Abstract
Clostridium acetobutylicum ATCC 824 is a solventogenic bacterium that grows heterotrophically on a variety of carbohydrates, including glucose, cellobiose, xylose, and lichenan, a linear polymer of β-1,3- and β-1,4-linked β-d-glucose units. C. acetobutylicum does not degrade cellulose, although its genome sequence contains several cellulase-encoding genes and a complete cellulosome cluster of cellulosome genes. In the present study, we demonstrate that a low but significant level of induction of cellulase activity occurs during growth on xylose or lichenan. The celF gene, located in the cellulosome-like gene cluster and coding for a unique cellulase that belongs to glycoside hydrolase family 48, was cloned in Escherichia coli, and antibodies were raised against the overproduced CelF protein. A Western blot analysis suggested a possible catabolite repression by glucose or cellobiose and an up-regulation by lichenan or xylose of the extracellular production of CelF by C. acetobutylicum. Possible reasons for the apparent inability of C. acetobutylicum to degrade cellulose are discussed.
Clostridium acetobutylicum ATCC 824 has been used for the industrial production of acetone, butanol, and ethanol by the fermentation of carbohydrates (8). This organism is able to utilize a wide range of carbohydrate substrates, including the polymers starch and xylan and saccharides such as glucose, xylose, and cellobiose. However, like the rest of the well-studied solvent-producing clostridial strains, it does not degrade cellulose, the main sugar component in lignocellulosics, which from an economic point of view are among the most interesting substrates for solvent production (20).
Cellulose is a highly recalcitrant substrate for enzymatic degradation because of its physical properties. Cellulose molecules are composed of chains of β-1,4-linked glucose units. The chains are insoluble and form fibrils in which cellulose chains are arranged in parallel bundles that are very stable due to interchain hydrogen bonds and Van der Waals interactions between the pyranose rings (23). The microbial degradation of cellulose is carried out by the concerted action of differentglycoside hydrolases (http://afmb.cnrs-mrs.fr/CAZY/index.html). According to their mode of action, cellulases are subdivided into endo- and exoglucanases (also called cellobiohydrolases). Endoglucanases (EC 3.2.1.4) randomly cleave the cellulose chains at exposed positions and create new ends, while exoglucanases (EC 3.2.1.91) degrade the polymeric chain from either the reducing or the nonreducing end, producing cellobiose as the main product. These two types of enzymes can be distinguished by their substrate specificities. Endoglucanases show a high level of activity on soluble cellulose derivatives, such as carboxymethylcellulose (CMC) and very low levels (or none at all) on microcrystalline cellulose, while exoglucanases show relatively high levels of activity on microcrystalline cellulose. During the efficient degradation of cellulose, both types of enzymes act synergistically, and all cellulolytic organisms known so far produce at least one, but in most cases several, of each type of glucanase (19).
Sequence analysis of the genome of C. acetobutylicum ATCC 824 has indicated the presence of a gene cluster containing 10 unidirectionally transcribed genes that are predicted to encode secreted proteins with cohesin or dockerin modules (24). These modules are typically found in components of a large extracellular complex that is specialized in the degradation of cellulose, the so-called cellulosome, produced by a number of anaerobic microorganisms, including clostridial species (32, 33). In an active cellulosome, glycoside hydrolases bind to a large nonenzymatic cellulose-binding scaffolding protein called cellulose-binding protein (Cbp) (5) or cellulose integrating protein (Cip) (9). Dockerin modules, usually located at the C termini of the enzymatic cellulosomal subunits, consist of two duplicated sequences of, on average, 22 amino acids each. Dockerin modules bind to cohesin modules, which are about 100 amino acids long, that are present in the C-terminal portion of Cbp. The gene clusters encoding cellulosomal subunits generally start with a gene encoding Cbp followed by a gene encoding a putative glycoside hydrolase family 48 protein, which for C. acetobutylicum has been annotated celF. Orthologs of CelF produced by several cellulolytic clostridia have been characterized as cellobiohydrolases (exocellulases) and identified as major components of the cellulosome (11, 14, 27).
For this study, we tested the growth of and cellulase activity production by C. acetobutylicum ATCC 824 on different carbon sources. This organism produced higher extracellular cellulolytic activities during growth on lichenan (a polymer of 1,3-1,4-β-linked glucose units) or xylose than on glucose or cellobiose. The production of extracellular CelF by C. acetobutylicum was determined by Western blotting with antibodies raised against Escherichia coli-produced CelF. We found that in lichenan- or xylose-grown cultures, a significantly larger amount of CelF was present than in glucose- or cellobiose-grown cultures.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
C. acetobutylicum ATCC 824 was kindly supplied by P. Soucaille (INSA, Toulouse, France). Stock cultures were maintained as spore suspensions in sterile 10% (vol/vol) glycerol at −20°C. Spore suspensions were heat shocked for 10 min at 75°C prior to inoculation. For the production of precultures, cells were grown overnight at 37°C in clostridial growth medium (28, 34). For growth experiments, the same medium was used, but with a 2% (wt/vol) concentration of one of the following carbon sources: glucose (Merck, Darmstadt, Germany), CMC (low viscosity), cellobiose, xylose, lichenan, or laminarin (all from Sigma). Avicel (Merck) was used at 6% (wt/vol) alone or supplemented with Celluclast 1.5L (a gift from Novozymes, Bagsvaerd, Denmark) at 2% (wt/wt). Celluclast 1.5L is a commercially available liquid cellulase prepared from the fungus Trichoderma reesei (2). Cultures were inoculated with 2% (vol/vol) overnight precultures. All experiments were performed anaerobically.
For vector construction and protein production, the E. coli strains XL1 Blue (Stratagene) and M15(pREP4) (Qiagen), respectively, were used. These strains were grown in Luria-Bertani broth as described previously (30), supplemented with ampicillin (50 μg/ml), isopropyl-β-d-thiogalactopyranoside (IPTG; 50 μg/ml), 5-bromo-4-chloro-3-indolyl-β-galactoside (X-Gal; 40 μg/ml), or kanamycin (50 μg/ml) when appropriate.
Transformation procedures, DNA manipulation, and PCR.
All general DNA manipulations in E. coli were performed essentially as previously described (30). Restriction endonucleases and modification enzymes were purchased from Roche Diagnostics, Eurogentec, or Qiagen. DNA isolation from E. coli was performed with a Wizard Plus SV miniprep kit (Promega Inc.). Genomic DNA from C. acetobutylicum ATCC 824 was isolated by the method of Pospiech and Neumann (26).
Oligonucleotides for PCR were purchased from Eurogentec. The DNA fragments containing the desired mutations were first cloned into pGEMT-Easy (Promega Inc.) and afterwards into the expression vector pQE60 (Qiagen). The mutations were verified by sequencing. The complete coding sequence from the celF gene was amplified by a PCR using the following primers based on the sequence of the celF gene (GenBank no. gi:15894198): forward primer CELF1, 5′-CGCCATGGTAAAGATAAGTAAGAA-3′, and reverse primer CELF3, 5′-GCGGATCCTTAACCTATTTTTGCAATTAATTTAG-3′, which contain NcoI and BamHI sites (underlined), respectively. The resulting PCR fragment of approximately 2.1 kb was cloned into vector pQE60, generating pWUR69. The celF gene, without a signal peptide sequence, was amplified with the CELF3 primer and the forward primer CELF2 (5′-CGCCATGGCTACAACTACAGATTCATCC-3′) (the NcoI site is underlined). The resulting PCR fragment of approximately 2 kb was cloned into pQE60, generating pWUR70. The NcoI sites were added in such a way that they allowed the in-frame fusion of the coding region to the pQE60 expression sequences. To obtain a six-histidine tag at the C-terminal end of the celF gene, we used primers CELF2 and CELF4 (5′ GCGGATCCACCTATTTTTGCAATTAATTTAG 3′) (the BamHI site is underlined) as a forward and a reverse primer, respectively, and the resulting fragment was cloned into pQE60, generating pWUR70-his.
Analytical methods.
Solvents and acids produced during fermentations were determined by high-performance liquid chromatography as previously described (15).
Preparation of culture samples for enzyme assays and Western blotting.
Cells were sedimented by centrifugation at 10,000 × g at 4°C for 15 min. The culture supernatant was collected, filtered through a 0.20-μm-pore-size sterile syringe filter (26-mm-diameter PES membrane; Corning Inc., New York, N.Y.), and concentrated approximately 100-fold by ammonium sulfate precipitation with 6.3 M (85% [wt/vol]) ammonium sulfate on ice. Low-molecular-weight compounds and medium components were removed from the concentrated material by dilution with 4 volumes of ice-cold 50 mM sodium citrate buffer, pH 5.7, followed by reconcentration by ultrafiltration through Macrosep devices with a molecular size cutoff of 10 kDa (low-protein-binding PES membrane; Pall Filtron, East Hill, N.Y.). The fractions obtained were used for enzymatic activity determinations or for Western blot assays.
Cellulase activity assays.
E. coli transformants grown on agar plates were tested for endoglucanase activity by a Congo red staining assay (14). Hydrolytic activities were determined as described previously (12). The following substrates (all from Sigma, unless indicated otherwise) were used at the indicated final concentrations: 0.5% (wt/vol) CMC, 0.2% (wt/vol) lichenan, 0.5% (wt/vol) laminarin, 0.5% (wt/vol) xylan, and 0.5% (wt/vol) Avicel (Merck) in 50 mM citrate buffer, pH 5.7. The substrates were incubated at 39°C in a water bath with the enzyme samples for 10 to 60 min, except for Avicel, laminarin, and CMC, which were incubated for periods of up to 30 h. The reducing sugars formed were measured by a previously described method using 3,5-dinitrosalicylic acid (4). One unit of activity corresponds to the formation of 1 μmol of reducing sugars (d-glucose) per min, unless stated otherwise. Protein concentrations in the samples were determined with the Bradford assay (Bio-Rad). Activity staining of gels, with xylan, lichenan, laminarin, and CMC as substrates, was performed as described previously (32a).
Immunological procedures.
Cell extracts of E. coli were loaded in a sodium dodecyl sulfate-10% polyacrylamide gel. The protein band corresponding to CelF was cut out of the gel, homogenized, mixed 1:1 with adjuvant (Specoll), and injected into a New Zealand White rabbit. The antiserum was collected 9 weeks after the first immunization. Western blot analyses were performed with the antiserum diluted 1,000 times according to a standard protocol (30).
Bioinformatics.
A search for catabolite response elements (CRE) was performed by use of the software package PATSCAN (13).
RESULTS
Growth of and production of cellulase activity by C. acetobutylicum on different substrates.
C. acetobutylicum was inoculated on media containing glucose, cellobiose, xylose, laminarin, lichenan, and Avicel as carbon sources. On glucose, cellobiose, xylose, or lichenan, C. acetobutylicum grew well, and after 4 days of fermentation the insoluble lichenan residue had disappeared completely. Growth of the cultures was monitored by optical density determinations at 600 nm, except for lichenan-grown cultures, for which the amount of protein in cell extracts was determined by obtaining growth curves comparable to those described previously (16).
Laminarin, a homopolymer composed of β-d-glucose monomers linked by β-1,3-glycosidic bonds, was not degraded by C. acetobutylicum, and even under different preculture conditions (precultures grown on glucose or xylose or inocula of up to 10% [vol/vol]), no growth was observed on this substrate. In addition, C. acetobutylicum was not able to grow on microcrystalline cellulose (Avicel) or CMC, although a fast and significant reduction in the viscosity of the medium was observed after inoculation of the medium containing the latter substrate, possibly due to a residual cellulase activity in the inoculum. When the medium containing Avicel was supplemented with a mixture of cellulolytic enzymes (Celluclast 1.5L), growth and solvent production were observed (Table 1). Due to the presence of insoluble Avicel in the medium, growth could not be determined by optical density measurements at 600 nm or by protein measurements, but only by product analysis. When CMC was supplemented with Celluclast 1.5L, no growth was observed and only residual amounts of products were found in the medium (Table 1).
TABLE 1.
Acids and solvents produced on different substrates at a concentration of 2% (wt/vol), except for Avicel, which was used at a concentration of 6% (wt/vol) and supplemented with Celluclast 1.5L (CL) at 2% (wt/wt), by C. acetobutylicum ATCC 824 after fermentation
| Growth substrate (time of incubation [h]) | Amt of product (g/liter)
|
||||
|---|---|---|---|---|---|
| Acetic acid | Butyric acid | Acetone | Butanol | Ethanol | |
| Glucose (48) | 1.0 | 1.1 | 0.8 | 3.4 | 0.3 |
| Cellobiose (72) | 1.4 | 2.9 | 0.1 | 2.4 | 0.2 |
| Xylose (72) | 1.9 | 4.3 | 0.2 | 1.3 | 0.2 |
| Lichenan (144) | 1.9 | 1.9 | 1.0 | 3.5 | 0.2 |
| Avicel (96) | 0.5 | 0.6 | 0.1 | 0.4 | 0.1 |
| Avicel + CL (216) | 2.0 | 1.7 | 2.4 | 7.2 | 0.8 |
| CMC + CL (72) | 0.3 | 0.9 | 0.1 | 0.4 | 0.0 |
The concentrations of acids (acetic and butyric acid) and solvents (acetone, butanol, and ethanol) in the extracellular medium were determined at the end of the fermentation period (Table 1). The highest production level of acids, which was coupled with a low level of solvent production, was found after the fermentation of xylose. This indicates that during fermentation of xylose, the cultures did not switch from the acidogenic to the solventogenic phase, a phenomenon known as “acid-crash.” A high level of solvent production was observed during growth on glucose and lichenan, although the fermentation was slower on the latter substrate (Table 1).
During the late exponential growth phase, cultures were collected, the extracellular medium was filtered, and the proteins were concentrated by ammonium sulfate precipitation and dialyzed. Hydrolytic activities considered typical for cellulose-degrading enzymes were determined for these concentrated medium samples. The highest levels of enzymatic activity were observed in the extracellular medium from lichenan-grown cultures, in which a low level of avicelase activity was detected (Avicel) (Table 2). In order to gain insight into the production of extracellular glycoside hydrolases, zymograms (with xylan, lichenan, CMC, and laminarin as substrates in sodium dodecyl sulfate-polyacrylamide gels) were created with the same extracellular medium samples as those used for the determination of enzymatic activities. As expected, samples from cultures grown on different substrates showed different activity patterns on the same zymogram, showing that the growth substrate influences the production of glycoside hydrolases. The trend observed in the zymogram assays corresponded exactly to the results obtained with the enzymatic assays (in zymograms containing xylan, CMC, or lichenan, the samples from cultures grown on lichenan and xylose showed the highest numbers of activity bands and also the most intense bands, and in zymograms with laminarin as the substrate, no activity bands were observed) (results not shown).
TABLE 2.
Cellulolytic activities in extracellular medium of cultures of C. acetobutylicum ATCC 824 grown on different substratesa
| Enzyme | Sp act on growth substrate (U/mg of protein)
|
|||
|---|---|---|---|---|
| Glucose | Cellobiose | Xylose | Lichenan | |
| CMCase | 0.06 | 0.09 | 0.3 | 1.1 |
| Laminarinase | 0.02 | 0.08 | 0.08 | 0.013 |
| Avicelase | <0.002 | <0.006 | <0.03 | 0.01 |
| PNPCase | 7.4 | 21.8 | 20.3 | 39.8 |
Samples were taken at the end of the exponential growth phase. One unit of activity corresponds to the formation of 1 μmol of reducing sugars per min, except for the activity on pNPC (p-nitrophenyl-β-d-cellobioside), for which it corresponds to 1 μmol of pNP released per min per mg of protein.
For an analysis of activities on methyl-umbelliferyl-cellobioside (MUC), a substrate typically used for the detection of exoglucanase activity, aliquots of the concentrated supernatant were spotted onto soft agar plates containing this substrate. Fluorescent halos, indicating hydrolysis of the β-1,4 link between the cellobiose and methyl-umbelliferyl groups of MUC, were observed only around the aliquots from lichenan-, and to a minor extent, xylose-grown cultures (Fig. 1). This MUC-degrading activity in the extracellular medium may be due to cellulases with exoglucanase activity or to the presence of other glycoside hydrolases that are active on this substrate, such as family 10 xylanases (encoded by genes CAP0116 and CAP0053).
FIG. 1.
MUC hydrolysis by concentrated extracellular medium samples from C. acetobutylicum grown on different substrates. Proteins (2.5 μg) from glucose (G), cellobiose (C), xylose (X), and lichenan (L) medium samples were loaded into each well. The blank well (B) contained 30 μl of citrate buffer (pH 5.7).
Expression of celF by C. acetobutylicum.
The genome of C. acetobutylicum contains a single gene (CAC0911, gi:15894198; annotated as celF) coding for a putative family 48 glycoside hydrolase that is located in the putative cellulosome gene cluster. The predicted protein, CelF, contains an N-terminal signal peptide of 35 amino acids, which is typical for gram-positive bacteria. The mature protein consists of a family 48 catalytic domain and a 22-amino-acid long dockerin module at its C terminus. The overall amino acid sequence, including its modular structure, is highly homologous to those of cellulosomal family 48 enzymes of cellulolytic clostridia, such as CelF from Clostridium cellulolyticum (58% identity and 73% similarity), the exoglucanase from Clostridium josui (61% identity and 74% similarity) (9, 14), and ExgS from Clostridium cellulovorans (58% identity and 71% similarity) (27).
The C. acetobutylicum celF gene was cloned into the expression vector pQE60 with or without the coding sequence for its predicted signal peptide, resulting in the plasmids pWUR69 and pWUR70, respectively. To facilitate purification, we fused the mature celF gene to a nucleotide sequence encoding a six-His tag at its C-terminal end, resulting in pWUR70-his. E. coli M15(pREP4) harboring any of these constructs produced a new protein of the expected size upon induction with IPTG. However, the strain harboring pWUR69 produced much smaller amounts of recombinant protein than the strains harboring pWUR70 or pWUR70-his (results not shown). For this reason, only the strains harboring the gene coding for the mature CelF protein with or without an extra His tag were used for further studies. The production of CMCase activity by E. coli M15 harboring either pWUR70 or pWUR70 was tested on agar plates as described previously (16). None of these strains showed CMC degradation on plates even after prolonged incubation (results not shown).
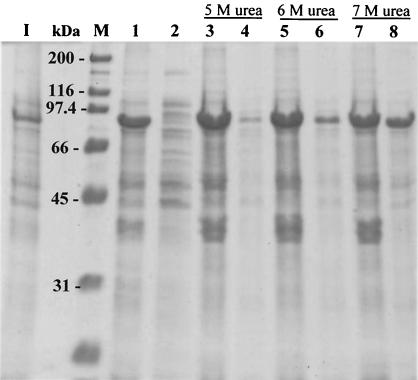
Under all growth temperatures (15 to 37°C) and induction conditions (0.05 to 1 mM IPTG) tested, E. coli-produced CelF and CelF-His were insoluble, and no recombinant protein was detectable in cell extracts of induced cells (Fig. 2). As a consequence, cellulase activity could not be detected in any of the fractions. CelF-His was purified to homogeneity under denaturing conditions (in a buffer containing 8 M urea), making use of the histidine tag. However, when standard dialysis or renaturation techniques were applied, CelF-His became insoluble as soon as the concentration of urea decreased. Despite considerable efforts dedicated to the solubilization of CelF or CelF-His (results not shown), no success was achieved and the proteins remained insoluble.
FIG. 2.
Expression of recombinant CelF containing a His tag at its C-terminal end by E. coli M15(pREP4)(pWUR70-his). Induced cells (3 h, 37°C, 1 mM IPTG; lane I) were resuspended in different buffers and sonicated, resulting in insoluble fractions and cell extracts that were separated by centrifugation. Lanes 1 and 2, insoluble fraction and cell extract, respectively, in 50 mM citrate buffer, pH 5.7; lanes 3 and 4, insoluble fraction and cell extract, respectively, in phosphate buffer with 5 M urea; lanes 5 and 6, insoluble fraction and cell extract, respectively, in phosphate buffer with 6 M urea; lanes 7 and 8, insoluble fraction and cell extract, respectively, in phosphate buffer with 7 M urea.
In order to investigate the production and localization of CelF by C. acetobutylicum, we raised polyclonal antibodies against the E. coli-overproduced CelF protein. C. acetobutylicum cultures grown on glucose, cellobiose, xylose, lichenan (the same cultures used for enzymatic activity determinations), or Avicel supplemented with Celluclast 1.5L were collected and concentrated, and the extracellular medium and cell extract samples were subjected to Western blot analyses. No proteins of the expected size reacting with the anti-CelF antibody were observed in any of the cell extracts tested when 10 μg of protein was loaded into each lane of the gel, indicating that CelF was not present in the cytoplasm. An analysis of the concentrated extracellular medium by Western blotting revealed the presence of an anti-CelF antibody-reacting protein of the expected size of mature CelF (approximately 75 kDa) (Fig. 3). This size corresponded to that of the recombinant CelF protein without a signal peptide present in cell extracts of E. coli harboring pWUR70 (results not shown). In cultures grown on xylose, lichenan, or Avicel supplemented with Celluclast 1.5L, the amount of CelF was significantly higher than that on glucose or cellobiose (Fig. 3). The anti-CelF antibody did not cross-react with any of the enzymes present in Celluclast 1.5L (results not shown). A diffuse band corresponding to a protein of approximately 70 kDa that cross-reacted with the anti-CelF antibody was present in almost all of the samples. This band appeared as well in Western blots for which were used the same protein samples and preimmunization serum from the same rabbit that was used for antibody production. A kinetic analysis showed that CelF was produced during the entire growth phase of C. acetobutylicum on glucose, although it decreased during the stationary phase (data not shown).
FIG. 3.
Western blot analysis of CelF with anti-CelF antibodies. CelF was detected in the extracellular growth medium of C. acetobutylicum cultures grown on glucose (G), cellobiose (C), xylose (X), lichenan (L), and Avicel plus Celluclast (A+CL). Five micrograms of protein was loaded into each lane.
The activities determined were lowest in cultures grown on glucose compared to the rest of the growth substrates tested, suggesting that a catabolite repression mechanism regulates the expression of glycoside hydrolase-encoding genes. The regulation of carbon metabolism in gram-positive bacteria is mediated by a global regulator termed catabolite control protein A (CcpA) via a well-studied mechanism (21). CcpA binds to a specific cis-acting DNA sequence known as the CRE, repressing or activating gene expression. In the genome of C. acetobutylicum ATCC 824, several genes encoding putative CcpA-like proteins are present (e.g., CAC3037 encodes a protein that is highly homologous to Bacillus subtilis CcpA [6]). When the genome sequence was screened for putative CRE sites by use of the consensus sequence WTGNAANCGNWNNCW (21), 43 hits were found, and when the consensus used was TGWNANCGNTNWCA (18), six additional hits were detected. Of the putative sites found, only those in which the CRE started within 200 bp with respect to the start codon were considered significant, based on earlier studies (7). Most of the hits found were located in the promoter region or coding sequences of genes involved in sugar metabolism. Putative CREs were found in the promoter regions of several genes encoding putative extracellular polymer-degrading enzymes (Table 3). However, no potential CRE sites were found in the putative cellulosomal genes, CAC0910 to CAC0919, CAC0561, and CAC3469, or in their immediate flanking regions.
TABLE 3.
Genes that encode putative extracellular proteins containing a glycoside hydrolase (GH) or a polysaccharide lyase (PL) catalytic domain and in which a putative CRE site has been detected in the promoter sequence or in the coding region at a distance of ±200 bp from the start codon
| Gene no. | Catalytic domaina | Positionb | Predicted function |
|---|---|---|---|
| CAC0706 | GH5 | −61 | Endo-1,4-β-glucanase (fused to two ricin-B-like domains) |
| CAC0812 | PL9 | −164 | Pectate lyase-related protein |
| CAC2252 | GH31 | +134 | α-Glucosidase |
| CAP0056 | PL9 | −43 | Pectate lyase |
The number beside the family letters indicates the family to which the catalytic domain belongs.
+ and −, location in the coding region or the promoter, respectively.
DISCUSSION
C. acetobutylicum ATCC 824 utilizes glucose, xylose, and lichenan, but not laminarin or cellulose, for growth and solvent production. Lichenan has a structure that resembles the β-glucans that are abundant in plant cell walls of cereals such as barley, rye, and wheat (25). This polymer is a substrate for several different enzymes, including lichenanases (EC 3.2.1.73) and 1,3-1,4-β-glucanases (EC 3.2.1.6), and is often degraded by cellulases of both the exo- and the endo- type (1). Laminarin is degraded mainly by laminarinases (EC 3.2.1.39), lichenanases, and 1,3-1,4-β-glucanases. All of these enzymes belong to family 16 of the glycoside hydrolases, but each class contains important unique sequences, especially in the region surrounding the strictly conserved catalytic residues (25). The genome of C. acetobutylicum ATCC 824 contains a single gene (CAC2807) that encodes a glycoside hydrolase from family 16 which does not belong to the cellulosome gene cluster and lacks dockerin modules. The protein encoded by this gene shows high homology to LicB from Clostridium thermocellum (54% identity and 70% similarity), which seems to be a cellulosomal component. When produced by E. coli, LicB showed a high level of activity on lichenan and no activity on laminarin (31). The absence of a gene coding for a canonical laminarinase from family 16 in the genome of C. acetobutylicum could explain the low laminarinase activity (Table 2) produced and the bacterium's inability to utilize laminarin.
Although C. acetobutylicum produced CMCase activity under certain conditions (Table 2), it did not grow on CMC, either because the hydrolysis products obtained could not be further converted due to the presence of the carboxymethyl groups or because the degradation rate was not fast enough to support growth. A similar inability to grow on CMC, even when the medium was supplemented with Celluclast 1.5L, was observed previously for Clostridium beijerinckii (17).
For lichenan-grown cultures, all of the enzymatic activities measured were the highest observed for all cultures, and a low but significant avicelase activity could be detected, suggesting an induced expression of cellulolytic enzymes by lichenan (Table 2). In a previous study, members of our laboratory could not detect extracellular avicelase activity in similarly grown C. acetobutylicum cultures (16). In the experiments described here, however, the protein concentrations were substantially higher, as the extracellular proteins were concentrated approximately 60-fold. In xylose-grown cultures, the activities were the second highest (Table 2) of those observed, and activity on MUC was detectable, although it was lower than that in lichenan-grown cultures (Fig. 1). Lee et al. (12) reported the production of endoglucanase (CMCase) and cellobiosidase (pNPCase) activities by C. acetobutylicum grown on various carbon sources (glucose, cellobiose, xylose, and mannose) in a chemostat.
A conserved feature between cellulosomes produced by clostridial species is the presence of a family 48 glycoside hydrolase enzyme with exoglucanase activity that seems to play an essential role in cellulosomal function. The only gene that possibly encodes a glycoside hydrolase from family 48 in the genome of C. acetobutylicum, celF, was overexpressed in E. coli. The resulting protein, CelF, was produced in inclusion bodies, always appearing in the insoluble fraction of the cell lysates, and therefore its enzymatic activity (CMCase or avicelase) could not be determined. Other clostridial cellulases from family 48, such as ExgS from C. cellulovorans (14) and CelF from C. cellulolyticum (27), have been produced by E. coli, for which denaturation-renaturation methods or optimized production conditions allowed solubilization of at least part of the corresponding protein. However, similar methods were unsuccessful in the case of CelF from C. acetobutylicum. Recently, it was shown that the fusion of a cellulose-binding domain to the catalytic domains increased the solubilities of cellulosomal and noncellulosomal cellulases of C. cellulovorans upon expression in E. coli (22), a strategy that looks promising for the solubilization of recombinant CelF.
Polyclonal antibodies raised against the E. coli-produced CelF protein were used to detect the production of this protein by C. acetobutylicum ATCC 824 grown on different substrates. On all substrates, CelF was specifically detected in the extracellular medium as a single band of the expected size (Fig. 3). CelF was found to be constitutively produced by C. acetobutylicum but was induced during growth on lichenan, xylose, or Avicel. Our results are consistent with those in which the expression of CelF by C. acetobutylicum grown on cellobiose was confirmed by N-terminal sequencing of the extracellular protein (29). In C. cellulovorans, the major cellulosomal components are constitutively expressed, and depending on the growth substrate, specific enzymes are induced and the compositions of the cellulosomes vary (10). In C. thermocellum, transcriptional regulation of the cellulosomal cel48A gene (encoding a cellulosomal subunit analogous to CelF in C. acetobutylicum) appears to be dependent not only on the growth substrate, but also on the growth rate (3), but whether this is the case in C. acetobutylicum still needs to be investigated.
In the genome of C. acetobutylicum, there is a gene encoding a CcpA-like regulator, CAC3037, and several CRE sites that are putative targets for CcpA. A search for CREs in the genome was performed by the use of well-characterized consensus sequences, and various hits were found both in the chromosome and in the megaplasmid. CRE sequences are present in many genes, including some that encode extracellular polymer-degrading enzymes (Table 3). No CRE sites were found in the promoter or in the coding regions of genes encoding putative cellulosomal subunits.
Despite the presence of genes involved in cellulose degradation in its genome and the production of cellulolytic enzymes, C. acetobutylicum ATCC 824 is not able to utilize cellulose for growth. In this study, we showed that CelF, a protein encoded by one of the genes present in the cellulosomal gene cluster, is produced and exported to the extracellular medium during growth on glucose, cellobiose, xylose, lichenan, or Avicel. On the last three substrates, there were larger amounts of CelF in the extracellular medium, which correlated with increased cellulolytic activities. However, at this stage, these activities cannot be assigned to CelF only, since other genes coding for different glycoside hydrolases are present in the genome. A functional study of the genes encoding proteins involved in (hemi)cellulose degradation, in particular the putative cellulosomal genes, will be an essential step toward understanding the reason for the lack of true cellulolytic properties by C. acetobutylicum ATCC 824 and the enhancement of the utilization of lignocellulosic substrates for the production of acetone, butanol, and ethanol by this strain.
Acknowledgments
This work was partially supported by an EU Marie Curie fellowship (grant number FAIR-CT96-5047) to A.M.L.-C.
REFERENCES
- 1.Amano, Y., M. Shiroishi, K. Nisizawa, E. Hoshino, and T. Kanda. 1996. Fine substrate specificities of four exo-type cellulases produced by Aspergillus niger, Trichoderma reesei, and Irpex lacteus on (1→3), (1→)-beta-d-glucans and xyloglucan. J. Biochem. 120:1123-1129. [DOI] [PubMed] [Google Scholar]
- 2.Claeyssens, M., and G. Aerts. 1992. Characterisation of cellulolytic activities in commercial Trichoderma reesei preparations: an approach using small chromogenic substrates. Biores. Technol. 39:143-146. [Google Scholar]
- 3.Dror, T. W., E. Morag, A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of the cellulosomal celS (cel48A) gene of Clostridium thermocellum is growth rate dependent. J. Bacteriol. 185:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghose, T. K. 1987. Measurement of cellulase activities. Pure Appl. Chem. 59:257-268. [Google Scholar]
- 5.Goldstein, M. A., M. Takagi, S. Hashida, O. Shoseyov, R. H. Doi, and I. H. Segel. 1993. Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J. Bacteriol. 175:5762-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol. Microbiol. 5:575-584. [DOI] [PubMed] [Google Scholar]
- 7.Hueck, C. J., W. Hillen, and M. H. Saier, Jr. 1994. Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res. Microbiol. 145:503-518. [DOI] [PubMed] [Google Scholar]
- 8.Jones, D. T., and D. R. Woods. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kakiuchi, M., A. Isui, K. Suzuki, T. Fujino, E. Fujino, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1998. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosugi, A., K. Murashima, and R. H. Doi. 2001. Characterization of xylanolytic enzymes in Clostridium cellulovorans: expression of xylanase activity dependent on the growth substrate. J. Bacteriol. 183:7037-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruus, K., W. K. Wang, J. Ching, and J. H. D. Wu. 1995. Exoglucanase activities of the recombinant Clostridium thermocellum CelS, a major cellulosome component. J. Bacteriol. 177:1641-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, S. F., C. W. Forsberg, and L. N. Gibbins. 1985. Cellulolytic activity of Clostridium acetobutylicum. Appl. Environ. Microbiol. 50:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leunissen, J. 2002. PATSCAN (flexible pattern scanner), v. 3.0. CMBI, University of Nijmegen, Nijmegen, The Netherlands.
- 14.Liu, C.-C., and R. H. Doi. 1998. Properties of exgS, a gene for a major subunit of the Clostridium cellulovorans cellulosome. Gene 211:39-47. [DOI] [PubMed] [Google Scholar]
- 15.López-Contreras, A. M., P. A. M. Claassen, A. Mooibroek, and W. M. d. Vos. 2000. Utilisation of saccharides in extruded domestic organic waste by Clostridium acetobutylicum ATCC 824 to produce acetone, butanol and ethanol. Appl. Microbiol. Biotechnol. 54:162-167. [DOI] [PubMed] [Google Scholar]
- 16.López-Contreras, A. M., A. A. Martens, N. Szijarto, H. Mooibroek, P. A. M. Claassen, J. van der Oost, and W. M. de Vos. 2003. Production by Clostridium acetobutylicum ATCC 824 of CelG, a cellulosomal glycoside hydrolase from family 9. Appl. Environ. Microbiol. 69:869-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Contreras, A. M., H. Smidt, J. van der Oost, P. A. M. Claassen, H. Mooibroek, and W. M. de Vos. 2001. Clostridium beijerinckii cells expressing Neocallimastix patriciarum glycoside hydrolases show enhanced lichenan utilization and solvent production. Appl. Environ. Microbiol. 67:5127-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luesink, E. J., R. E. M. A. van Herpen, B. P. Grossiord, O. P. Kuipers, and W. M. de Vos. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30:789-798. [DOI] [PubMed] [Google Scholar]
- 19.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell, W. J. 1998. Physiology of carbohydrate to solvent conversion by clostridia. Adv. Microb. Physiol. 39:31-130. [DOI] [PubMed] [Google Scholar]
- 21.Miwa, Y., A. Nakata, A. Ogiwara, M. Yamamoto, and Y. Fujita. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 28:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murashima, K., A. Kosugi, and R. H. Doi. 2003. Solubilization of cellulosomal cellulases by fusion with cellulose-binding domain of noncellulosomal cellulase engd from Clostridium cellulovorans. Proteins 50:620-628. [DOI] [PubMed] [Google Scholar]
- 23.Nishiyama, Y., J. Sugiyama, H. Chanzy, and P. Langan. 2003. Crystal structure and hydrogen bonding system in cellulose Ialpha from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 125:14300-14306. [DOI] [PubMed] [Google Scholar]
- 24.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Marakova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubios, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Planas, A. 2000. Bacterial 1,3-1,4-β-glucanases: structure, function and protein engineering. Biochim. Biophys. Acta 1543:361-382. [DOI] [PubMed] [Google Scholar]
- 26.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 27.Reverbel-Leroy, C., S. Pages, A. Belaich, J.-P. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roos, J. W., J. K. McLaughlin, and E. T. Papoutsakis. 1985. The effect of pH on nitrogen supply, cell lysis, and solvent production in fermentations of C. acetobutylicum. Biotechnol. Bioeng. 27:681-694. [DOI] [PubMed] [Google Scholar]
- 29.Sabathe, F., A. Belaich, and P. Soucaille. 2002. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 217:15-22. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schimming, S., W. Schwarz, and W. L. Staudenbauer. 1991. Properties of a thermoactive β-1,3-1,4-glucanase (lichenase) from Clostridium thermocellum expressed in Escherichia coli. Biochem. Biophys. Res. Commun. 177:447-452. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 32a.Schwarz, W. H., K. Bronnemeier, F. Grabnitz, and W. L. Staundenbauer. 1987. Activity staining of cellulases in polyacrylamide gels containing mixed linkage beta-glucans. Anal. Biochem. 164:72-77. [DOI] [PubMed] [Google Scholar]
- 33.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 34.Wiesenborn, D. P., E. T. Papoutsakis, and F. B. Rudolph. 1988. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl. Environ. Microbiol. 54:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]