Abstract
Polygalacturonase-inhibiting proteins (PGIPs) are extracellular plant inhibitors of fungal endopolygalacturonases (PGs) that belong to the superfamily of Leu-rich repeat proteins. We have characterized the full complement of pgip genes in the bean (Phaseolus vulgaris) genotype BAT93. This comprises four clustered members that span a 50-kb region and, based on their similarity, form two pairs (Pvpgip1/Pvpgip2 and Pvpgip3/Pvpgip4). Characterization of the encoded products revealed both partial redundancy and subfunctionalization against fungal-derived PGs. Notably, the pair PvPGIP3/PvPGIP4 also inhibited PGs of two mirid bugs (Lygus rugulipennis and Adelphocoris lineolatus). Characterization of Pvpgip genes of Pinto bean showed variations limited to single synonymous substitutions or small deletions. A three-amino acid deletion encompassing a residue previously identified as crucial for recognition of PG of Fusarium moniliforme was responsible for the inability of BAT93 PvPGIP2 to inhibit this enzyme. Consistent with the large variations observed in the promoter sequences, reverse transcription-PCR expression analysis revealed that the different family members differentially respond to elicitors, wounding, and salicylic acid. We conclude that both biochemical and regulatory redundancy and subfunctionalization of pgip genes are important for the adaptation of plants to pathogenic fungi and phytophagous insects.
Molecular diversification is crucial in plant-pathogen interactions. Pathogens encounter a vast number of hosts, against which polymorphic molecular weapons have been evolved (Gassmann et al., 2000; Idnurm and Howlett, 2001). Conversely, plants rely for defense on a finely honed innate surveillance apparatus consisting of highly polymorphic recognition molecules (Bergelson et al., 2001). For example, plants recognize cell wall-degrading enzymes produced by pathogenic (micro)organisms and inhibit their enzymatic activity to hamper the invasion process and the release of nutrients necessary for pathogen growth. Among the inhibitors of cell wall-degrading enzymes so far identified (Bellincampi et al., 2004, and references therein), polygalacturonase-inhibiting proteins (PGIPs) have been clearly shown to play a role in defense (De Lorenzo and Ferrari, 2002). Endopolygalacturonases (PGs; EC 3.2.1.15) cleave the α-(1→4) linkages between d-GalUA residues in homogalacturonan and cause separation of cells and maceration of host tissue. The importance of these enzymes in pathogenesis has been demonstrated for fungi and bacteria (De Lorenzo and Ferrari, 2002; Oeser et al., 2002) and has been proposed for nematodes (Jaubert et al., 2002). PGs from salivary glands are considered a main cause of plant damage by phytophagous bugs (Heteroptera: Miridae) and other insects (Girard and Jouanin, 1999; Wheeler, 2001; Boyd et al., 2002). To accommodate pathogenesis in a variety of different conditions and on various hosts, pathogens produce a variety of PG isoenzymes (De Lorenzo et al., 2001), which often exhibit polymorphism in different isolates or races (Caprari et al., 1993; Daroda et al., 2001; Poinssot et al., 2003).
Against the many PGs produced by pathogens, plants have evolved many PGIPs. In addition to reducing the aggressive potential of PGs, PGIPs favor the formation of long-chain oligogalacturonides (OGs) that are able to induce defense responses (De Lorenzo et al., 2001). Consistent with their role in defense, PGIPs are ubiquitous in flowering plants and are up-regulated in response to stress-related signals, wounding, and fungal and insect attack (De Lorenzo et al., 2001, and references therein; Li et al., 2003; Ndimba et al., 2003). The overexpression of PGIPs in transgenic plants limits fungal colonization (Powell et al., 2000; Ferrari et al., 2003), thereby demonstrating the defensive potential of these proteins.
The Leu-rich repeat (LRR) structure of PGIPs is responsible for the molecular interaction with PGs. The LRR structure is shared by many plant proteins involved in recognition, such as the majority of the resistance (R) gene products (Martin et al., 2003), and several receptors involved in development, perception of hormones (Becraft, 2002; Szekeres, 2003), or elicitors (Gomez-Gomez et al., 2001), defense responses against insects (Szekeres, 2003), and bacterial and fungal symbiosis (Kistner and Parniske, 2002). The 10 LRRs of PGIPs are of the extracytoplasmic type (LxxLxLxxNxLT/SGxIPxxLxxLxx) and are organized to form two β-sheets, B1 and B2 (Di Matteo et al., 2003). It has been shown that a single amino acid variation in B1 confers the ability to recognize a novel PG (Leckie et al., 1999), implying that diversity in the LRR domain is crucial for determining recognition specificity toward pathogens.
PGIPs are encoded by small gene families in many plant species (De Lorenzo et al., 2001; Li et al., 2003). So far only the Arabidopsis gene family, which comprises two tandemly duplicated genes, has been studied in detail and shown to encode functionally redundant proteins that are up-regulated through different signal transduction pathways in response to fungal infection (Ferrari et al., 2003).
Gene duplication, divergence, and selection pressure are major mechanisms shaping gene families, and family sizes are often the result of selection for useful functions. PGIP families provide an interesting case of molecular evolution and adaptation. Their structure is the result of coevolution with PGs of pathogenic organisms. Moreover, PGIPs, like PGs, are subject to both functional constraints and selection pressure for diversification.
In this study, we have studied the significance of diversity in the bean (Phaseolus vulgaris) pgip family, which has been mapped in the B2 linkage group of the bean core map (Freyre et al., 1998). We report here that the full complement of pgip genes in the bean genotype BAT93 is represented by four clustered genes. We show that diversification of paralogous genes is associated with different modes of regulation, as well as with functional redundancy and subfunctionalization for recognition of PGs of fungi and phytophagous insects.
RESULTS
Isolation of pgip Genes of Bean Genotypes BAT93 and Pinto
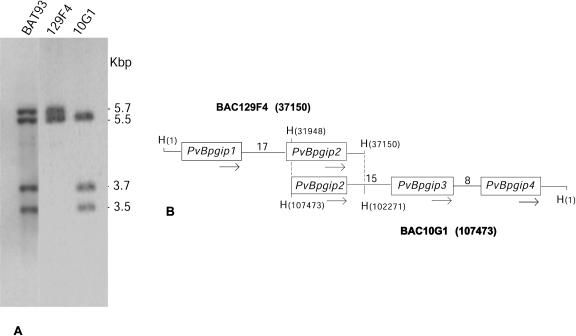
To investigate intra- and intergenotype variation in bean pgip genes, the full complements of pgip genes of the Mesoamerican genotypes BAT93 and Pinto were characterized. From a BAT93 bacterial artificial chromosome (BAC) library, two overlapping clones (129F4 and 10G1) were isolated that contained all pgip genes, as shown by a Southern blot probed with a fragment corresponding to the portion encoding the mature protein of the previously characterized Pinto pgip1 gene (Leckie et al., 1999; Fig. 1A). Sequencing of the two clones showed the presence of four intronless pgip open reading frames (ORFs) oriented in the same direction and indicated as PvBpgip1 and PvBpgip2, on the basis of their similarity to previously characterized bean pgip genes (Fig. 1B), and PvBpgip3 and PvBpgip4. The two latter genes have never been described before. The Pvpgip locus spans about 50 kb and does not comprise additional pgip-related sequences. Distances between the four genes (about 17, 15, and 8 kb) are much larger than those observed between members of the Arabidopsis and soybean pgip families (about 500 bp and 3,000 bp, respectively).
Figure 1.
Genomic organization of the pgip gene family in bean genotype BAT93. A, Southern-blot analysis of genomic DNA from line BAT93 and plasmid DNA from 129F4 and 10G1 BAC clones; DNA was double digested with EcoRI-HindIII and probed with a fragment corresponding to the portion of the PvPpgip1 ORF encoding the mature protein. The 5.5-kb hybridizing fragment is present in both BAC clones. B, Schematic representation of the arrangement of the PvBpgip genes as determined by sequencing the 129F4 and 10G1 BAC clones. The two clones overlap for about 5.5 kb; this region includes PvBpgip2. Numbers between pgip members represent distances in kb. Arrows indicate the direction of the coding region from ATG to stop codon. H, HindIII restriction site. Numbers in brackets indicate the position of the terminal HindIII sites in each BAC clone, where numbering starts from the HindIII site following the T7 promoter sequence of pBeloBAC11. In the overlapping region, the positions of the corresponding HindIII sites in the two BAC clones are also indicated.
Pinto exhibited a Southern-blot hybridization pattern identical to that of BAT93 (data not shown). Characterization of cDNAs of Pinto Pgip1 and Pgip2, hereon indicated as PvPpgip1 and PvPpgip2.1, has been described previously (Leckie et al., 1999). To isolate additional genes, a phage genomic library was screened with the pgip-specific probe described above. Among 34 positive clones analyzed by double digestion with EcoRI and HindIII, clones Pt38, Pt10, and Pt21 exhibited specific hybridization to three (5.7, 3.7, and 3.5 kb) of the four EcoRI/HindIII fragments observed in Southern-blot analysis of genomic DNA. The hybridizing inserts of the three phage clones, individually subcloned and sequenced, each contained a pgip-related ORF (PvPpgip2.2, PvPpgip3, and PvPpgip4). PvPpgip2.2 differed from PvPpgip2.1 in four synonymous substitutions in the coding region and a 2-nucleotide (nt) insertion (AA) in the 3′ untranslated region (UTR), likely due to sequence variation between the Pinto seeds used for the construction of the cDNA and genomic libraries. Pinto is a commercial class of dry bean that includes a number of bean varieties.
Sequence Diversity of Pvpgip Genes and Their Encoded Products
The products encoded by the isolated Pvpgip genes share the typical PGIP topology, which includes a signal peptide for secretion (domain A), a 52-amino acid N-terminal domain B, a domain C comprising 10 imperfect LRRs, and a C-terminal 24-amino acid domain D. All exhibit the eight Cys residues, which are conserved in all PGIPs: four are located in domain B, one in the 10th LRR, and three in the C-terminal domain D (Fig. 2). Numbering of pgip nucleotide sequences reported in this work differs from that in previous articles, where +1 indicated the A of the first of two in-frame putative translation initiation codons present in the Pvpgip1 ORF (Toubart et al., 1992; Devoto et al., 1998; Leckie et al., 1999). Since the first Met codon is absent in all the other Pvpgip genes, the start of the ORFs is now assigned to the second ATG codon of Pvpgip1. Numbering of PGIP amino acid residues has also been changed in this article and starts from the first residue of the mature protein, which corresponds to residue 30 (a Glu) described in a previous article by Leckie et al. (1999).
Figure 2.
Variability of bean PGIPs. Deduced amino acid sequences of BAT93 PGIPs (PvBPGIP1-4) are aligned. Numbering is referred to the PvBPGIP1 sequence and starts from the first residue of the mature protein. Domains are on the basis of crystallografic analysis (Di Matteo et al., 2003). The xxLxLxx region is boxed. Empty spaces indicate gaps. Cys residues are in bold; putative glycosylation sites are underlined and in italics. Sites that vary in Pinto PvPPGIPs are highlighted in gray; corresponding Pinto residues are indicated beyond the vertical line on the right. The 3-amino acid deletion that distinguish BAT93 PvBPGIP2 from Pinto PvPPGIP2 is indicated by dashes and is also highlighted in gray.
Nucleotide identity/amino acid divergence among BAT93 and Pinto pgip genes and their encoded products are summarized in Table I. On the basis of their similarity, in both bean genotypes the four paralogous genes form two pairs (Pvpgip1/Pvpgip2 and Pvpgip3/Pvpgip4), which may represent functionally distinct classes of PGIPs. The presence of two insertions, one located in the signal peptide and one in the eighth LRR, characterizes the products of the pair Pvpgip3/Pvpgip4 (Fig. 2). The two deduced proteins lack the first putative N-linked glycosylation site (at position 35), which has been shown to be occupied by a typical complex N-glycan in Pinto PvPPGIP2 (Mattei et al., 2001). In a comparison with the products of the paralogous Pvpgip3 genes, both Pinto and BAT93 PvPGIP4 show an additional Cys residue in domain D, as well as two single amino acid deletions in the seventh and eighth LRRs (Fig. 2).
Table I.
Nucleotide identities and amino acid variationsa among pgip genes of bean genotypes Pinto and BAT93
| PvPpgip1 | PvPpgip2.1 | PvPpgip2.2 | PvPpgip3 | PvPpgip4 | PvBpgip1 | PvBpgip2 | PvBpgip3 | PvBpgip4 | |
|---|---|---|---|---|---|---|---|---|---|
| PvPpgip1 | – | 8 + 0 | 8 + 0 | 66 + 3 | 79 + 1 | 1 + 0 | 7 + 3 | 67 + 3 | 79 + 1 |
| PvPpgip2.1 | 97.4% | – | 0 + 0 | 65 + 3 | 76 + 1 | 9 + 0 | 0 + 3 | 66 + 3 | 76 + 1 |
| PvPpgip2.2 | 97.6% | 99.6% | – | 65 + 3 | 76 + 1 | 9 + 0 | 0 + 3 | 66 + 3 | 76 + 1 |
| PvPpgip3 | 82.5% | 83.2% | 82.3% | – | 30 + 2 | 67 + 3 | 65 + 6 | 1 + 0 | 30 + 2 |
| PvPpgip4 | 79.8% | 80.5% | 80.3% | 94.9% | – | 80 + 1 | 74 + 4 | 31 + 2 | 0 + 0 |
| PvBpgip1 | 99.9% | 97.3% | 97.7% | 82.4% | 79.7% | – | 8 + 3 | 68 + 3 | 80 + 1 |
| PvBpgip2 | 97.8% | 99.6% | 100% | 83.0% | 80.5% | 97.7% | – | 66 + 6 | 74 + 4 |
| PvBpgip3 | 82.4% | 83.0% | 82.8% | 99.9% | 94.8% | 82.3% | 82.9% | – | 31 + 2 |
| PvBpgip4 | 79.8% | 80.5% | 80.3% | 94.9% | 100% | 79.6% | 80.5% | 94.8% | – |
Only sequences corresponding to mature proteins have been considered. For each comparison, normal characters (bottom left part of the table) indicate nucleotide identities excluding indels. Numbers in bold characters (top right) indicate total amino acid replacements and indels, respectively.
Intergenotype comparison of corresponding ORF sequences of Pinto and BAT93 shows that, whereas Pvpgip4 genes are identical, Pvpgip1 and Pvpgip3 show a single nonsynonymous 1-nt replacement at codons 265 and 9, respectively. Instead, a 9-nt deletion distinguishes BAT93 PvBpgip2 from Pinto PvPpgip2.2; the resulting amino acid deletion includes residue 224 (previously indicated as 253) that has been shown to be crucial for recognition of FmPG (Leckie et al., 1999; Fig. 2).
Redundant and Unique Functions of Paralogous Pvpgip Genes and Intergenotype Variation
The effect of sequence variation on PGIP function was investigated. The inhibitor activities of Pinto PvPPGIP1 and PvPPGIP2 toward several fungal PGs have been described previously (Desiderio et al., 1997; Leckie et al., 1999). The proteins encoded by the four BAT93 PvBpgip genes and the Pinto PvPpgip3 and PvPpgip4 were expressed in Nicotiana benthamiana using a vector based on potato virus X (PVX; Baulcombe et al., 1995). Western-blot analysis of crude protein extracts from plants inoculated with each construct was carried out to demonstrate the presence of a PGIP-specific signal with a molecular mass of 39 kD, which was absent in extracts from noninfected plants or from plants infected with the empty vector (data not shown).
Because the presence of multiple PGIPs may respond to the need of inhibiting PGs in different microenvironments, such as those created, for example, by altered ion fluxes during the plant defense response, or the need of inhibiting PGs of diverse organisms harmful to plants, we tested the activity of the PvPGIPs purified from N. benthamiana protein extracts at two different pH values (4.7 and 7.0), and against PGs of fungi and insects. The two plant bugs Lygus rugulipennis Poppius and Adelphocoris lineolatus (Goeze; Heteroptera: Miridae) were examined in this study.
The tested PGs differed in their activity: PGs of Colletotrichum acutatum (CaPG) and Botrytis cinerea (BcPG) showed activity at both pH 4.7 and 7.0; PGs of Aspergillus niger (AnPG), Fusarium moniliforme (FmPG), and Stenocarpella maydis (SmPG) showed activity only at pH 4.7, while PGs of both L. rugulipennis and A. lineolatus were active only at pH 7.0. Our analyses showed that PGIPs encoded by paralogous genes all exhibited different recognition abilities and that inhibitory activities of all BAT93 PvBPGIPs, except PvBPGIP2, were identical to the corresponding Pinto proteins (Table II). Only PvPGIP3 of both BAT93 and Pinto were unable to inhibit AnPG, and only Pinto PvPPGIP2 inhibited FmPG, because the 3-amino acid deletion encompassing residue 224 abolishes the ability of BAT93 PvBPGIP2 to inhibit this enzyme. All eight PvPGIPs inhibited CaPG, and SmPG, albeit with different strength. Notably, inhibitory activity toward PGs of insects was exhibited only by PvPGIP3 and PvPGIP4, which are weak inhibitors of fungal PGs when compared to PvPGIP1 and PvPGIP2 (Table II).
Table II.
Inhibitory activitiesa of PGIPs of bean genotypes BAT93 and Pinto against PGs of fungi and insects
| Polygalacturonase | pHb | Relative Activityc | PvBPGIP1 PvPPGIP1 | PvBPGIP2 | PvPPGIP2 | PvBPGIP3 PvPPGIP3 | PvBPGIP4 PvPPGIP4 |
|---|---|---|---|---|---|---|---|
| Aspergillus niger | 4.7 | 1 | 140 | 10 | 1 | ∞ | 250 |
| 7.0 | 0 | - | - | - | - | - | |
| Fusarium moniliforme | 4.7 | 1 | ∞ | ∞ | 9 | ∞ | ∞ |
| 7.0 | 0 | - | - | - | - | - | |
| Stenocarpella maydis | 4.7 | 1 | 7.5 | 4.5 | 5 | 260 | 65 |
| 7.0 | 0 | - | - | - | - | - | |
| Colletotrichum acutatum | 4.7 | 1 | 16 | 12 | 12 | 230 | 1,200 |
| 7.0 | 0.8 | 16 | 15 | 13 | 690 | 1,400 | |
| Botrytis cinerea | 4.7 | 1 | 200 | 16 | 2.5 | 100 | 70 |
| 7.0 | 1.1 | ∞ | 17 | 4 | ∞ | 120 | |
| Lygus rugulipennis | 4.7 | 0 | - | - | - | - | - |
| 7.0 | 1 | ∞ | ∞ | ∞ | 490 | 1,950 | |
| Adelphocoris lineolatus | 4.7 | 0 | - | - | - | - | - |
| 7.0 | 1 | ∞ | ∞ | ∞ | 2,100 | 1,954 |
Values indicate the amount (in ng) of PGIP that determines 50% inhibition of 1 agarose plate unit (at pH 4.7) of PG. The symbol ∞ indicates >6 μg.
Enzyme activities were assayed in 20 mm Na acetate pH 4.7 or 7.0.
Activity at pH 4.7 is taken as a reference (=1).
The xxLxxLxx-Encoding Region Is the Most Variable Portion of Pvpgip Genes
Because the structural and functional viability of PGIPs likely derives from a positive diversifying evolution toward the many PGs produced by pathogens, we evaluated the type of selection acting on the bean pgip family by using the method that compares the number of nonsynonymous (amino acid changing; Ka) and synonymous (silent; Ks) substitutions per site. In the absence of selection on codon usage, Ks represents the frequency of neutral substitutions. With no selection pressure being present, Ka/Ks ratios are predicted to be 1, whereas in the case of positive diversifying and purifying selection, ratios >1 and <1, respectively, are obtained (Li, 1997). Pairwise comparisons between all Pvpgip members, except Pvpgip3/Pvpgip4, produced Ks that significantly exceeded Ka (Ka/Ks < 0.35; P < 0.05), indicating that purifying rather than diversifying selection has acted on the gene as a whole. For Pvpgip3/Pvpgip4 instead Ka was not significantly different from Ks and was significantly higher (P < 0.05) than that obtained for Pvpgip1/Pvpgip2 (Table III). We hypothesize that the latter pair has been subject to a stronger purifying selection pressure.
Table III.
Ka and Ks values in pgip genes of bean genotype BAT93
| Complete Gene
|
Domain B
|
Region C-Outa
|
xxLxLxx
|
xx(L)x(L)xxb
|
Domain D
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pinto | BAT93 | Pinto | BAT93 | Pinto | BAT93 | Pinto | BAT93 | Pinto | BAT93 | Pinto | BAT93 | ||
| Pvpgip1/2 | Ka | 0.010 | 0.011 | 0.000 | 0.000 | 0.002 | 0.005 | 0.023 | 0.018 | 0.033 | 0.026 | 0.053 | 0.053 |
| Ks | 0.051 | 0.052 | 0.040 | 0.040 | 0.043 | 0.043 | 0.086 | 0.087 | 0.127 | 0.129 | 0.043 | 0.043 | |
| Ka/Ks | 0.196 | 0.211 | 0.000 | 0.000 | 0.046 | 0.116 | 0.267 | 0.206 | 0.259 | 0.201 | 1.232 | 1.232 | |
| Pvpgip1/3 | Ka | 0.124 | 0.124 | 0.098 | 0.107 | 0.114 | 0.112 | 0.192 | 0.192 | 0.233 | 0.235 | 0.062 | 0.062 |
| Ks | 0.426 | 0.431 | 0.444 | 0.447 | 0.383 | 0.386 | 0.399 | 0.405 | 0.382 | 0.394 | 0.641 | 0.641 | |
| Ka/Ks | 0.291 | 0.287 | 0.220 | 0.239 | 0.297 | 0.290 | 0.481 | 0.474 | 0.609 | 0.596 | 0.096 | 0.096 | |
| Pvpgip1/4 | Ka | 0.159 | 0.156 | 0.096 | 0.096 | 0.130 | 0.128 | 0.280 | 0.276 | 0.357 | 0.352 | 0.177 | 0.177 |
| Ks | 0.484 | 0.486 | 0.443 | 0.443 | 0.443 | 0.447 | 0.519 | 0.511 | 0.510 | 0.497 | 0.558 | 0.558 | |
| Ka/Ks | 0.328 | 0.320 | 0.216 | 0.216 | 0.293 | 0.286 | 0.539 | 0.540 | 0.700 | 0.708 | 0.317 | 0.317 | |
| Pvpgip2/3 | Ka | 0.124 | 0.124 | 0.098 | 0.107 | 0.112 | 0.106 | 0.192 | 0.199 | 0.233 | 0.246 | 0.082 | 0.082 |
| Ks | 0.407 | 0.412 | 0.363 | 0.364 | 0.401 | 0.404 | 0.355 | 0.361 | 0.320 | 0.330 | 1.031 | 1.031 | |
| Ka/Ks | 0.304 | 0.300 | 0.269 | 0.293 | 0.279 | 0.262 | 0.540 | 0.551 | 0.728 | 0.745 | 0.079 | 0.079 | |
| Pvpgip2/4 | Ka | 0.158 | 0.154 | 0.096 | 0.096 | 0.127 | 0.121 | 0.285 | 0.284 | 0.363 | 0.366 | 0.170 | 0.170 |
| Ks | 0.459 | 0.465 | 0.361 | 0.361 | 0.462 | 0.466 | 0.447 | 0.461 | 0.410 | 0.426 | 0.983 | 0.983 | |
| Ka/Ks | 0.344 | 0.331 | 0.265 | 0.265 | 0.274 | 0.259 | 0.637 | 0.616 | 0.885 | 0.859 | 0.172 | 0.172 | |
| Pvpgip3/4 | Ka | 0.048 | 0.048 | 0.017 | 0.025 | 0.022 | 0.022 | 0.118 | 0.113 | 0.159 | 0.154 | 0.107 | 0.107 |
| Ks | 0.073 | 0.073 | 0.000 | 0.000 | 0.097 | 0.097 | 0.071 | 0.073 | 0.085 | 0.087 | 0.040 | 0.040 | |
| Ka/Ks | 0.657 | 0.657 | NAc | NAc | 0.226 | 0.226 | 1.661 | 1.547 | 1.870 | 1.770 | 2.675 | 2.675 | |
| Range Ka/Ks | 0.196–0.657 | 0.211–0.657 | 0.000–0.269 | 0.000–0.293 | 0.046–0.297 | 0.116–0.290 | 0.267–1.661 | 0.206–1.547 | 0.259–1.870 | 0.201–1.770 | 0.079–2.675 | 0.079–2.675 | |
| Average Ka/Ks | 0.353 | 0.351 | 0.194 | 0.202 | 0.235 | 0.239 | 0.687 | 0.655 | 0.841 | 0.813 | 0.761 | 0.761 | |
Region C-out represents the region encoding domain C outside of the xxLxLxx.
xxLxLxx-encoding region without the triplets for the conserved aliphatic residues.
NA, Not applicable.
Because different regions in a gene can be subject to different selection pressures, Ka/Ks ratios were also calculated separately for the regions of the pgip ORF encoding the N-terminal domain B (region B), the LRR domain C excluding the xxLxLxx motifs that includes the β-sheet B1 (region C-out), the portion corresponding to the 10 xxLxLxx motifs with and without the conserved hydrophobic residues (xxLxLxx and xx(L)x(L)xx regions, respectively), and the C-terminal domain D (region D). This analysis revealed that patterns of substitutions are not equivalent among the different protein regions (Table III). In regions B and C-out, Ka values were significantly lower than Ks values (Ka/Ks < 0.30; P < 0.05), suggesting purifying selection. In the xx(L)x(L)xx region instead all Ka values were significantly higher (P < 0.01) than those of regions B and C-out and not significantly different from Ks values (Ka/Ks ratios approximately 1). Variability in domain D appeared to be intermediate between that of the B and C-out regions and that of the xx(L)x(L)xx region. This analysis therefore shows that the xxLxLxx region is the most variable portion of the Pvpgip genes but provides no evidence for positive selection.
The Members of the Pvpgip Family Are Differentially Regulated
We also investigated whether the presence of multiple genes might respond to the need of a differential expression in different stress situations. Because the high degree of identity at the nucleotide level between the pgip genes makes difficult the design of gene-specific probes for northern-blot analysis, a reverse transcription (RT)-PCR based approach was used to study their regulation in Pinto. Primers specific for PvPpgip1, PvPpgip2 (both PvPpgip2.1 and PvPpgip2.2), PvPpgip3, and PvPpgip4 were synthesized (see “Materials and Methods”); they included, as the last base at the 3′ end, a mismatched nucleotide specific for the individual gene members.
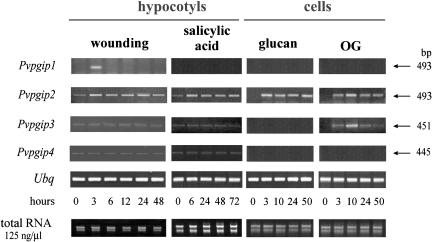
Analyses were carried out on RNA extracted from bean hypocotyls at different times after wounding, or treatment with salicylic acid (SA) or water (control), and from suspension-cultured cells after treatment with OGs or glucan from Phytophthora megasperma f. sp. glycinea or water (control). PCR amplification followed the RT of total RNA using the appropriate antisense primer. Conditions for PCR were optimized to show induction relative to basal levels (time 0).
Transcripts of PvPpgip1 accumulated 3 h after wounding and returned to basal levels after 6 h; the other treatments had no effects. Even with a high number of amplification cycles and at an RNA template concentration higher than that used for the analysis of the other PvPpgip genes, amplification of PvPpgip1 never occurred in RNA samples extracted at time 0 from both untreated tissues and suspension-cultured cells. Transcripts of PvPpgip2 were induced by all the treatments examined. PvPpgip3 transcripts were induced in suspension-cultured cells in response to OGs but not to glucan, and did not vary in hypocotyls upon wounding or SA treatment, whereas PvPpgip4 transcripts were not induced by any treatment (Fig. 3).
Figure 3.
Differential expression of Pinto pgip genes in response to stress-related stimuli. RT-PCR was performed on total RNA extracted from bean tissues or cells at different times after the indicated treatments, using the gene-specific primers described in “Materials and Methods.” Amplification products were analyzed by electrophoresis on agarose gel; the expected size of amplified fragment for each experiment is indicated by the arrow on the right. Conditions for PCR were optimized to show induction relative to basal levels (time 0). Comparison of band intensities is meaningful only within a single data set. Analyses of the water-treated controls showed no induction (data not shown).
Analysis of Sequences Upstream and Downstream of the pgip-Coding Regions
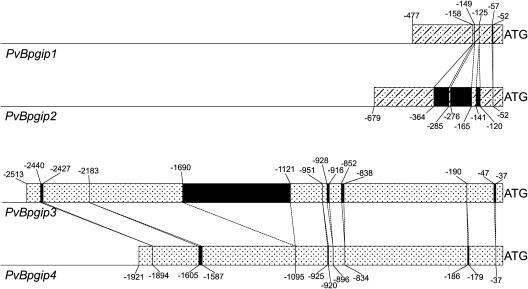
The observed difference in their regulation of expression reflects differences in the sequence of the Pvpgip regulatory regions. PvBpgip1 and PvBpgip2 share 93.4% identity in 500 bp of the 5′ region proximal to the ATG, with several insertions/deletions (indels; Fig. 4), and 97% identity in the proximal 400-bp region downstream of the stop codon. Putative 3′ UTR, deduced by comparison with the Pinto cDNA sequences, are of about 100 bp and differ by only 3 nt. The 5′ region proximal to the ATG of PvBpgip1 shares about 99% identity with the corresponding region of the Saxa Pvpgip1 (Toubart et al., 1992), with two deletions of 27 bp and 48 bp at positions −1,109 and −928, while the 3′ UTRs differ by 2 nt.
Figure 4.
Schematic representation of the 5′ flanking region of the four BAT93 pgip genes. Regions sharing sequence similarity are indicated by dots-dashes in PvBpgip1 and PvBpgip2 or dots in PvBpgip3 and PvBpgip4. Black boxes indicate insertions. The A of the ATG codon corresponds to +1.
Regions sharing sequence similarity between PvBpgip3 and PvBpgip4 span about 2,500 bp upstream of the translation initiation codon (95.1% identity), with an indel of 569 bp and a few additional short ones, and about 600 bp at the 3′ of the translation stop codon (96% identity). No similarity was observed in the flanking regions between the pairs PvBpgip1/PvBpgip2 and PvBpgip3/PvBpgip4. PvBpgip3 and PvBpgip4 abruptly diverge from PvBpgip1 and PvBpgip2 upstream of nt −21 and immediately downstream of the translation stop codon, with only a few short nucleotide stretches conserved up to 150 nt after the TAA.
A number of sequences with significant similarity to known cis-acting elements are present in the 1.5-kb region upstream from the start codon of all four PvBpgip genes. In particular, several W-box elements, which have been identified in the promoters of several defense genes and are known to mediate transcriptional responses to pathogen-derived elicitors (Eulgem et al., 2000), can be observed. Notably, PvBpgip1 and PvBpgip2 contain six (−84, −97, −332, −352, −486, −502) and five (−79, −538, −727, −736, −764) W-boxes in the proximal 500 bp and 800 bp from the start codon, respectively, whereas PvBpgip3 contains a single W-box at −1,220 and PvBpgip4 contains none.
Regions immediately upstream of the ATG and downstream of the TAA of the cloned Pinto pgip genes were analyzed and compared to the corresponding regions of the BAT93 genes. We sequenced 1,607 bp, 1,786 bp, and 357 bp of the 5′ regions, and 581 bp, 896 bp, and 131 bp of 3′ regions of PvPpgip2.2, PvPpgip3, and PvPpgip4, respectively. In the 5′ regions, PvPpgip2.2 and PvPpgip4 were identical to the corresponding BAT93 genes, while PvPpgip3 showed a few substitutions (98.6% identity). In the 3′ region PvPpgip2 showed one single base deletion and one substitution, PvPpgip3 had three substitutions and two small deletions (1 nt and 11 nt), and PvPpgip4 showed no variations.
DISCUSSION
In this study, we have structurally and functionally characterized the pgip gene family of bean. The family consists of four clustered genes; the arrangement and the similarities among them suggest that they derive from a common ancestor by a sequence of duplication-divergence-duplication events. Duplication was at close proximity to the parent sequence, rather than to ectopic chromosomal location, similar to what was observed for nucleotide binding site-LRR genes (Baumgarten et al., 2003). The first duplication event might have generated the ancestors of the two pairs Pvpgip1/Pvpgip2 and Pvpgip3/Pvpgip4, each of which independently underwent diversification, with limited genetic rearrangement within the coding regions represented by point mutations and small insertions/deletions. Subsequently, duplication of genomic segments of about 2,000 bp and 4,000 bp may have generated Pvpgip1 and Pvpgip2, and Pvpgip3 and Pvpgip4, respectively. The extent of nucleotide identity within the two gene pairs, including the regulatory regions (95% to 97.6%), suggests that the two recent duplications occurred approximately at the same time. The observation that Gmpgip3 of soybean (Glycine max), a species of the Phaseoleae tribe very close to bean, groups with Pvpgip1 and Pvpgip2 (De Lorenzo et al., 2001), suggests that these two genes are probably closer to the ancestral gene.
The finding that a large number of families of genes have arisen in eukaryotes during evolution through duplication and have persisted for longer periods of time than expected by classical models (Lynch et al., 2001) is drawing attention into the dynamics of duplicate genes. Only a few studies on gene families having a known function have been carried out. A key question is whether duplicate genes are elements of developmental stability and conservation of function or of evolutionary innovation. Once structural genomic information about a family is available, an understanding of whether changes in gene numbers and structure have adaptive significance generally requires information about function and phenotypes. In many cases, genetics is of little help in the analysis of many gene families because null mutation or deletion of one copy may have no phenotype due to compensation by duplicated genes. The difficult task of designing appropriate experiments to test the adaptive role of each gene relies mainly on biochemical and regulatory information (Brenner, 2000; Meyer, 2003). The results presented in this article show that partial regulatory and functional redundancy, as well as recruitment into diverse biochemical functions and regulatory patterns (biochemical and regulatory subfunctionalization), may explain why four bean pgip genes have passed through the effective sieve of natural selection.
Functional redundancy is apparent for recognition of BcPG, CaPG, and SmPG, while evidence for subfunctionalization emerges when a wider spectrum of PGs is considered. For example, in the Pinto pgip family, only PvPPGIP2 inhibits FmPG. Moreover, in both Pinto and BAT93, only PvPGIP3 is unable to inhibit AnPG, only PvPGIP2 and PvPGIP4 inhibit BcPG at pH 7.0, and, remarkably, only PvPGIP3 and PvPGIP4 inhibit PGs of insects. Sequence diversification between the gene pairs Pvpgip1/Pvpgip2 and Pvpgip3/Pvpgip4 therefore corresponds to functionally distinct classes of PGIPs, one devoted to high-affinity recognition of fungal PGs, the other exerting a weak activity against fungal PGs but showing inhibition of PGs of phytophagous insects. The biochemical functions of these two gene pairs therefore are not interchangeable. Inhibition of PGs by PGIPs is here reported for sap-sucking insects such as mirids and was previously reported for a weevil by a PGIP from orange (Citrus sinensis) exocarp (flavedo; Doostdar et al., 1997). Inhibition of PGs of L. rugulipennis and A. lineolatus however is not exerted by Arabidopsis PGIPs (our unpublished data), suggesting that the function of inhibiting PGs of insect has been acquired (or lost) more than once during evolution. Since PGIP expression is induced in response to insect feeding (Li et al., 2003), the possibility exists that the weak inhibitory activity of PGIPs against PGs of insects has a relevance in defense. The possible adaptive significance of the presence of PvPGIP1 is not obvious. Although its weak inhibitory activity may be due to relaxation of purifying selection, it is possible that the limited and simplified experimental conditions examined were not suitable to reveal its main function.
Our studies may help understand how the ability to recognize a pathogenicity factor that continually varies to escape recognition is maintained in a plant. No evidence for adaptive sequence evolution in pgips was obtained by determining Ka/Ks ratios, which are considered as suitable indicators to evaluate whether variability is the result of random drift (Ka/Ks = 1) or adaptive evolution (Ka/Ks > 1). Ka/Ks ratios estimated for the different regions of the pgip ORFs and the whole genes were either lower than 1 (for regions B, C-out, and D) or around 1 (for the xxLxLxx-encoding region that includes the β-sheet B1). However, frequency of nonsynonymous substitutions significantly higher than those in the other portion of the ORF indicated a higher variability of the xxLxLxx region, consistent with the hypervariability observed in many R genes (Meyers et al., 1998; Hulbert et al., 2001; Lehmann, 2002) and the role of this region in ligand recognition (Warren et al., 1998; Leckie et al., 1999). Maximum likelihood models of codon evolution in protein-coding DNA sequences (Nielsen and Yang, 1998) have been applied to pgip genes from several dicots to propose that positive selection acting at a handful of sites is responsible for the evolution of these genes (Stotz et al., 2000). Unfortunately, this model cannot be applied at this stage to analyze the intraspecific evolution of the bean pgip family, due to the low number of members and the low degree of variability of paralogs and homologs so far characterized.
However, positive selection has likely acted during evolution of bean pgip genes, as in most cases the limited variation between PvPGIPs results in significant changes of their recognition ability. A gain of function is associated with the difference between Pinto PvPPGIP1 and PvPPGIP2 (recognition of FmPG), between PvPGIP3 and PvPGIP4 (recognition of AnPG), and between the paralogous pairs PvPGIP1/PvPGIP2 and PvPGIP3/PvPGIP4 (recognition of PGs of insects).
The characteristics of the PGIP-PG interaction may explain why limited changes can drastically modify PGIP specificity. A stable interaction between the two proteins likely requires a network of multiple and relatively weak contacts, which may be maintained during the evolution, and only one or very few strong contacts that lock the complex (Di Matteo et al., 2003). This limited number of locking contacts may be different in different PGIP-PG interactions and involve any of the solvent-exposed side chains of the nonconserved and nonstructural residues located in or close to the concave surface of PGIP. Different though overlapping subsets of residues may therefore be critical for binding different PGs (Federici et al., 2001; Di Matteo et al., 2003). The LRR structure of PGIP is particularly amenable to a tuning of the surface interaction properties with a very limited number of amino acid changes.
The amino acid sites 224 and 271 differ in all four paralogous Pvpgip genes; the former shows intergenotype variation in PvPGIP2. Replacements at position 271 do not significantly change the ability to recognize FmPG and AnPG (Leckie et al., 1999). Instead, replacements at 224 determine gain/loss of recognition of FmPG both between paralogous genes (PvPPGIP1 versus PvPPGIP2; Leckie et al., 1999) and between corresponding genes of different genotypes (PvBPGIP2 versus PvPPGIP2). Surprisingly, neither position is among those that are proposed to evolve adaptively in PGIPs, according to models of codon evolution (Stotz et al., 2000).
Duplication and diversification of Pvpgip genes result not only in a diversification of their biochemical function, but also in a diversification of their regulation. PvPpgip3 responds to OGs but not to fungal glucan or SA or wounding, while Pvpgip4 responds to none of these treatments. PvPpgip1 responds only to wounding, in agreement with the results of the functional analysis of its promoter in Nicotiana tabacum (Devoto et al., 1998). Remarkably, PvPpgip2, which encodes the most efficient inhibitor of fungal PGs so far characterized, is the only family member that is up-regulated by all the stress stimuli examined. The presence of at least one W-box element in the 5′ regulatory regions correlates with the inducibility of Pvpgip1, Pvpgip2, and Pvpgip3 by stress-related stimuli.
Strategies of exploitation of pgip genes appear to differ in different plant species; for example, SA-dependent up-regulation of pgip expression occurs in bean but not in Arabidopsis or Brassica napus (Ferrari et al., 2003; Li et al., 2003). Interestingly, in both bean and Arabidopsis, OGs are elicitors of the expression of pgip genes in a specific manner because they induce only some of the family members (Atpgip1, Pvpgip2, and Pvpgip3). The ability to switch on different pgip members in response to different stress-related signals therefore appears to be a common feature in plants and, like the biochemical subfunctionalization, is likely to have adaptive significance because it ensures the expression of at least one PGIP if a pathogen blocks or avoids the activation of a particular defense transduction pathway.
In conclusion, by using both a structural and functional genomic approach, we have shown that the presence of multiple pgip genes in bean likely reflects the need to adapt both their regulation and recognition features to combat more efficiently pathogenic fungi and phytophagous insects. Our observations will help to define the adaptive role of the multiple PGIPs present in bean. Furthermore, they pave the way to molecular anatomy studies to identify the amino acid replacements with specific adaptive significance and, therefore, to shed light into the basis of the recognition properties of plant LRR proteins.
MATERIALS AND METHODS
Plant Material
Seeds of Phaseolus vulgaris genotypes BAT93 and Pinto were obtained from Dr. V. Geffroy (Institut de Biotechnologie des Plantes, Orsay, France) and from a local merchant, respectively. Seeds were germinated and grown for 4 to 5 d in moist sterile vermiculite and maintained at 24°C with a 16-h-light period. Wounding and treatment with SA were performed as described previously (Bergmann et al., 1994).
Calli of Pinto bean were obtained by incubation of axenic bean hypocotyls as described previously (Salvi et al., 1990). Suspension-cultured cells were established and grown in Schenk and Hindebrand liquid medium containing 17 g L−1 Suc, 1 g L−1 naphthylacetic acid, 0.2 g L−1 kinetin, in the dark at 26°C, under continuous agitation. When the packed cell volume (obtained by sedimenting cells for 30 min at RT) of 5 mL of suspension-cultured cells was 1.2 mL, cells were subcultured (packed cell volume = 0.4 mL). Four days after subculture, OGs (degree of polymerization = 9–18) or glucan were added to final concentration of 50 μg mL−1. Control cells were treated with distilled water. Cells were collected 0, 4, 10, 24, and 50 h after treatments. All tissues and cells were frozen at −70°C prior to RNA extraction. Three independent experiments were carried out for each treatment.
Nucleic Acid Manipulation and Sequence Analysis
DNA manipulation, PCR, and cloning were performed according to standard procedures (Sambrook et al., 1989). For DNA sequencing, the Dye Terminators cycle sequencing kit with AmpliTaq DNA polymerase (Applied Biosystems, Monza, Italy) was used. Sequence reactions were analyzed using an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). Nucleotide and amino acid sequence analysis was carried out using the programs in the GCG package (Devereux et al., 1984) and DNAMAN (Lynnon BioSoft, Vaudreuil, Quebec). Scans of promoter sequences for putative regulatory elements were performed using the PlantCARE (http://sphinx.rug.ac.be:8080/PlantCARE/) database algorithm (Rombauts et al., 1999). Ka and Ks nucleotide substitutions per site were estimated using the program K-estimator 5.5 (Comeron, 1999). A 2 × 2 contingency table G test was used to test for the significance of differences in Ka and Ks values (Zhang et al., 1997). The G-test was also applied to verify the homogeneity of the distribution for the different values observed between the six pairwise comparisons and between the different sequence regions. The expected values of the Ka and Ks were computed considering the sequence of the gene coding for the mature protein.
Screening of Genomic Libraries
A λ phage and a BAC library were screened using as a probe a radiolabeled 732-bp DNA fragment, corresponding to most of the PvPpgip1 coding region (Toubart et al., 1992). The BAC library was prepared from bean genotype BAT93 into the HindIII site of pBeloBAC11. Screening was performed using the same conditions used for the λ phage library. The phage genomic library, constructed in Lambda FIX II vector from Pinto bean, was obtained from Stratagene (La Jolla, CA). The library was plated, and a total of approximately 1 × 106 recombinant plaque-forming units were screened. Plaque hybridization was carried out at high stringency as described in Sambrook et al. (1989). BAC DNA fragments containing pgip gene sequences were subcloned into plasmid vectors for subsequent analysis. Similarly, phage DNA from selected positive clones was prepared, and EcoRI fragments showing positive signals in Southern-blot experiments were subcloned in pBlueScript II SK+ for further analysis.
Preparation of Vectors for Expression Using PVX
The coding regions of pgip genes from BAT93 and Pinto were amplified by PCR using sequence-specific oligonucleotides, including restriction sites for ClaI and SalI at 5′ and 3′ ends, respectively, to facilitate cloning. The amplified fragments were digested with ClaI and SalI and cloned into the pPVX201 expression vector. The plasmids obtained were used to inoculate Nicotiana benthamiana plants directly using 30 μg of DNA/plant as described by Baulcombe et al. (1995).
Preparation of PGIPs
PGIPs were purified from leaves of PVX-infected N. benthamiana plants. Leaves were homogenized in 1 m NaCl (2 mL/g), incubated with gentle shaking for 1 h at 4°C, and centrifuged 20 min at 10,000g. Supernatants were filtered through Miracloth (Calbiochem, San Diego). PvPGIP1 and PvPGIP2 of Pinto or BAT93 were purified as described by Leckie et al. (1999). For purification of PvPGIP3 and PvPGIP4, samples were brought to 25% (NH4)2SO4, incubated at 4°C, and centrifuged. Proteins were precipitated from supernatant with 85% (NH4)2SO4, recovered by centrifugation, and redissolved in 0.5 m NaCl, 20 mm Na acetate, pH 4.7. For PvPGIP3 purification, protein samples were subjected to chromatography on a desalting Sephadex G-25 Superfine column equilibrated with 100 mm sodium acetate, pH 4.7, and subsequently on a cation exchange (SP-Sepharose) column. Bound proteins were eluted with a 40-min linear gradient of 0 to 0.5 m NaCl in 100 mm sodium acetate, pH 4.7, at a flow rate of 1 mL min−1. For PvPGIP4 purification, the dissolved proteins were diluted about 10-fold and loaded on a diethylaminoethyl cellulose (DE52; Whatman, Kent, UK) column preequilibrated with 20 mm acetate, pH 4.7. The nonabsorbed proteins were loaded on a Sepharose-Aspergillus niger PGII column preequilibrated with 20 mm acetate, pH 4.7; bound PvPGIP4 was eluted with 1× phosphate-buffered saline (10 mm sodium phosphate, 150 mm NaCl, pH 7.4). SDS-PAGE and immunoblotting were performed as described previously (Desiderio et al., 1997). Polyclonal antibodies raised against PGIP purified from bean pods were used for immunoblotting experiments.
Preparation and Assay of Polygalacturonases and PGIPs
Colletotrichum acutatum isolate SHK788 (from lupin) and Stenocarpella maydis isolate PPRI #6353 (from maize) were a kind gift of Dr. Berger (University of Pretoria, South Africa). These are the same strains used in a previous work (Ferrari et al., 2003), where C. acutatum was indicated as Colletotrichum gleosporioides according to Yang and Sweetingham (1998); this fungal isolate has been now renamed by Dr. Berger based on the study of Talhinhas et al. (2002). Botrytis cinerea strain B05-10 was a kind gift of Dr. P. Tudzynski (Institut fur Botanik Westfalische Wilhelms-Universitat, Munster, Germany). Fungi were grown for 20 d on potato dextrose agar (Oxoid, Milan) at 24°C under constant light. Conidia of C. acutatum (5 × 10−5 mL−1) or mycelium of S. maydis (1 cm2) were harvested, used to inoculate Czapek-Dox medium (NaNO3 2 g L−1, K2HPO4 1 g L−1, MgSO4 0.5 g L−1, KCl 0.5 g L−1, FeSO4 10 mg L−1, pH 7.0), and supplemented with 1% pectin. Cultures were incubated in a rotary shaker at 180 rpm and 21°C for 12 d, and filtrates were used for the PG activity assay. PGII of A. niger was prepared as described by Cervone et al. (1987), and PG of Fusarium moniliforme expressed in Saccharomyces cerevisiae was prepared as described previously (Caprari et al., 1996). Lygus rugulipennis Poppius and Adelphocoris lineolatus (Goeze) were field collected from alfalfa (Medicago sativa) and laboratory reared on fresh green beans and sunflower kernels (25 ± 2°C, 70% ± 10% relative humidity, 14-h-light:10-h-dark photoperiod). For total protein extracts, males and females of either species were homogenized in 1 m NaCl, 25 mm Na acetate, pH 4.7 (2 mL/g) in a mortar; the homogenate was incubated with gentle shaking for 1 h at 4°C and centrifuged 20 min at 10,000g. Supernatants were filtered through Miracloth and frozen.
Enzymatic activity of PGs (expressed in agarose plate units) and inhibitory activity of PGIPs were measured using a modified agarose diffusion assay in the presence of 20 mm Na acetate, pH 4.7 or 7.0, as described by Ferrari et al. (2003).
Elicitor Preparation
Glucan, defined as those oligosaccharides, prepared by partial acid hydrolysis of isolated mycelia walls of Phytophthora megasperma f. sp. glycinea, that elute at the void volume of a low-resolution P-2 column (void β-glucan), was kindly provided by Dr. M. Hahn (University of Georgia, Athens). Elicitor-active OGs were prepared as described by Bellincampi et al. (1996).
RT-PCR Analysis
Total RNA was isolated using RNeasy kit (Qiagen USA, Valencia, CA) according to manufacturer's instructions. After extraction, RNA samples were treated with RNase-free DNase I by incubation at 37°C for 30 min, extracted with one volume of phenol/chloroform, and the RNA precipitated by adding 0.1 volume 3 m sodium acetate and 2.5 volumes of 100% ethanol and stored at −70°C. Prior to each experiment RNA aliquots were centrifuged, washed with 70% ethanol, and resuspended in sterile diethylpyrocarbonate-treated water. RNA concentration was determined both spectrophotometrically and by densitometric analysis of RNA bands following agarose gel electrophoresis. RT-PCR was carried out on total RNA (300 ng) using Ready-To-Go RT-PCR beads (Amersham Biosciences, Buckinghamshire, UK), according to manufacturer's instructions, in a Perkin Elmer 9600 thermal cycler (Perkin-Elmer Applied Biosystems, Foster City, CA). Extensive trials indicated that the number of amplification cycles and the initial amount of RNA template were critical parameters to avoid reaction plateau. Oligonucleotide primers (sense and antisense, respectively) were as follows: 5′-TCTTTGAGAACTGCACT and 5′-CGTCGAATGTGATTCCTC for Pvpgip1, 5′-TCTTTGAGCACTGCACA and 5′-CGTCGAATGTGATTCCGA for Pvpgip2.1/2.2, 5′-CCAAACTCCGTTTTCTCTAC and 5′-CCCTCTAAGTCCTTCGACT for Pvpgip3, and 5′-CCAAACTCGGTTTTATCCGA and 5′-CCCTCTAAGTTCTTCGACC for Pvpgip4.
The PCR conditions consisted of 35 cycles as follows: 94°C 1 min, 60°C 1 min, 72°C 1 min. The specificity of the primers was assessed in separate PCR experiments using, as a template, recombinant plasmid DNA containing the appropriate pgip cDNAs. A dilution series, ranging from 2 pg to 20 ng, confirmed that each oligonucleotide pair specifically amplified the correct pgip gene at 58°C and 60°C (data not shown). Negative control experiments, where the reverse transcriptase step was omitted, showed no amplification, demonstrating that specific amplification was due to initial RNA templates and not to contaminating DNA fragments. For each treatment, three independent experiments were performed.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ786408 to AJ786411.
Acknowledgments
We thank F. Cervone for helpful discussion and S. Benedettelli for statistical analysis of Ka and Ks values.
This work was supported by the Giovanni Armenise-Harvard Foundation, by MIUR (Ministero dell'Istruzione, dell'Università e della Ricerca; grants PRIN [Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale] 2002 and FIRB [Fondo per gli Investimenti della Ricerca di Base] 2001), by the Institute Pasteur-Fondazione Cenci Bolognetti, and by European Community Grants (grant nos. ICA4–CT–2000–30033 and QLK1–CT–2000–00811).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044644.
References
- Baulcombe D, Chapman S, Santa Cruz S (1995) Jellyfish green fluorescent protein as a reporter for virus infections. Plant J 7: 1045–1053 [DOI] [PubMed] [Google Scholar]
- Baumgarten A, Cannon S, Spangler R, May G (2003) Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW (2002) Receptor kinase signaling in plant development. Annu Rev Cell Dev Biol 18: 163–192 [DOI] [PubMed] [Google Scholar]
- Bellincampi D, Camardella L, Delcour JA, Desseaux V, D'Ovidio R, Durand A, Elliot G, Gebruers K, Giovane A, Juge N, et al (2004) Potential physiological role of plant glycosidase inhibitors. Biochim Biophys Acta 1696: 265–274 [DOI] [PubMed] [Google Scholar]
- Bellincampi D, Cardarelli M, Zaghi D, Serino G, Salvi G, Gatz C, Cervone F, Altamura MM, Costantino P, De Lorenzo G (1996) Oligogalacturonides prevent rhizogenesis in rol B transformed tobacco explants by inhibiting auxin-induced expression of the rol B gene. Plant Cell 8: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D (2001) Evolutionary dynamics of plant R-genes. Science 292: 2281–2285 [DOI] [PubMed] [Google Scholar]
- Bergmann C, Ito Y, Singer D, Albersheim P, Darvill AG, Benhamou N, Nuss L, Salvi G, Cervone F, De Lorenzo G (1994) Polygalacturonase-inhibiting protein accumulates in Phaseolus vulgaris L. in response to wounding, elicitors, and fungal infection. Plant J 5: 625–634 [DOI] [PubMed] [Google Scholar]
- Boyd DW Jr, Cohen AC, Alverson DR (2002) Digestive enzymes and stylet morphology of Deraeocoris nebulosus (hemiptera: Miridae), a predacious plant bug. Ann Entomol Soc Am 95: 395–401 [Google Scholar]
- Brenner S (2000) Biochemistry strikes back. Trends Biochem Sci 25: 584. [DOI] [PubMed] [Google Scholar]
- Caprari C, Bergmann C, Migheli Q, Salvi G, Albersheim P, Darvill A, Cervone F, De Lorenzo G (1993) Fusarium moniliforme secretes four endopolygalacturonases derived from a single gene product. Physiol Mol Plant Pathol 43: 453–462 [Google Scholar]
- Caprari C, Mattei B, Basile ML, Salvi G, Crescenzi V, De Lorenzo G, Cervone F (1996) Mutagenesis of endopolygalacturonase from Fusarium moniliforme: histidine residue 234 is critical for enzymatic and macerating activities and not for binding to polygalacturonase-inhibiting protein (PGIP). Mol Plant Microbe Interact 9: 617–624 [DOI] [PubMed] [Google Scholar]
- Cervone F, De Lorenzo G, Degrà L, Salvi G (1987) Elicitation of necrosis in Vigna unguiculata Walp. by homogeneous Aspergillus niger endo-polygalacturonase and by α-D-galacturonate oligomers. Plant Physiol 85: 626–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM (1999) K-estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15: 763–764 [DOI] [PubMed] [Google Scholar]
- Daroda L, Hahn K, Pashkoulov D, Benvenuto E (2001) Molecular characterization and in planta detection of Fusarium moniliforme endopolygalacturonase isoforms. Physiol Mol Plant Pathol 59: 317–325 [Google Scholar]
- De Lorenzo G, D'Ovidio R, Cervone F (2001) The role of polygacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol 39: 313–335 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G, Ferrari S (2002) Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr Opin Plant Biol 5: 295–299 [DOI] [PubMed] [Google Scholar]
- Desiderio A, Aracri B, Leckie F, Mattei B, Salvi G, Tigelaar H, Van Roekel JS, Baulcombe DC, Melchers LS, De Lorenzo G, et al (1997) Polygalacturonase-inhibiting proteins (PGIPs) with different specificities are expressed in Phaseolus vulgaris. Mol Plant Microbe Interact 10: 852–860 [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberlei P, Smithies O (1984) A comprehensive set of sequence analysis programs for the Vax. Nucleic Acids Res 12: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Leckie F, Lupotto E, Cervone F, De Lorenzo G (1998) The promoter of a gene encoding PGIP (polygalacturonase-inhibiting protein) of Phaseolus vulgaris L. is activated by wounding but not by elicitors or pathogen infection. Planta 205: 165–174 [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, De Lorenzo G, Tsernoglou D, Cervone F (2003) The crystal structure of PGIP (polygalacturonase-inhibiting protein), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci USA 100: 10124–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doostdar H, McCollum TG, Mayer RT (1997) Purification and characterization of an endo-polygalacturonase from the gut of West Indies sugarcane rootstalk borer weevil (Diaprepes abbreviatus L.) larvae. Comp Biochem Physiol 118B: 861–867 [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Federici L, Caprari C, Mattei B, Savino C, Di Matteo A, De Lorenzo G, Cervone F, Tsernoglou D (2001) Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase-inhibiting protein). Proc Natl Acad Sci USA 98: 13425–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G (2003) Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15: 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyre R, Skroch PW, Geffroy V, Adam-Blondon A-F, Shirmohamadali A, Johonson WC, Llaca V, Nodari RO, Pereira PA, Tsai S-M, et al (1998) Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP map. Theor Appl Genet 97: 847–856 [Google Scholar]
- Gassmann W, Dahlbeck D, Chesnokova O, Minsavage GV, Jones JB, Staskawicz BJ (2000) Molecular evolution of virulence in natural field strains of Xanthomonas campestris pv. vesicatoria. J Bacteriol 182: 7053–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Jouanin L (1999) Molecular cloning of cDNAs encoding a range of digestive enzymes from a phytophagous beetle, Phaedon cochleariae. Insect Biochem Mol Biol 29: 1129–1142 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Bauer Z, Boller T (2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39: 285–312 [DOI] [PubMed] [Google Scholar]
- Idnurm A, Howlett BJ (2001) Pathogenicity genes of phytopathogenic fungi. Mol Plant Pathol 2: 241–255 [DOI] [PubMed] [Google Scholar]
- Jaubert S, Laffaire JB, Abad P, Rosso MN (2002) A polygalacturonase of animal origin isolated from the root-knot nematode Meloidogyne incognita. FEBS Lett 522: 109–112 [DOI] [PubMed] [Google Scholar]
- Kistner C, Parniske M (2002) Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci 7: 511–518 [DOI] [PubMed] [Google Scholar]
- Leckie F, Mattei B, Capodicasa C, Hemmings A, Nuss L, Aracri B, De Lorenzo G, Cervone F (1999) The specificity of polygalacturonase-inhibiting protein (PGIP): a single amino acid substitution in the solvent-exposed β-strand/β-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. EMBO J 18: 2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann P (2002) Structure and evolution of plant disease resistance genes. J Appl Genet 43: 403–414 [PubMed] [Google Scholar]
- Li R, Rimmer R, Yu M, Sharpe AG, Seguin-Swartz G, Lydiate D, Hegedus DD (2003) Two Brassica napus polygalacturonase inhibitory protein genes are expressed at different levels in response to biotic and abiotic stresses. Planta 217: 299–308 [DOI] [PubMed] [Google Scholar]
- Li W-H (1997) Molecular Evolution. Sinauer, Sunderland, MA
- Lynch M, O'Hely M, Walsh B, Force A (2001) The probability of preservation of a newly arisen gene duplicate. Genetics 159: 1789–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G (2003) Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 54: 23–61 [DOI] [PubMed] [Google Scholar]
- Mattei B, Bernalda MS, Federici L, Roepstorff P, Cervone F, Boffi A (2001) Secondary structure and post-translational modifications of the leucine-rich repeat protein PGIP (polygalacturonase-inhibiting protein) from Phaseolus vulgaris. Biochemistry 40: 569–576 [DOI] [PubMed] [Google Scholar]
- Meyer A (2003) Molecular evolution: duplication, duplication. Nature 421: 31–32 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW (1998) Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndimba BK, Chivasa S, Hamilton A, Simon WJ, Slabas AR (2003) Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 3: 1047–1059 [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z (1998) Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeser B, Heidrich PM, Muller U, Tudzynski P, Tenberge KB (2002) Polygalacturonase is a pathogenicity factor in the Claviceps purpurea/rye interaction. Fungal Genet Biol 36: 176–186 [DOI] [PubMed] [Google Scholar]
- Poinssot B, Vandelle E, Bentejac M, Adrian M, Levis C, Brygoo Y, Garin J, Sicilia F, Coutos-Thevenot P, Pugin A (2003) The endopolygalacturonase 1 from Botrytis cinerea activates grapevine defense reactions unrelated to its enzymatic activity. Mol Plant Microbe Interact 16: 553–564 [DOI] [PubMed] [Google Scholar]
- Powell AL, van Kan J, ten Have A, Visser J, Greve LC, Bennett AB, Labavitch JM (2000) Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol Plant Microbe Interact 13: 942–950 [DOI] [PubMed] [Google Scholar]
- Rombauts S, Dehais P, Van Montagu M, Rouze P (1999) PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27: 295–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi G, Giarrizzo F, De Lorenzo G, Cervone F (1990) A polygalacturonase-inhibiting protein in the flowers of Phaseolus vulgaris L. J Plant Physiol 136: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Stotz HU, Bishop JG, Bergmann CW, Koch M, Albersheim P, Darvill AG, Labavitch JM (2000) Identification of target amino acids that effect interactions of fungal polygalacturonases and their plant inhibitors. Physiol Mol Plant Pathol 56: 117–130 [Google Scholar]
- Szekeres M (2003) Brassinosteroid and systemin: two hormones perceived by the same receptor. Trends Plant Sci 8: 102–104 [DOI] [PubMed] [Google Scholar]
- Talhinhas P, Screenivasaprasad S, Neves-Martins J, Oliveira H (2002) Genetic and morphological characterization of Colletotrichum acutatum causing anthracnose of lupins. Phytopathology 92: 986–995 [DOI] [PubMed] [Google Scholar]
- Toubart P, Desiderio A, Salvi G, Cervone F, Daroda L, De Lorenzo G, Bergmann C, Darvill AG, Albersheim P (1992) Cloning and characterization of the gene encoding the endopolygalacturonase-inhibiting protein (PGIP) of Phaseolus vulgaris L. Plant J 2: 367–373 [DOI] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW (1998) A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10: 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler AGJ (2001) Biology of the Plant Bugs (Hemiptera: Miridae). Cornell University Press, Ithaca, NY
- Yang HA, Sweetingham MW (1998) The taxonomy of Colletotrichum isolates associated with lupin anthracnose. Aust J Agric Res 49: 1213–1223 [Google Scholar]
- Zhang J, Kumar S, Nei M (1997) Small-sample tests of episodic adaptive evolution: a case study of primate lysozymes. Mol Biol Evol 14: 1335–1338 [DOI] [PubMed] [Google Scholar]