.
Single-Base Mutation of TBSV P19
Mutations were introduced into the TBSV P19 gene cloned in pENTR4 using the QuikChange kit (Stratagene, La Jolla, CA). Primers 603 (GGAAAGTTGTATTTAAGGGATATCTCAGATACG) and 604 (CGTATCTGAGATATCCCTTAAATACAACTTTCC) introduce a single base change that replaces the Arg residue at position 72 with Gly. Primers 605 (GGAAACTTCATTGTACGGAGTCCTTGGATCTTGG) and 606 (CCAAGATCCAAGGACTCCGTACAATGAAGTTTCC) were used to replace Arg 85 with Gly. The mutant P19 genes were recombined into pAS-attR for yeast two-hybrid screening, and into pLX222-attR, a binary vector, for transformation into A. tumefaciens strain LBA4404 and infiltration into plants for transient expression.
Silencing Assays
A 295-nt fragment at the N terminus of the NbALY615 gene (EMBL accession no. AJ697695) was amplified with oligonucleotide primers (ACTCCATGGATCTCTCGACGATT and TGAGCATGCTCGAGTTTGAAATGAGGAGC) to add 5′ NcoI and 3′ XhoI sites. Similarly, a 345-nt fragment at the N terminus of the NbALY617 gene (EMBL accession no. AJ697696) was amplified with oligonucleotide primers (CCCTCGAGATGGCTGAGGCAGCTTTGGATATG and GGGGGTACCGAGAAGAGCTCCTTGATATCC) to add 5′ XhoI and 3′ KpnI sites. The ALY fragments were ligated in tandem into the TRV vector (TRV-GFPc; MacFarlane and Popovich, 2000) to replace the GFP gene and to create the construct TRV-615/617. The construction of TRV-PDS was described before (Yu et al., 2003). In vitro transcripts of these constructs were mixed with TRV RNA1 and inoculated to plants as described before (MacFarlane and Popovich, 2000). For uninfected controls, plants were mock-inoculated using tap water. Plants were grown for around 3 weeks to allow VIGS to become established.
Silencing of the NbALY615 and NbALY617 genes was assessed by a qualitative RT-PCR assay. Leaf samples were collected and RNA isolated using the RNeasy kit as directed by the manufacturer (Qiagen, Valencia, CA). RNA samples were reverse transcribed using an oligo(dT) primer and MMLV reverse transcriptase according to standard procedures. PCR was carried out for 28 cycles, shown by experimentation to be within the range of exponential amplification. NbALY615-specific primers (TGCAGAGGCAGCTATCAAGA and TGGAGCAGGATTTGGATTTC) amplified a 142-nt fragment of the gene located outside of the region carried by TRV-615/617. Similarly, NbALY617-specific primers (GGAAGCCGATGAAGATTGAA and GATCAGTTCTGGGACCTCCA) amplified a 111-nt fragment of the gene located outside of the region carried by TRV-615/617. As a control, primers (AGCTGAGGGGAGGAATG and GCAACCTAGAAACCACC) from the Nicotiana tabacum ubiquitin gene (accession no. U66264) were used that also amplified a 250-nt region of the N. benthamiana ubiquitin gene.
Silencing of ALY was established in N. benthamiana plants expressing GFP. Upper leaves of these plants were then infiltrated with an Agrobacterium culture carrying a binary vector with a 35SP/GFP/NosT cassette to induce silencing of the GFP transgene as described before (Voinnet and Baulcombe, 1997). GFP expression was monitored by UV illumination, and at 8 d postinfiltration, leaf samples were collected and processed for mRNA and siRNA extraction and analyzed by northern blotting using digoxygenin-labeled GFP-specific probes using procedures described before (Canto et al., 2002).
For overexpression studies, nontransgenic N. benthamiana plants were infiltrated with a culture of Agrobacterium containing the binary plasmid pGPTV(+35,+NosT) expressing GUS (Canto et al., 2002) either alone or mixed with other cultures expressing ALY2, ALY1-GFP, ALY2-GFP, P19, or P19-mRFP. Expression of ALY-GFP and P19-mRFP was confirmed by confocal fluorescence microscopy. Infiltrated tissue was collected after 6 d and RNA samples were extracted and analyzed as described above.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ697695 and AJ697696.
Supplementary Material
Acknowledgments
We thank Maud Swanson for excellent technical assistance. We thank John Brown for careful reading of the manuscript, Klaus Salchert and Csaba Koncz for the Arabidopsis cDNA library, David Baulcombe for seeds of the 16c line of N. benthamiana expressing GFP, Elisa Izaurralde for the clone of Drosophila ALY, and Adrian Whitehouse for the human ALY clone. We also thank Herman Scholthof and Jozsef Burgyan for providing us with antibodies against P19.
This work was supported by the Scottish Executive Environment and Rural Affairs Department and by The Royal Society (ESEP grant no. 12822 to J.F.U. and S.A.M.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046086.
References
- Anandalakshmi R, Marathe R, Ge X, Herr JM Jr, Mau C, Mallory A, Pruss G, Bowman L, Vance VB (2000) A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290: 142–144 [DOI] [PubMed] [Google Scholar]
- Brown JWS, Shaw PJ (1998) Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell 10: 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn L, Munnerlyn A, Grosschedl R (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev 11: 640–653 [DOI] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto T, Cillo F, Palukaitis P (2002) Generation of siRNAs by T-DNA sequences does not require active transcription or homology to sequences in the plant. Mol Plant Microbe Interact 15: 1137–1146 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Cary SM, Parks TD, Dougherty WG (1989) A second proteinase encoded by a plant potyvirus genome. EMBO J 8: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H (2003) RNA interference: travelling in the cell and gaining functions? Trends Biochem Sci 19: 39–46 [DOI] [PubMed] [Google Scholar]
- Chen IB, Sciabica KS, Sandri-Goldin RM (2002) ICP27 interacts with the RNA export factor Aly/REF to direct Herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J Virol 76: 12877–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Sheng J, Hind G, Handa AK, Citovsky V (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for vital cell-to-cell movement. EMBO J 19: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M, Desvoyes B, Turina M, Noad R, Scholthof HB (2000) Genetic dissection of tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and systemic invasion. Virology 266: 79–87 [DOI] [PubMed] [Google Scholar]
- Denli AM, Hannon GJ (2003) RNAi: an ever-growing puzzle. Trends Biochem Sci 28: 196–201 [DOI] [PubMed] [Google Scholar]
- Gillespie T, Boevink P, Haupt S, Roberts AG, Toth R, Valentine T, Chapman S, Oparka KJ (2002) Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of Tobacco mosaic virus. Plant Cell 14: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govier DA, Kassanis B (1974) A virus-induced component of plant sap needed when aphids acquire potato virus Y from purified preparations. Virology 61: 420–426 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Vionnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small anti-sense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Havelda Z, Hornyik C, Crescenzi A, Burgyan J (2003) In situ characterisation of Cymbidium ringspot tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J Virol 77: 6082–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiriart E, Farjot G, Gruffat H, Nguyen MVC, Sergeant A, Mante E (2003) A novel nuclear export signal and a REF interacting domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J Biol Chem 278: 335–342 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC (2001) Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285: 71–81 [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Cronin S, Carrington JC (1997) Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 228: 251–262 [DOI] [PubMed] [Google Scholar]
- Lakatos L, Szittya G, Silhavy D, Burgyán J (2004) Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J 23: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo PS, Ochoa AG, Gascon S (1978) Alpha-Galactosidase (melibiase) from Saccharomyces carlsbergensis: structural and kinetic properties. Arch Biochem Biophys 191: 316–324 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000. b) The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE (2000. a) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev 14: 1098–1108 [PMC free article] [PubMed] [Google Scholar]
- Luis-Areteaga M, Rodriguez-Cerezo E, Fraile A, Saez E, Garcia-Arenal F (1996) Different tomato bushy stunt virus strains cause disease outbreaks on solanaceous crops in Spain. Phytopathology 86: 535–542 [Google Scholar]
- MacFarlane SA, Popovich AH (2000) Expression of foreign proteins in roots from tobravirus vectors. Virology 267: 29–35 [DOI] [PubMed] [Google Scholar]
- Maule A, Leh V, Lederer C (2002) The dialogue between viruses and hosts in compatible interactions. Curr Opin Plant Biol 5: 279–284 [DOI] [PubMed] [Google Scholar]
- Moissiard G, Voinnet O (2004) Viral suppression of RNA silencing in plants. Mol Plant Pathol 5: 71–82 [DOI] [PubMed] [Google Scholar]
- Momose F, Basler CF, O'Neill RE, Iwamatsu A, Palese P, Nagata K (2001) Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J Virol 75: 1899–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-W, Faure-Rabasse S, Robinson MA, Desvoyes B, Scholthof HB (2004) The multifunctional plant viral suppressor of gene silencing P19 interacts with itself and an RNA binding host protein. Virology 323: 49–58 [DOI] [PubMed] [Google Scholar]
- Qiu W, Park J-W, Scholthof HB (2002) Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol Plant Microbe Interact 15: 269–280 [DOI] [PubMed] [Google Scholar]
- Ratcliff FG, MacFarlane SA, Baulcombe DC (1999) Gene silencing without the gene: RNA-mediated cross protection between viruses. Plant Cell 11: 1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Foseca M, Izaurralde E (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci USA 98: 1030–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof HB, Morris TJ, Jackson AO (1995. b) Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7: 1157–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof HB, Scholthof KBG, Kikkert M, Jackson AO (1995. a) Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology 213: 425–438 [DOI] [PubMed] [Google Scholar]
- Shi XM, Miller H, Verchot J, Carrington JC, Bowman Vance V (1997) Mutations in the region encoding the central domain of helper component-proteinase (HC-Pro) eliminate potato virus X/potyviral synergism. Virology 231: 35–42 [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J (2002) A viral protein suppresses RNA silencing and binds RNA silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21: 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soellick TR, Uhrig JF (2001) Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol 2: research 0052.1–0052.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko S, Inze D, Van Montagu M, Kushnir S (2001) Arabidopsis coactivator ALY-like proteins, DIP1 and DIP2, interact physically with the DNA-binding domain of the Zn-finger poly(ADP-ribose) polymerase. J Exp Bot 52: 1375–1380 [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E (2000) REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turina M, Omarov R, Murphy JF, Bazaldua-Hernandez C, Desvoyes B, Scholthof HB (2003) A newly identified role for Tomato bushy stunt virus P19 in short distance spread. Mol Plant Pathol 4: 67–72 [DOI] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115: 799–811 [DOI] [PubMed] [Google Scholar]
- Virbasius C-MA, Wagner S, Green MR (1999) A human nuclear-localised chaperone that regulates dimerization, DNA binding and transcriptional activity of bZIP proteins. Mol Cell 4: 219–228 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC (1997) Systemic signalling in gene silencing. Nature 389: 553. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103: 157–167 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe DC (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Weidman MK, Sharma R, Raychaudhuri S, Kundu P, Tsai W, Dasgupta A (2003) The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res 95: 75–85 [DOI] [PubMed] [Google Scholar]
- Ye K, Malinina L, Patel DJ (2003) Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426: 874–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Fan B, MacFarlane SA, Chen Z (2003) Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol Plant Microbe Interact 16: 206–216 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo M-J, Straesser K, Katahira J, Hurt E, Reed R (2000) The protein ALY links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.