Abstract
The abundance, activity, and diversity of ammonia-oxidizing bacteria (AOB) were studied in prepared microcosms with and without microphytobenthic activity. In the microcosm without alga activity, both AOB abundance, estimated by real-time PCR, and potential nitrification increased during the course of the experiment. AOB present in the oxic zone of these sediments were able to fully exploit their nitrification potential because NH4+ did not limit growth. In contrast, AOB in the alga-colonized sediments reached less than 20% of their potential activity, suggesting starvation of cells. Starvation resulted in a decrease with time in the abundance of AOB as well as in nitrification potential. This decrease was correlated with an increase in alga biomass, suggesting competitive exclusion of AOB by microalgae. Induction of N limitation in the oxic zone of the alga-colonized sediments and O2 limitation of the majority of AOB in darkness were major mechanisms by which microalgae suppressed the growth and survival of AOB. The competition pressure from the algae seemed to act on the entire population of AOB, as no differences were observed by denaturing gradient gel electrophoresis of amoA fragments during the course of the experiment. Enumeration of bacteria based on 16S rRNA gene copies and d-amino acids suggested that the algae also affected other bacterial groups negatively. Our data indicate that direct competitive interaction takes place between algae and AOB and that benthic algae are superior competitors because they have higher N uptake rates and grow faster than AOB.
Nitrification proceeds in two steps and is carried out by two different types of chemolithoautotrophic organisms, distinguishable by their substrate utilization. The oxidation of NH4+ to NO2− is performed by ammonia-oxidizing bacteria (AOB) belonging to the β- and γ-proteobacteria (36), whereas the oxidation of NO2− to NO3− is performed by nitrite-oxidizing bacteria. Nitrification plays a key role in the N cycle, as this process facilitates N removal from the ecosystem through denitrification. The rate-limiting step in this coupled nitrification-denitrification process is the oxidation of NH4+ to NO2−, and AOB are therefore important regulators of the availability of combined nitrogen in the ecosystem.
In aquatic sediments, the activity of AOB is restricted to the upper few millimeters where O2 is present (19, 20). In shallow water systems, this habitat is also subject to colonization by benthic microalgae, which can form dense mats on the sediment surface if light is present. Under these conditions, the algae can have a significant impact on chemical components central to the metabolism of AOB, i.e., O2 and N concentrations as well as pH and dissolved inorganic C (26, 38, 40). Reduced N loading to estuaries, a consequence of intervention against eutrophication, can be expected to promote microalga colonization of the sediment surface (for an example, see reference 7). This colonization will alter the niches occupied by AOB and probably lead to changes in the abundance, activity, and maybe also diversity of the AOB community, since physiological properties are known to vary considerably among different strains of nitrifying bacteria (23, 48, 51). These changes may influence the capacity of the estuarine sediments for removing N via coupled nitrification-denitrification, and knowledge of interactions between AOB and benthic microalgae is therefore relevant when it comes to understanding the effects of changing the N load to estuaries. Studies directly addressing interactions between AOB and benthic microalgae are few, however, and have focused mainly on the coupled nitrification-denitrification process, but the conclusions drawn from these studies seem to be contradictory. Several studies (2, 20, 42, 46) have proposed that benthic microalgae may stimulate coupled nitrification-denitrification by increasing the availability of O2 via oxygenic photosynthesis and thereby stimulating aerobic NH4+ oxidation. In contrast, other studies (10, 33, 50) have proposed that benthic microalgae may inhibit coupled nitrification-denitrification by reducing N availability for nitrifying and denitrifying bacteria. However, a statistical analysis of field denitrification data from 18 European estuaries suggests a general trend towards lower coupled nitrification-denitrification rates in sediment with microalgal production (40). This conclusion is further supported by laboratory studies showing that microalgae do in fact inhibit coupled nitrification-denitrification, probably through the induction of N limitation of nitrifying bacteria (40). Hence, induction of N limitation of the AOB community in the oxic zone is an apparent paradox. Ammonia for nitrification is mainly supplied from the suboxic sediment strata (6, 20), and microsensor studies have shown maximum nitrification activity at the oxic/suboxic interface (6), which is usually well below the photic zone (39). Recently, Risgaard-Petersen (40) suggested that N limitation below the photic zone may be induced by the growth of heterotrophic bacteria stimulated by alga exudates.
The present study complements the work of Risgaard-Petersen (40) by focusing more specifically on the population ecology of AOB during a microalga colonization process. We investigate how the growth, abundance, and potential and actual activity of AOB populations are affected by microalga colonization of estuarine sediments. We also analyze whether initiation of N limitation is likely to be responsible for a decrease in AOB abundance. In this context, we investigate whether the elevated growth of bacteria is likely to be responsible for the initiation of N limitation and growth suppression of AOB below the photic zone in the sediment, as proposed previously (40). Furthermore, we investigate whether alga colonization leads to the selection of specific AOB species adapted to the chemical microenvironment created by the microalgae.
The study was performed in experimental microcosms incubated in 12:12-h light-dark (LD) cycles. For 21 days, we monitored the abundance, diversity, and activity of AOB as well as bacterial abundance by using a combination of molecular, 15N, and microsensor (O2 and NO3− plus NO2− [NOx−]) techniques. The data were compared with a similar data set obtained in parallel dark-incubated microcosms. Due to difficulties in constructing the membrane of the NOx− microscale sensors, we did not succeed in measuring all parameters simultaneously in the same experiment. The present study, therefore, integrates the results of two separate experiments designed according to identical protocols.
MATERIALS AND METHODS
Preparation of experimental microcosms.
The sediment used for the microcosms was collected in Norsminde Fjord, Denmark, and sieved through a 1-mm-wide mesh screen to remove large animals and shell fragments. The sediment used for AOB activity measurements in intact cores, enumeration of bacteria and AOB, and diversity studies of AOB (experiment 1) was collected in February 2002, and the sediment used for NOx− profile measurements (experiment 2) was collected in May 2002. Both experiments were initiated immediately after sampling. The parameters measured in the two experiments as well as the sampling frequencies are listed in Table 1.
TABLE 1.
Parameters measured in experiments 1 and 2
| Parameter | Experiment 1 | Experiment 2 | Sampling day(s) |
|---|---|---|---|
| Nitrification potential | |||
| Time series | Xa | 0, 5, 12, 21 | |
| Effect of N load | X | 21 | |
| Coupled nitrification-denitrification and O2 and nutrient fluxes | X | 21 | |
| Bacterial abundance | X | 0, 21 | |
| Alga biomass and species composition | X | 21 | |
| O2 porewater profiles | X | X | 5, 12, 21 |
| NOx− porewater profiles | X | 21 | |
| Diversity, phylogeny, and abundance of AOB | X | 0, 5, 12, 21 |
X, parameter was measured in the indicated experiment.
Sediment cores were prepared by adding approximately 250 ml of sieved sediment to Plexiglas tubes (inner diameter, 55 mm) with a height of 300 or 100 mm. The 100-mm-tall cores were used for microsensor studies, and the surface of the sediment was aligned with the rim of the tube. Black plastic was wrapped around the tubes from the bottom to the sediment surface to prevent microalgae from colonizing the core walls. Half of the cores were placed in a transparent aquarium with filtered (1-μm pore size; Millipore) seawater and exposed to a 12:12-h LD cycle (irradiance, 140 μmol of photons m−2 s−1, provided by 400-W HPI-T+ mercury lamps from Phillips). These cores are referred to as alga sediments. The other half of the cores were immersed in a darkened reservoir also containing 20 liters of filtered seawater. These cores are referred to as alga-free sediments. Stirring of the water overlying the sediment inside the 300-mm-tall cores was performed with Teflon-coated magnetic stir bars, positioned approximately 6 cm above the sediment surface. The setups were placed in a temperature-controlled room at 19°C, and cores were incubated for 21 days. The reservoir water was constantly aerated and renewed every 3 or 4 days. Nitrate and NH4+ concentrations in the reservoir water were 15 and <1 μM, respectively.
Abundance, diversity, and phylogeny of AOB.
DNA was extracted from 400 to 600 mg of sediment from the upper 3 mm of three replicate cores from each treatment by using a commercial kit (Fast DNA spin kit for soil; Qbiogene, Inc.) according to the manufacturer's instructions.
The abundance of AOB was estimated by real-time quantitative PCR (16) with a light cycler (Roche Diagnostics GmbH, Mannheim, Germany). Standard SYBR Green detection was performed by using the LightCycler-FastStart DNA Master SYBR Green I kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. By this approach, the formation of all double-stranded DNA is detected. The primer set amoA-1F-amoA-2R-TC targeting the gene encoding subunit A of the ammonia monooxygenase (amoA) (Table 2) was used in a 20-μl setup with 3.5 mM MgCl, 1.25 μM concentrations of each primer, and 1× Mastermix (Roche Diagnostics GmbH). The cycling program was as follows: 1 min of initial denaturation at 95°C, followed by 40 cycles of 5 s of denaturation at 95°C, 20 s of annealing at 57°C, and 45 s of elongation at 72°C. Fluorescence was detected after each cycle at 84°C to avoid detection of primer dimers. As the detection by SYBR Green is not fragment specific, amplicons were subsequently checked by electrophoresis to verify fragment length and quality of the amplicons. An external standard curve based on known gene copy numbers of amoA was constructed from 8 dilutions ranging from 3 × 102 to 3 × 107 gene copies/reaction. Fluorescence schemes were collected in the computer for subsequent analysis. The slope of the standard curve was −3.243 cycles/log [amoA]. The Fit Points method included in the LightCycler software (Roche Diagnostics GmbH) was used to estimate the crossing point (Ct), and the concentration of gene copy numbers was subsequently calculated based on the standard curve.
TABLE 2.
Primer sequences
| Primer | Nucleotide sequence (5′-3′) | Target | Reference(s) |
|---|---|---|---|
| amoA-1F | GGG GTT TCT ACT GGT GGT | amoA gene of betaproteobacteria AOB | 44 |
| amoA-2R-TC | CCC CTC TGC AAA GCC TTC TTC | amoA gene of betaproteobacteria AOB | 31, 44 |
| amoA-1F-clamp | (CGCCGCGCGGCGGGCGGGGCGGGGGCACGGGGG)-GGG GTT TCT ACT GGT GGTa | amoA gene of betaproteobacteria AOB | 31 |
| Eub 338 F | ACT CCT ACG GGA GGC AGC | Bacteria | 1 |
| Univ 907 R | CCG TCA ATT CCT TTR AGT TT | Bacteria | 30 |
The clamp portion of the sequence is shown in parentheses.
The obtained gene copy estimates were corrected for sample dilution, extraction efficiency, and sample size before comparison. A minimum of four external standard dilutions of amoA fragments was included in each round of PCR to verify the setup.
PCR preceding cloning, sequencing, and denaturing gradient gel electrophoresis (DGGE) was performed as described by Nicolaisen and Ramsing (31) with primers amoA-1F and amoA-2R-TC. For DGGE-PCR, a GC tail was added to the forward primer (amoA-1F-clamp) (Table 2). DGGE was performed as described previously (31) with a denaturing gradient of 35 to 70%. Partial amoA sequences were cloned (TOPO TA cloning kit; Invitrogen) and subsequently sequenced as described previously (31). Obtained sequences were manually aligned in SeqPup, version 0.6 (http://iubio.bio.indiana.edu/soft/molbio/seqpup/java/seqpup-doc.html), against a selection of pure culture sequences available from the GenBank database.
Phylogenetic analysis was implemented on 450 aligned nucleotides by using the distance matrix and maximum-parsimony algorithms in PAUP, version 4.0 (Sinauer Associates), with the default settings. Bootstrap values (100 replicates) were calculated based on both algorithms.
Accession numbers.
Sequences retrieved during this study are available in GenBank under accession no. AY616014 to AY616021. Kysing Fjord sequences are available under accession no. AF489632 to AF489642.
Abundance of bacteria.
We used two different proxies for net bacterial growth in the upper 3 mm of the sediment: net changes in d-amino acids in the total hydrolyzable amino acid (THAA) pool and net changes in the abundance of genes encoding 16S rRNA. d-Amino acids are known to be bacterium-specific components of peptidoglycan (27). The alternative production of d-amino acids by racemization is a slow process, which takes ∼104 to ∼106 years at 0°C (3). The net increase in the total hydrolyzable d-amino acid pool during the experiment can therefore be assumed to reflect bacterial growth. 16S rRNA is prokaryote specific, and the net increase in the abundance of genes encoding 16S rRNA likewise reflects bacterial growth.
Sediment samples (0.9 to 2.0 g) from three replicate cores from each treatment for THAA extractions were sampled in 20-ml injection vials (La-Pha-Pack, Langerwehe, Germany) containing 10 ml of 6 N HCl. Vials were capped with aluminum ColorSeal caps mounted with butyl/PTFE septa, after which sediment and acid were mixed thoroughly and the headspace was replaced with N2. The vials with sediment-HCl samples were stored upside down at 5°C. Blanks were prepared as samples, leaving out the sediment. Hydrolysis of the sediment-HCl mixture and sample neutralization were performed according to the method of Guldberg et al. (15). d-Isomers of aspartate (Asp), glutamate (Glu), serine (Ser), and alanine (Ala) were measured as dissolved free amino acids by using the method of Mopper and Furton (29) with the modifications described by Guldberg et al. (15).
The quantification of 16S rRNA genes was performed by real-time PCR with universal 16S rRNA gene primers (338F and 907R) (Table 2) targeting most known bacteria. The protocol for real-time PCR and gene copy estimation was similar to the procedure used for quantification of AOB, with the exceptions that the MgCl2 concentration was adjusted to 4 mM and the external standard curve was based on known copy numbers of 16S rRNA genes in dilutions ranging from 3 × 102 to 3 × 107 gene copies/reaction with a slope of −4.0104 cycles/log [16S rRNA].
Biomass and composition of the alga community.
Sediment from the upper 3 mm of three replicate cores was sampled from each treatment for chlorophyll a (Chl a) determination. Sediment was transferred to glass vials containing 10 ml of acetone, and after 24 h of extraction, Chl a was quantified by photometry (25). Samples of approximately 1 ml were collected at the end of the experiment for determination of the composition and biomass of the alga community, transferred to vials, and preserved in 1 ml of 5% (vol/vol) glutaraldehyde. Determination of the biomass and composition of the alga community was performed by Bio/Consult A/S, Aabyhøj, Denmark, by using the procedures of Utermöhl (52). The alga biovolume was estimated by use of the appropriate geometric formulas (12) using the linear dimensions of the cells (n = 10 for every unit) measured during counting. The carbon content of the alga cells was calculated from the biovolume according to the method of Edler (12).
Potential nitrification activity.
Potential nitrification was estimated from the production of NOx− in NH4+-enriched (approximately 500 μM) slurries (17) prepared from sediment samples of three replicate cores from each treatment. The concentration of NOx− in the samples was determined by using the vanadium chloride reduction method (9) on an NOx analyzer (model 42c; Thermo Environmental Instruments). In experiment 1, changes in the nitrification potential were followed in parallel with the AOB abundance measurement.
N limitation.
In experiment 2, we investigated the effects of N limitation on the nitrification potential in cores with and without algae as follows. The alga cores were preincubated as described above, with the exception that half of the cores were incubated with additional 100 μM NO3− in the water column. The dark-incubated (alga-free) cores were immersed in a tank with filtered seawater without additional N. After 21 days of incubation, potential nitrification in the top 3 mm of 3 cores from each treatment was measured as described above.
Coupled nitrification-denitrification and DIN fluxes.
Coupled nitrification-denitrification was measured in intact cores by using the 15N isotope pairing technique (32) as described by Dalsgaard et al. (11). The exchange rates of NOx−, NH4+, and O2 were likewise measured as described by Dalsgaard et al. (11). In the alga sediment, fluxes and denitrification were measured both in light and in darkness (n = 5 for each treatment). In the alga-free sediment, fluxes and denitrification were measured only in darkness (n = 5). Incubations were performed in two sessions. Fluxes were measured first, and after an equilibrium period of 20 h, the denitrification measurements were performed. All measurements were initiated 4 h after a change in light regimen. An abundance of 15N2 (29N2 and 30N2) gas was measured by chromatography and mass spectrometry (RoboPrep-G+ in line with Tracermass; Europa Scientific) (41). The 15N atom% of NO3− was estimated by mass spectrometry after biological reduction to N2 (43). NO3− plus NO2− was determined as described above. NH4+ was determined by the salicylate-hypochlorite method (8) and analyzed automatically on a robotic sample processor (RSP-5051; Tecan AG) in line with a spectrophotometer (M330; Camspec Ltd.). O2 was measured by Winkler titration (14).
Oxygen and NOx− profiles.
Oxygen concentration profiles were measured with Clark-type microsensors (37). Concentration profiles of NOx− were measured with an NOx− biosensor (24) equipped with an electrophoretic sensitivity control to optimize the sensitivity of the sensor (21). Prior to profile measurements, the cores were placed in filtered seawater in a temperature-controlled (19°C) container. The water was aerated to ensure stirring and constant O2 concentration during measurements. For the LD-incubated sediment, 3 to 5 depth profiles of NOx− and O2 were measured both in light and in darkness. Light was provided by a halogen lamp (irradiance, 140 μmol of photons m−2 s−1). For the sediment incubated in the dark, 3 depth profiles of each species were measured only in darkness. The profiles of O2 and NOx− production rates were obtained by modeling the experimental data by the numerical method described by Berg et al. (6).
Statistical analysis.
Effects of the treatment (algae versus no algae) and time effects on the measured parameters were evaluated by analysis of variance or Student's t test. All analyses were performed by using the SAS system for Windows (release 8.02; SAS Institute).
RESULTS
Enumeration and potential activity of AOB.
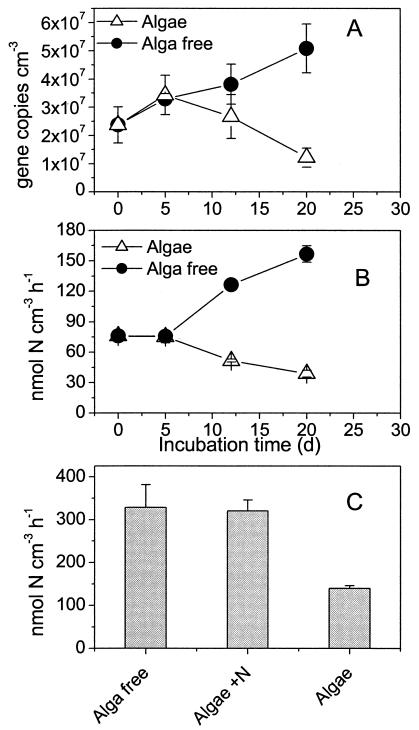
A significant decrease in amoA gene copy numbers was seen in the alga sediment from day 5 to 21 (Fig. 1A) (P = 0.023). The decay rate was 0.07 ± 0.02 day−1. In contrast, the number of amoA genes increased significantly in the alga-free sediment (P = 0.006). The specific net growth rate of amoA genes in this sediment was 0.04 ± 0.01 day−1. By the end of the experiment, the number of amoA gene copies was approximately four times higher in the alga-free sediment than in the alga sediment. The nitrification potential in the alga sediment and the alga-free sediment followed a similar trend, displaying a significant decrease and increase (Fig. 1B) (P < 0.0001), respectively, during the course of the experiment. The nitrification potential in the alga-free sediment was approximately four times higher than the activity in the alga sediment at the end of the experiment (Fig. 1B). The ratio of potential nitrification to gene copy number was 2.9 ± 0.2 fmol of N/gene copy/h in the alga-free sediment and 2.7 ± 0.3 fmol of N/gene copy/h in the alga sediment. There was no indication of changes in this ratio during the course of the experiment or of differences between the treatments (P > 0.3).
FIG. 1.
(A) Abundance of β-proteobacterial AOB amoA as measured by real-time PCR in samples from the upper 3 mm of the sediment in experiment 1; (B) nitrification potential measured in the samples from the upper 3 mm of the sediment in experiment 1; (C) nitrification potential in the upper 3 mm of the sediment at different N loadings measured after 21 days of incubation in experiment 2. Alga free refers to sediment incubated in the dark, algae refers to sediment incubated in LD cycles, and Algae + N refers to sediment incubated in LD cycles with 100 μM NO3− in the water column. Error bars represent standard errors of the means (n = 3).
The nitrification potential was lowest in the alga sediments incubated without supplementary NO3− in the water column, whereas the potential was the same in the alga-free sediment and the alga sediment incubated with 100 μM NO3− in the water column (Fig. 1C) (alpha = 0.05 by analysis of variance supplemented with Tukey comparisons). The potential nitrification activity measured in experiment 2 (Fig. 1C) was about two times higher than the activity measured in experiment 1 for the same treatments (Fig. 1B), which may reflect differences in the initial size of the nitrifying bacterial populations.
Diversity and phylogeny of AOB.
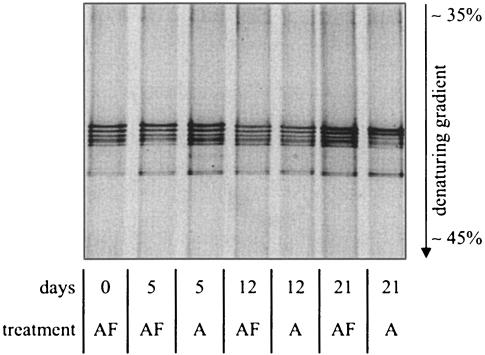
DGGE of amoA amplicons showed identical band patterns during the course of the experiment. Furthermore, identical band patterns were observed at both treatments, indicating no loss of community members due to alga colonization (Fig. 2). After 21 days of incubation, the relative intensity of the bands in the alga-colonized sediment changed slightly, with the two uppermost bands being relatively more intense than the three lower bands compared to previous samples. This could indicate a possible selection for organisms representing these two bands during the alga colonization process.
FIG. 2.
DGGE profile of PCR-amplified amoA fragments from the alga and alga-free sediments on days 0, 5, 12, and 21 after initiation of the experiment. A, alga sediment; AF, alga-free sediment.
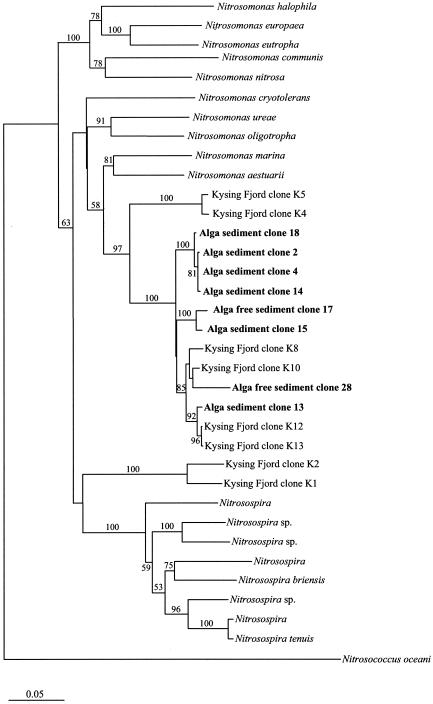
All sequences melted at denaturant concentrations of <46%, indicating that the origin of all detected sequences was the Nitrosomonas genus (31). Partial amoA sequence analysis revealed similar branching patterns based on both algorithms used (Fig. 3). Furthermore, the branching pattern obtained was similar to those of previously published amoA-based trees (for examples, see reference 35). All retrieved sequences were mutually highly similar and very closely related to amoA sequences previously retrieved from the nearby site, Kysing Fjord (30). Together, these sequences comprised a distinct cluster (bootstrap values of 97 and 85% based on distance matrix and maximum-parsimony settings, respectively) within the Nitrosomonas marina cluster (as defined by Purkhold et al. [36]), supporting the findings from the DGGE profile of Nitrosomonas-like sequences only. Even though bootstrap values above 50% for the clustering of the retrieved sequences within the N. marina cluster were only obtained with the distance matrix method, pure culture representatives, N. marina and Nitrosomonas aestuarii, are both retrieved from marine environments (22), supporting the clustering of the retrieved sequences within this group.
FIG. 3.
Phylogenetic distance matrix tree reflecting the phylogenetic affiliation of the retrieved amoA gene sequences compared with a selection of database sequences. Distance matrix bootstrap values (100 replicates) for branches are reported. Missing bootstrap values indicate that the branching was not recovered in the majority of bootstrap replicates by the distance matrix method. Kysing Fjord clones were retrieved in a previous study (30). The bar indicates 5% estimated sequence divergence.
Bacterial growth estimated from d-amino acids and 16S rRNA.
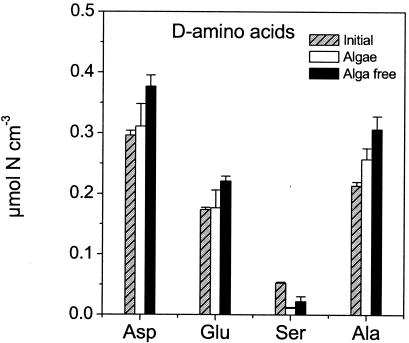
During the course of the experiment, there was a significant (Fig. 4) (P < 0.01) increase in d-Asp (128%), d-Glu (128%), and d-Ala (143%) in the alga-free sediment, whereas d-Ser decreased to 41% of the initial level (Fig. 4) (P < 0.0001). In the alga sediment, there was no significant increase in d-Asp, d-Glu, or d-Ala (P > 0.08), whereas d-Ser decreased significantly (Fig. 4) (P < 0.0001) to 21% of its initial content during incubation.
FIG. 4.
d-Amino acids in the THAA pool measured in the upper 3 mm of the sediment in samples from the initial sediment pool and samples from the alga-free and alga sediments after 21 days of incubation. Error bars represent standard errors of the means (n = 3).
At present, we do not have any explanations for the divergent patterns of d-Ser relative to the other d-amino acids. At the end of the experiment, all measured total hydrolyzable d-amino acids were highest in the alga-free cores, suggesting the highest bacterial numbers in this sediment compared to the alga sediment. In the initial samples, 16S rRNA genes numbered 2.41 × 1010± 5.43 × 109 gene copies cm−3, which was not significantly different from the number of 16S rRNA genes in the alga-free sediment (2.65 ×1010± 2.32 × 109 gene copies cm−3) or in the alga sediment (2.09 × 1010± 5.40 × 109 gene copies cm−3) after 21 days of incubation. Although the data indicated highest gene copy numbers of 16S rRNA genes in the alga-free sediment at the end of the experiment, Student's t test gave no statistical evidence for this hypothesis (P > 0.04).
Chl a and composition of the alga community.
Chl a increased significantly (P = 0.001) from 21 ± 0.5 to 76 ± 8 μg of Chl a cm−3 in the alga cores during the course of the experiment, and the net growth rate was 0.07 ± 0.01 day−1. In the sediment without algae there was no difference in Chl a content from the beginning to the end of the experiment (data not shown). The alga community was composed mainly of pennate diatoms (Table 3) and was almost entirely dominated by diatoms of the Gyrosigma and Pleurosigma genera, which constituted almost 97% of the phototrophic biomass. Gyrosigma balticum, Gyrosigma fasciola, Gyrosigma limosum, Pleurosigma aestuarii, Pleurosigma angulatum, and two unidentified Gyrosigma species were found. All diatoms observed possessed a raphe on one or both frustule halves and were judged to be motile (M. Temponeras, Bio/Consult A/S, personal communication).
TABLE 3.
Abundance and carbon content of the microalgae after 21 days of incubationa
| Organism(s) | Abundance (104 cells cm−3) | C content (μmol cm−3) |
|---|---|---|
| Cyanobacteria | 1 (0.2) | 4.0 (0.8) |
| Nitzschia sp. | 3.2 (0.64) | 5.8 (1.2) |
| Other pennate diatoms (20-50 μm) | 4.8 (0.96) | 0.3 (0.1) |
| Gyrosigma | 42 (8.4) | 311 (62.3) |
Numbers in parentheses represent standard errors.
Coupled nitrification-denitrification and fluxes of DIN and O2.
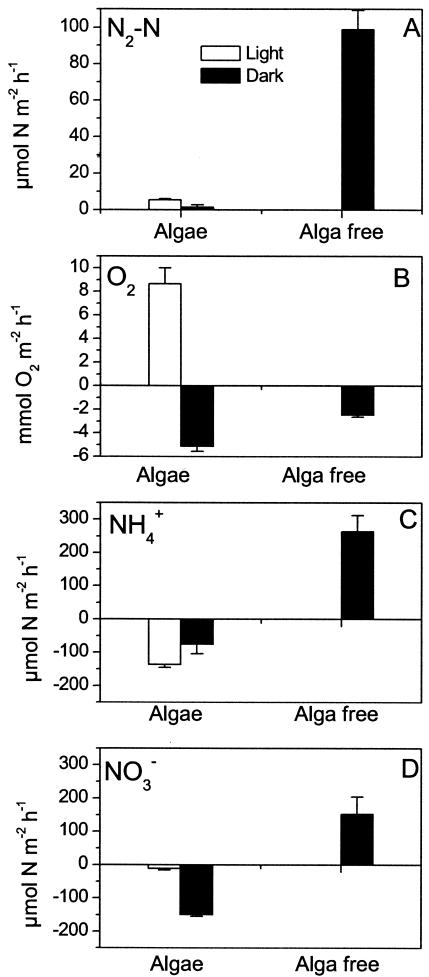
The coupled nitrification-denitrification rate was more than 16 times higher in the alga-free sediment than in the alga sediment (Fig. 5A) (P < 0.0001). In the alga sediment, the coupled nitrification-denitrification rate was approximately two times higher in light than in darkness. However, the activity was extremely low (<2 μmol of N m−2 h−1) in darkness. O2 uptake in the alga sediments during darkness was more than two times higher than in the alga-free sediment, and this difference was highly significant (Fig. 5B) (P < 0.001). NH4+ was taken up exclusively from the water column by the alga sediment, whereas NH4+ was released to the water column from the alga-free sediment (Fig. 5C). The latter sediment also exported NOx−, whereas the alga sediment took up NOx− both in darkness and in light (Fig. 5D).
FIG. 5.
Coupled nitrification-denitrification (A), O2 flux (B), NH4+ flux (C), and NO3− plus NO2− flux (D) in alga-free and alga sediments after 21 days of incubation. Error bars represent standard errors of the means (n = 5).
In the alga-free sediments, 40% of the NOx− produced was denitrified and 60% was released to the water column. Nitrification, as estimated from these opposing NO3− fluxes from the nitrification zone, was 250 ± 37 μmol m−2 h−1. Assuming a similar partition of upward and downward NO3− fluxes from the nitrification zone in the alga sediments (i.e., 40% of the produced NO3− is denitrified and 60% is assimilated), nitrification in the alga-colonized sediment was 13 ± 1 μmol m−2 h−1 in light and 4 ± 2 μmol m−2 h−1 in darkness. Ammonification, estimated from the denitrification rate and DIN fluxes measured in the alga-free sediments (40), was 512 ± 72 μmol m−2 h−1. Assimilation of N in the alga sediment, estimated as ammonification plus DIN uptake minus the loss of nitrogen via denitrification (40), was 656 ± 73 μmol m−2 h−1 in light and 746 ± 78 μmol m−2 h−1 in darkness. The two assimilation rates were not significantly different.
NOx− and O2 porewater profiles.
In the alga-free sediment, NOx− accumulated in the oxic zone of the sediment and was depleted well below the oxic-suboxic interface at a depth of 2 mm (Fig. 6A). An NOx− peak of 36 μM was observed above the oxic-suboxic interface. The profile indicated a flux of NOx− from the sediment toward the water column. The zones of net NOx− production and consumption appeared to be separated by the oxic-suboxic interface, with nitrification occurring in the entire oxic zone and NOx− consumption occurring in the suboxic sediment strata (Fig. 6B). Nitrification was highest immediately above the oxic-suboxic interface, suggesting that the majority of the nitrifying bacteria developed in the deepest part of the oxic zone to obtain optimal conditions for the supply of both NH4+ and O2. Volume-specific O2 and NOx− consumption was almost constant throughout the oxic and suboxic zones, respectively. In the alga sediment, both NOx− consumption and production were observed in the oxic zone during illumination (Fig. 6D). Aerobic NOx− consumption was located in the net O2-producing zone and may therefore be attributed to microphytobenthic assimilation. Below the assimilation zone, a constant volume-specific net nitrification was observed in the remainder of the oxic zone, whereas NOx− was consumed in the suboxic strata below and was depleted at a depth of approximately 2.5 mm (Fig. 6C). There was no indication of aerobic net NOx− production in the alga sediment in darkness. NOx− was exclusively consumed in the oxic and suboxic sediment strata (Fig. 6E and F), and NOx− penetrated 0.8 mm into the sediment, i.e., only 0.2 mm deeper than O2. Volume-specific O2 consumption activity was highest at the very surface of the sediment, reflecting respiration of the alga biomass.
FIG. 6.
Microprofiles of O2 (open circles) and NOx− (black squares) concentrations in alga-free sediment (A), illuminated alga sediment (C), and darkened alga sediment (E), measured after 21 days of incubation. Error bars represent standard errors of the means (n = 3 to 4), and the line represents the fitted profile. Consumption and production profiles are shown for O2 (white area) and NOx− (gray area) in alga-free sediment (B), illuminated alga sediment (D), and darkened alga sediment (F). The values are estimated from the porewater profiles.
Both depth-integrated and specific nitrification activity were significantly higher in the alga-free sediment than in the alga sediment (Table 4) (P < 0.0001). Depth-integrated and volume-specific anaerobic NOx− consumption were likewise higher in the alga-free sediment than in the alga sediment (Table 4) (P < 0.001). Depth-integrated and volume-specific O2 consumption rates were higher in the alga sediment than in the alga-free sediment by a factor of approximately two to three. However, only differences in depth-integrated O2 consumption were statistically significant (Table 4) (P = 0.038).
TABLE 4.
Depth-integrated and volume-specific NOx− and O2 transformation rates as estimated from numerical modeling of NOx− and O2 porewater profiles (n = 4 for Algae Light treatment and n = 3 for the remainder)
| Parameter | Result with treatmenta:
|
||
|---|---|---|---|
| No algae (n = 3) | Algae with light (n = 4) | Algae with darkness (n = 3) | |
| NOx− production (nmol cm−2 h−1) | 21.09 (1.88) | 5.82 (0.74) | 0 |
| Vol-specific NOx− production (nmol cm−3 h−1) | 285.9 (32.4) | 53.5 (5.2) | 0 |
| Aerobic NOx− consumption (nmol cm−2 h−1) | 0 | 4.05 (0.27) | 4.51 (0.58) |
| Anaerobic NOx− consumption (nmol cm−3 h−1) | 11.30 (0.70) | 3.58 (0.55) | 1.03 (0.16) |
| Vol-specific anaerobic NOx− consumption (nmol cm−3 h−1) | 91.87 (4.56) | 35.04 (11.1) | 41.43 (15.4) |
| O2 production (nmol cm−2 h−1) | −123 (22.4) | 132 (37.9) | −296.5 (43.0) |
| Vol-specific O2 consumption (nmol cm−3 h−1) | 1,066 (222) | ND | 4,318 (1,353) |
Standard errors of the means are given in parentheses. ND, not determined.
The data in Fig. 6 reflect porewater chemistry after 21 days of incubation. However, O2 profiles changed significantly during the course of both experiments 1 and 2. In experiment 1, the O2 penetration depth was 1.5 mm in the alga-free sediment after 7 days of incubation and 1 mm after 21 days. In the same time interval, the O2 penetration depth in the alga cores changed from 0.9 mm in darkness and 2.8 mm in light to 0.4 and 1.6 mm, respectively. The pattern was similar in experiment 2.
DISCUSSION
Growth and death of AOB.
In the present study, we have shown that the presence of active benthic microalgae promotes the death of AOB, as indicated by the parallel decrease in abundance of the amoA genes and nitrification potential in the alga sediment, while similar parameters increased in the alga-free sediment during the experiment (Fig. 1A and B). However, the death of AOB in the alga sediments did not result in measurable changes in the cell-specific activity or loss of members of the AOB population during the experiment. The change in relative DGGE band intensities that was observed after 21 days (Fig. 2) could be indicative of a slight change in the AOB population structure in the alga sediment, suggesting that specific organisms were better adapted than others to chemical or biological factors initiated during the alga colonization process. Caution should be taken, however, when relating DGGE band intensities to organism abundance in situ, due to the biases that are often introduced in the preceding PCR (53).
The AOB abundance (Fig. 1A and B) in the alga-colonized sediment was inversely correlated (R2 = 0.79, P = 0.0001) with the alga biomass, measured as Chl a. This suggests that the decrease in the abundance of AOB with time can be explained by the principle of competitive exclusion, i.e., one organism directly or indirectly inhibiting the growth of the other. As indicated in the following calculation, this inhibition can be linked to an ability of the algae to reduce the metabolic activity of the AOB. In the alga sediment, the average diurnal nitrification was 8.5 μmol m−2 h−1 at the end of the experiment. With a mean diurnal O2 penetration depth of 1 mm (see previous section), this corresponds to a specific activity of 8.5 nmol cm−3 h−1, assuming that nitrification took place in the entire oxic zone. This is approximately 20% of the nitrification potential measured in the top 3 mm of this sediment on day 21 (Fig. 1B). The nitrification rate in the oxic zone of the alga-free sediment was 250 μmol m−2 h−1. With an O2 penetration depth of 1 mm, this corresponds to a specific activity of 250 nmol cm−3 h−1, which is approximately 160% of the nitrification potential in the upper 3 mm of this sediment (Fig. 1B). A similar pattern was observed in experiment 2. The volume-specific net nitrification estimated from the porewater profiles was 35 and 0%, respectively, of the potential measured in the upper 3 mm of the alga sediment in light and darkness, respectively (Fig. 1C and 6D and Table 4). In the alga-free sediment, the volume-specific net nitrification in the oxic zone was 87% of the potential activity (Fig. 1C) and the maximum activity near the oxic-suboxic border was 160% of the potential measured in the upper 3 mm of the sediment. Based on model simulations of the metabolism of Nitrosomonas strains, Poughon et al. (35) estimated that 75% of maximum NH4+ oxidation serves for maintenance alone. Given that the nitrification potential approximates the average maximum NH4+ oxidation rate of the population (5), the average activity of the AOB population in the intact alga sediment was thus only 30% of its maintenance requirement. In the alga-free sediment, however, the activity of the AOB exceeded the maintenance requirements of the population. This may explain differences in growth patterns of AOB in sediment with and without algae. The death of AOB in the alga sediment, as inferred from the decrease in the number of amoA genes and the nitrification potential may in this context be explained as the result of cell starvation. Batchelor et al. (4) observed a similar decrease in cells of starved Nitromonas europaea in a culture experiment.
Factors responsible for decreases in the population of AOB.
In a previous study, it was suggested that N limitation of nitrifying bacteria in alga-colonized sediments was the major reason for reduced coupled nitrification-denitrification (40). The results of the present study support this hypothesis. In experiment 2, we observed similar nitrification potentials in alga-free and alga sediment incubated with 100 μM NOx−, whereas the nitrification potential in alga sediments incubated without additional water column NO3− was only about 50% of the activity measured in the alga-free cores (Fig. 1C). Risgaard-Petersen (40) observed a similar but less pronounced response to NO3− additions. Furthermore, the indication of lower actual nitrification than potential nitrification in the alga sediments (see above) is an indication of NH4+ limitation of AOB (47), since the nitrification potential is estimated in slurries containing an excess of NH4+. Thus, in line with previous studies (10, 40) the results of the present study point to N limitation as a major mechanism reducing the number of AOB in alga-colonized sediments.
N limitation of the AOB community in the oxic zone is an apparent paradox given the fact that NH4+ for nitrification is mainly supplied from the suboxic sediment strata (6, 20), well below the photic zone of the sediment. Thus, in principle, AOB could avoid N limitation by developing maximum densities at the oxic-suboxic interface. However, according to our NOx− sensor measurements, an AOB distribution such as this was only observed in the alga-free sediments (Fig. 6). This may suggest N limitation throughout the oxic zone in the alga sediments. This hypothesis is further supported by the observation that net NOx− production in the oxic zone in light was less than 35% of the potential activity (Fig. 1C and 6). Risgaard-Petersen (40) likewise provided evidence for N limitation of AOB below the photic zone and suggested that benthic microalgae, via their release of photosynthetates (49), stimulated the growth of heterotrophic bacteria. According to this theory, N assimilation of these bacteria should be responsible for the decline in AOB in the lower part of the oxic zone. We found no evidence, however, of elevated bacterial net growth in the alga-colonized sediments compared to the alga-free sediment. The 16S rRNA real-time PCR data suggested no significant differences between the two types of sediment, and d-amino acid data even suggested the lowest bacterial net growth rates in the alga-colonized sediments (Fig. 4). The NOx− microsensor measurements likewise showed the lowest volume-specific activity of anaerobic NOx− reduction (Table 4) in the alga-colonized sediment, suggesting a lower density of denitrifying bacteria in the presence of algae. Oxygen uptake rates in darkness were highest in alga sediments, however, indicating the highest heterotrophic activity in these sediments. The inconsistency between bacterial abundance and O2 uptake rates suggests that the elevated O2 uptake can be attributed to algal respiration. Hence, the data obtained in the present study do not support the hypothesis of Risgaard-Petersen (40) stating that N limitation of the AOB population is initiated by heterotrophic bacterial growth stimulated by the release of easily accessible carbon by benthic microalgae.
We propose that direct competitive interaction takes place between algae and AOB and that depression and possible N limitation of AOB below the photic zone is caused by the algae, which may exploit the deeper parts of the oxic zone through vertical migration. Pennate diatoms dominated the alga-colonized sediments, and the morphology of these algae indicates that the majority of the population was motile. The diatoms observed possessed a raphe on one or both frustule halves, which according to Hoek et al. (18), is used for locomotion.
Are benthic microalgae better competitors than AOB?
Given that direct competitive interaction takes place between AOB and benthic microalgae, the alga must grow faster than AOB and their N uptake must exceed the cell-specific NH4+ oxidation rate of the AOB or, alternatively, algal N uptake may have a much higher impact on the N availability for AOB than NH4+ oxidation has on the availability of N for algae. The present data set indicates that all of these criteria were fulfilled.
The net growth rate of AOB in the alga-free sediment was 0.04 day−1, which is close to the growth rate reported for N. marina (0.03 to 0.17 day−1) when grown in a chemostat at 20°C (13). Since NH4+ was not limiting AOB growth in this sediment, as indicated by the NH4+ efflux and agreement between actual and potential nitrification rates, we must assume that the estimated net growth rate was near the optimum of the population. The growth rate of the algae, estimated from the increase in Chl a with time was nearly two times higher (0.07 day−1).
The maximum cell-specific NH4+ oxidation activity of AOB was 5.9 fmol of N cell−1 h−1 as judged from the ratio between amoA gene copies and the nitrification potential (Fig. 1) and assuming two amoA gene copies per cell (28, 34). This is comparable with data reported for N. marina (0.9 to 4.9 fmol of N cell−1 h−1) (13). The cell-specific algal N uptake rate estimated from the abundance of the dominant alga group (Gyrosigma sp.) in the upper 3 mm of the sediment and the assimilation estimated from the flux and denitrification data (Fig. 6) was 560 fmol of N cell−1 h−1. Thus, alga cells exhibited uptake rates nearly 2 orders of magnitude higher than the maximum cell-specific NH4+ oxidation rate of AOB. In addition, the algae may take up both NH4+ and NOx− (Fig. 6) while AOB take up only NH4+ and release NO2−. Thus, algal N uptake may have a much higher impact on the N availability for AOB than NH4+ oxidation has on the availability of N for algae.
In addition to superiority with respect to growth and N uptake, the algae may initiate O2 limitation of the majority of the AOB through their respiration. In both experiments 1 and 2, there were large diurnal fluctuations in the O2 penetration depth in the alga cores, and during darkness, the O2 penetration depth was lower in these cores than in alga-free cores. These diurnal fluctuations in O2 penetration significantly affected the growth conditions of AOB. The NOx− microsensor experiments performed in experiment 2 showed that net nitrification in alga sediments was absent in darkness and only present below a depth of 0.5 mm in light (Fig. 6D and F). Thus, the major fraction of AOB in the alga-colonized sediments were only able to maintain a detectable metabolism in periods when oxygenic photosynthesis allowed O2 to penetrate deeper than the superficial zone, corresponding to only 12 h per day in the present study. During the remaining 12 h, the population was O2 limited.
In the present study, we investigated the growth of AOB in sediments colonized by benthic microalgae. Our data show that microalgae, via their assimilation and alterations of the chemical microenvironment, create conditions that prevent AOB from maintaining their population size. In addition, our data indicate that not only the AOB population is affected. Bacteria with the ability to reduce NOx− anaerobically, e.g., denitrifiers, seem to be negatively affected by the presence of active microalgae as well. This observation needs further documentation, however, for instance, through measurements of the denitrification potential. We propose that direct competitive interaction takes place between algae and AOB and that the benthic algae are superior competitors because they have higher N uptake rates and grow faster than AOB. In addition, via their respiration, the algae may induce O2 limitation of the AOB population. The overall consequence of this interaction is a reduction in the capacity of the sediment for removing N via coupled nitrification-denitrification upon alga colonization. Risgaard-Petersen (40) showed that this was in fact the case for natural sediments. As this reduction is linked to the initiation of N limitation of the bacterial populations, the reduction of coupled nitrification-denitrification is most severe during the summer months when N availability is low (45, 50).
Acknowledgments
We thank Anna Haxen, Kitte Gerlich Lauridsen, Marlene Venø Skjœrbœk, Rikke O. Holm, and Pernille Vester Thykier for assistance in the laboratory and Preben Sørensen for constructing the NOx− sensors.
This study was supported by grants from the Danish Natural Science Research Council (contract no. 51-00-0320 to N.R.-P. and contract no. 21-01-0475 to B.A.L.).
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16s rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, S. M., and S. B. Joye. 2001. Enhancement of coupled nitrification-denitrification by benthic photosynthesis in shallow estuarine sediments. Limnol. Oceanogr. 46:62-74. [Google Scholar]
- 3.Bada, J. L. 1982. Racemization of amino-acids in nature. Interdiscip. Sci. Rev. 7:30-46. [Google Scholar]
- 4.Batchelor, S. E., M. Cooper, S. R. Chhabra, L. A. Glover, G. Stewart, P. Williams, and J. I. Prosser. 1997. Cell density-regulated recovery of starved biofilm populations of ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 63:2281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belser, L. W. 1979. Population ecology of nitrifying bacteria. Annu. Rev. Microbiol. 33:309-333. [DOI] [PubMed] [Google Scholar]
- 6.Berg, P., N. Risgaard-Petersen, and S. Rysgaard. 1998. Interpretation of measured concentration profiles in sediment pore water. Limnol. Oceanogr. 43:1500-1510. [Google Scholar]
- 7.Borum, J., and K. Sand-Jensen. 1996. Is total primary production in shallow coastal marine waters stimulated by nitrogen loading? Oikos 76:406-410. [Google Scholar]
- 8.Bower, C. E., and T. Holm-Hansen. 1980. A Salicylate-hypoclorite method for determining ammonia in seawater. Can. J. Fish. Aquat. Sci. 37:794-798. [Google Scholar]
- 9.Braman, R. S., and S. A. Hendrix. 1989. Nanogram nitrite and nitrate determination in environmental and biological-materials by vanadium(III) reduction with chemi-luminescence detection. Anal. Chem. 61:2715-2718. [DOI] [PubMed] [Google Scholar]
- 10.Cabrita, M. T., and V. Brotas. 2000. Seasonal variation in denitrification and dissolved nitrogen fluxes in intertidal sediments of the Tagus estuary, Portugal. Mar. Ecol. Ser. 202:51-65. [Google Scholar]
- 11.Dalsgaard, T., et al. 2000. Protocol handbook for NICE—Nitrogen Cycling in Estuaries: a project under the EU research programme: Marine Science and Technology (MAST III), Silkeborg. National Environmental Research Institute, Silkeborg, Denmark.
- 12.Edler, L. 1979. Recomendations for marine biological studies in the Baltic sea, vol. 5. Phytoplankton and chlorophyll. Baltic Marine Biologists, Uppsala, Sweden.
- 13.Glover, H. E. 1985. The relationship between inorganic nitrogen oxidation and organic-carbon production in batch and chemostat cultures of marine nitrifying bacteria. Arch. Microbiol. 142:45-50. [Google Scholar]
- 14.Grasshoff, K., M. Erhardt, and K. Kremling. 1983. Methods of seawater analysis, 2nd ed. Verlag Chemie, Weinheim, Germany.
- 15.Guldberg, L. B., K. Finster, N. O. G. Jorgensen, M. Middelboe, and B. A. Lomstein. 2002. Utilization of marine sedimentary dissolved organic nitrogen by native anaerobic bacteria. Limnol. Oceanogr. 47:1712-1722. [Google Scholar]
- 16.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Gen. Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 17.Henriksen, K. 1980. Measurement of in situ rates of nitrification in sediment. Microb. Ecol. 6:329-337. [DOI] [PubMed] [Google Scholar]
- 18.Hoek, C. V. D., D. G. Mann, and H. M. Jahns. 1995. Algae, an introduction to phycology. Cambridge University Press, Cambridge, United Kingdom.
- 19.Jensen, K., N. P. Revsbech, and L. P. Nielsen. 1993. Microscale distribution of nitrification activity in sediment determined with a shielded microsensor for nitrate. Appl. Environ. Microbiol. 59:3287-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, K., N. P. Sloth, N. Risgaard-Petersen, S. Rysgaard, and N. P. Revsbech. 1994. Estimation of nitrification and denitrification from microprofiles of oxygen and nitrate in model sediment systems. Appl. Environ. Microbiol. 60:2094-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjœr, T., L. H. Lars, and N. P. Revsbech. 1999. Sensitivity control of ion-selective biosensors by electropheretically mediated analyte transport. Anal. Chim. Acta. 391:57-63. [Google Scholar]
- 22.Koops, H. P., B. Bottcher, U. C. Moller, A. Pommerening Roser, and G. Stehr. 1991. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J. Gen. Microbiol. 137:1689-1699. [Google Scholar]
- 23.Koops, H. P., and A. Pommerening-Roser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 24.Larsen, L. H., T. Kjœr, and N. P. Revsbech. 1997. A microscale NO3− biosensor for environmental applications. Anal. Chem. 69:3527-3531. [DOI] [PubMed] [Google Scholar]
- 25.Lorenzen, C. J. 1967. Determination of chlorophyll and pheo-pigments—spectrophotometric equations. Limnol. Oceanogr. 12:343-346. [Google Scholar]
- 26.Lorenzen, J., L. H. Larsen, T. Kjaer, and N. P. Revsbech. 1998. Biosensor determination of the microscale distribution of nitrate, nitrate assimilation, nitrification, and denitrification in a diatom-inhabited freshwater sediment. Appl. Environ. Microbiol. 64:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madigan, M. T., J. M. Martinko, and J. Parker. 1997. Brock biology of microorganisms. Prentice Hall, Inc., Upper Saddle River, N.J.
- 28.McTavish, H., J. A. Fuchs, and A. B. Hooper. 1993. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J. Bacteriol. 175:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mopper, K., and K. C. Furton. 1991. Extraction and analysis of polysaccharids, chiral amion acids, and safe extractable lipids from marine POM. Geophys. Monogr. 63:151-161. [Google Scholar]
- 30.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 31.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen, L. P. 1992. Denitrification in sediments determined from nitrogen isotope pairing. FEMS Microbiol. Ecol. 86:357-362. [Google Scholar]
- 33.Nielsen, L.P. 1993. Denitrification in microbial gradient communities, p. 153-156. In R. Guerrero and C. Pedros-Alio (ed.), Trends in microbial ecology. Spanish Society for Microbiology, Barcelona, Spain.
- 34.Norton, J. M., J. M. Low, and G. Martin. 1996. The gene encoding ammonia monooxygenase subunit A exists in three nearly identical copies in Nitrosospira sp. NpAV. FEMS Microbiol. Lett. 139:181-188. [DOI] [PubMed] [Google Scholar]
- 35.Poughon, L., C. G. Dussap, and J. B. Gros. 2001. Energy model and metabolic flux analysis for autotrophic nitrifiers. Biotechnol. Bioeng. 72:416-433. [PubMed] [Google Scholar]
- 36.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Revsbech, N. P. 1989. An oxygen microelectrode with an guard cathode. Limnol. Oceanogr. 34:474-478. [Google Scholar]
- 38.Revsbech, N. P., and B. B. Jorgensen. 1983. Photosynthesis of benthic microflora measured with high spatial-resolution by the oxygen microprofile method—capabilities and limitations of the method. Limnol. Oceanogr. 28:749-756. [Google Scholar]
- 39.Revsbech, N. P., J. Nielsen, and P. K. Hansen. 1988. Benthic promary production and oxygen profiles, p. 70-83. In T. H. Blackburn and J. Sørensen (ed.), Nitrogen cycling in coastal marine environments. SCOPE. John Wiley & Sons, Inc., New York, N.Y.
- 40.Risgaard-Petersen, N. 2003. Coupled nitrification-denitrification in autotrophic and heterotrophic estuarine sediments: on the influence of benthic microalgae. Limnol. Oceanogr. 48:93-105. [Google Scholar]
- 41.Risgaard-Petersen, N., and S. Rysgaard. 1995. Nitrate reduction in sediments and waterlogged soils measured by 15N techniques, p. 287-296. In K. Alef and P. Nannipieri (ed.), Methods in applied soil microbiology. Academic Press Inc., London, United Kingdom.
- 42.Risgaard-Petersen, N., S. Rysgaard, L. P. Nielsen, and N. P. Revsbech. 1994. Diurnal variation of denitrification and nitrification in sediments colonized by benthic microphytes. Limnol. Oceanogr. 39:573-579. [Google Scholar]
- 43.Risgaard-Petersen, N., S. Rysgaard, and N. P. Revsbech. 1993. A sensitive assay for determination of N-14/N-15 isotope distribution in No3. J. Microbiol. Methods 17:155-164. [Google Scholar]
- 44.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rysgaard, S., P. B. Christensen, and L. P. Nielsen. 1995. Seasonal variation in nitrification and denitrification in estaurine sediment colonized by benthic microalgae and bioturbating infauna. Mar. Ecol. Prog. Ser. 126:111-121. [Google Scholar]
- 46.Rysgaard, S., N. Risgaard-Petersen, L. P. Nielsen, and N. P. Revsbech. 1993. Nitrification and denitrification in lake and estuarine sediments measured by the 15N dilution technique and isotope pairing. Appl. Environ. Microbiol. 59:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rysgaard, S., N. Risgaard-Petersen, N. P. Sloth, K. Jensen, and L. P. Nielsen. 1994. Oxygen regulation of nitrification and denitrification in sediments. Limnol. Oceanogr. 39:1643-1652. [Google Scholar]
- 48.Schramm, A., D. de Beer, J. C. van den Heuvel, S. Ottengraf, and R. Amann. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, D. J., and G. J. C. Underwood. 2000. The production of extracellular carbohydrates by estuarine benthic diatoms: the effects of growth phase and light and dark treatment. J. Phycol. 36:321-333. [Google Scholar]
- 50.Sundback, K., A. Miles, and E. Goransson. 2000. Nitrogen fluxes, denitrification and the role of microphytobenthos in microtidal shallow-water sediments: an annual study. Mar. Ecol. Ser. 200:59-76. [Google Scholar]
- 51.Suwa, Y., T. Sumino, and K. Noto. 1997. Phylogenetic relationships of activated sludge isolates of ammonia oxidizers with different sensitivities to ammonium sulfate. J. Gen. Appl. Microbiol. 43:373-379. [DOI] [PubMed] [Google Scholar]
- 52.Utermöhl, H. 1958. Zur vervollkomnung der quantitativen Phytoplankton metodik. Mitt. Int. Ver. Limnol. 9:1-38. [Google Scholar]
- 53.Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-219. [DOI] [PubMed] [Google Scholar]