Abstract
Sludges derived from wastewater treatment are foul-smelling, biologically unstable substances. As well as containing numerous pathogenic microorganisms, they also consist of organic matter that can be used as agricultural fertilizer. Legislation nevertheless requires sludges to be virologically tested prior to spreading by the counting of infectious enterovirus particles. This method, based on culture of enterovirus on BGM cells, is lengthy and not very sensitive. The aim of this study was to propose an alternative method of genome quantification for all enteroviruses that is applicable to verifying the elimination of viruses in complex samples such as sludges. Our complete protocol was compared to the official method, consisting of enterovirus enumeration with the most probable number of cythopathic unit (MPNCU) assay through the study of four stabilization procedures: liming, composting, heat treatment, and mesophile anaerobic digestion. Enterovirus quantities at the start of the stabilization procedures were between 37 and 288 MPNCU/g on the one scale and between 4 and 5 log genome copies/g on the other. It was shown that all procedures except mesophile anaerobic digestion were highly effective in the elimination of enterovirus particles and genomes in wastewater sludges. Reduction of viruses by mesophile anaerobic digestion was by only 1 log (infectious particles and genomes). In conclusion, stabilization processes can indeed be checked by virological quality control of sludges with gene amplification. However, the infectivity of genomes needs to be confirmed with cell culture or a correlation model if the virological risk inherent in the agricultural use of such sludges is to be fully addressed.
Sludges derived from wastewater treatment are foul-smelling, biologically unstable substances. They contain numerous pathogenic microorganisms, mostly of fecal origin. They also consist of organic matter that can be put to agricultural use as fertilizer. Agricultural use, in the form of spreading on the land, is the principal means by which sludge is disposed of and can only be of lasting value if the sludge has undergone treatment by biological, chemical, thermal, or other suitable processes to diminish its capacity for fermentation and eliminate any health risk related to such use. With the aim of reducing the risk of viral contamination associated with spreading such sludges, French legislation (decree of 8 January 1998 related to the landing of sewage sludge on agricultural soils) requires that microbiological testing be carried out for validation of stabilization processes. The virological testing method currently specified (appendix 5, table 6b of the decree) is based on the counting of enterovirus particles in culture on buffalo green monkey (BGM) cells with the most probable number of cytopathic units (MPNCU) method. This method is lengthy and not very sensitive.
We have recently developed a virological quality control method for testing sludges from water treatment plants based on gene amplification with TaqMan technology (16) and on a viral extraction technique compatible with both PCR and cell culture (17). Hitherto, false-negative responses have been averted by spiking all negative specimens with the RNA standard for a second run, an impractical, laborious, and very expensive solution. The aim in this study was to improve our previous technique for genome quantification (16, 17) by including an internal PCR quality control and to evaluate the efficacy, in terms of viral decontamination, of four sludge treatment processes (liming, composting, mesophile anaerobic digestion, and heat treatment) used in plants in different parts of France. The efficacy of these processes in eliminating viruses was evaluated by comparing enterovirus infectious particle quantities before and after sludge stabilization. The counting method specified in the legislation was then carried out in parallel with our technique for genome quantification, the TaqMan reverse transcription (RT)-PCR method.
MATERIALS AND METHODS
Lime stabilization process.
Liming is used in two wastewater treatment plants, plants 1 and 2. Their capacities are 600,000 and 120,000 equivalent inhabitants, respectively. Here, the process consists of the addition of quicklime at a proportion of 50% dry matter to the biological sludge, which has previously undergone thickening and dehydration. The mixture is homogenized in a twin-screw mixer-aerator equipped with a piston pump, which allows the limed sludge to be sent to the storage silo. After cooling, the limed sludge has a pH between 12.5 and 13.
Composting process.
Aerobic composting is used in one treatment plant whose capacity varies along with a seasonal influx of tourists: 12,500 equivalent inhabitants out of season and 80,000 equivalent inhabitants in season. The process here consists of aerating a mixture of dehydrated sludge and ground tree bark, which is then left to compost for 3 or 4 weeks. The maturing compost is next put through a sifter to homogenize it, and the resulting product is left to compost further for a minimum of 3 months in maturation racks. Temperatures of between 20 and 68°C are reached during this process.
Mesophile anaerobic digestion.
Mesophile anaerobic digestion is used in one treatment plant with a capacity of 200,000 equivalent inhabitants. Thickened sludge is sent to two digesters, one primary and the other secondary. The process of anaerobic fermentation takes place in the primary digester for 14 days at 36°C. The function of the secondary digester is storage, and the sludge that it contains is stirred intermittently for 9 days. The secondary digester allows the particles to be completely methanized, separates out and discharges floating matter, and facilitates a certain degree of thickening of the sludge. The total time that the sludge spends in the digesters is therefore 23 days.
Heat treatment.
Heat treatment is used in one treatment plant with a capacity of 7 million equivalent inhabitants. In this plant, sludge first undergoes mesophile anaerobic digestion and is then thickened and heated to 195°C in a pressure vat at a pressure of 19 to 21 bars for 100 min.
Sludge samples.
For the composting and mesophile anaerobic digestion processes, this study did not involve following a single batch of sludge through the different stages of the processes. The time elapsing between the beginning and end of processing was from 3 weeks to several months for composting and 23 days for anaerobic digestion. It was therefore more practical for us to carry out monthly sampling at several stages of the process, which in any case reflected the state of progress in the stabilization procedure. The samples most commonly described as preprocessing by staff at the plants did not, in fact, comprise primary sludge derived directly from wastewater. In general, they consisted of primary sludge that had undergone dehydration or thickening immediately before the stabilization process.
Liming stabilization process.
Two types of sludge were sampled at monthly intervals during the course of the liming stabilization process: upstream dehydrated sludge (18 to 20% dry matter) and limed sludge (30 to 33% dry matter); 500-g samples of the dehydrated sludge were taken leaving the centrifuge, and 500-g samples of limed sludge were taken from the storage silo. Twelve samples for each sludge type and for each treatment plant, making a total of 48 samples, were analyzed over a 1-year period (January 2000 to December 2000).
Composting process.
Five types of sample, each of 1 kg, corresponding to the different stages of the process, were taken at monthly intervals for the study: dehydrated sludge (18 to 20% dry matter), mixture of sludge and coproduct (29 to 30% dry matter), compost before sifting (38 to 40% dry matter), compost after sifting (45%), and the mature end product (60 to 80% dry matter). They were sent by post (4 to 5 days transit time) and were stored at +4°C (for a maximum of 3 days) if analysis could not be carried out on reception. This treatment plant was followed up for a period of 10 months, and a total of 49 sludge samples were analyzed.
Mesophile anaerobic digestion.
Samples of 1 liter were taken at monthly intervals of sludge entering the primary digester (thickened sludge, 3.5 to 6% dry matter) and sludge leaving the secondary digester (digested sludge, 2 to 3% dry matter). They were dispatched to the laboratory by post (48 h) and stored at +4°C (for a maximum of 3 days) if analysis could not be carried out on reception. This treatment plant was followed up for a period of 10 months, and a total of 20 sludge samples were analyzed.
Heat treatment.
Samples of 1 liter of sludge entering (digested and thickened sludge, 3 to 4% dry matter) and leaving (treated sludge, 1 to 2% dry matter) the pressure vat were taken. They were dispatched to the laboratory by post (48 h) and stored at +4°C (for a maximum of 3 days) if analysis could not be carried out on reception. This treatment plant was followed up for a period of 7 months, and 14 sludge samples were analyzed.
Test sample.
The exact test sample was the equivalent of 5 g (dry matter) for sludge whose siccity was known at the time of analysis, which was the case for sludge from the liming process. The siccities of the sludges from the other processes reached us 1 month after analysis. Accordingly, for the digestion and heat treatment processes, 50 ml of sludge entering the process and 100 ml of the end product were sampled; and for the composting process, 25 g (dry matter) of dehydrated sludge, 25 g of the mixture of sludge and coproduct, 10 g of sludge before and after sifting, and 6 g of the mature compost were sampled. The results were subsequently adjusted to 1 g of dry matter once the values for siccity were available.
Virus elution.
Virus was eluted from sludge with a technique adapted from that described by Ahmed and Sorensen (2). To a sludge volume yielding 5 g of dry matter, 100 ml of 10% beef extract (pH 8; LP029B; Oxoid) was added, the mixture was stirred at 700 oscillations/min for 30 min with the Flask Shaker SF-1 (Sigma-Aldrich, France), and then centrifuged at 5,000 × g for 1 h at 4°C. The supernatant, adjusted to pH 7.2 (if necessary), constituted the extract.
Virus concentration.
For virus concentration, we used polyethylene glycol 6000 precipitation as described by Lewis and Metcalf (12); 8% (wt/vol) polyethylene glycol 6000 (in a phosphate solution at pH 7.2) was added to each extract. After rigorous agitation, the mixture was kept at 4°C overnight and then centrifuged at 10,000 × g for 90 min at 4°C. The pellet, suspended in 12 ml of phosphate buffer (pH 7.2), constituted the concentrate; as a final step, the pellet was decontaminated by adding 0.33 volume of chloroform.
Virus counting by means of cell culture.
Infectious enteroviruses were counted by inoculating decontaminated concentrates into in vitro buffalo green monkey cell cultures in 96-well microplates. All cultures were inoculated in duplicate, with 40 wells for each dilution. Each well was filled with 50 μl of inoculum and 200 μl of nutritive medium (minimal essential medium; Life Technologies) with 5% newborn calf serum containing 1.5 × 105 cells/ml. The cells were incubated at 37°C in 5% CO2 for 5 days.
Viral density was determined from the cytopathogenic effects observed after duplicate inoculation of cell layers with three successive fivefold dilutions of a sample. After confirmation by transfer of 50-μl portions of the supernatants to new microplates, the mean viral concentration of the samples was estimated by the most-probable-number method with the software described by Maul (15). Thus, each viral concentration was determined from a combination of the positive responses observed in the 40 wells inoculated for each of three successive fivefold dilutions. The final result for each sample analyzed was expressed as the geometric mean of the concentrations calculated for two independent replicates. The results were expressed in MPNCU per milliliter of concentrate and then converted to MPNCU per gram (dry weight) of sludge in order to take account of sludge dryness.
Extraction of viral RNA.
Enterovirus RNA was extracted from 400 μl of concentrate with an RNeasy mini kit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. However, a modified lysis buffer containing 2% (wt/vol) polyvinylpyrrolidone 40,000 (Sigma, France) was used (16, 17).
Primers and probes for quantification by multiplex fluorogenic RT-PCR.
The primers and probes used for enterovirus amplification were as already published: Ev1 (5′-GATTGTCACCATAAGCAGC-3′), Ev2 (5′-CCCCTGAATGCGGCTAATC-3′), and Ev-probe (5′-FAM-CGGAACCGACTACTTTGGGTGTCCGT-TAMRA-phosphor-3′; FAM is 6-carboxyfluorescein, and TAMRA is 6-carboxytetramethylrhodamine) (16, 17).
For absolute quantification, an enterovirus RNA standard representing the 5′ noncoding region of enterovirus RNA was synthesized in vitro by plasmid cloning and in vitro transcription with cDNA from Mahoney type 1 poliovirus and primers Ev1Clon (5′-TGGCCAATCGAATTCGCTTTA-3′) and Ev2Clon (5′-CTACATAAGGATCCTCCGGCC-3′) (16, 17).
Internal positive control.
An exogenous internal positive control was introduced into each reaction well in order to prevent false-negative results. This was an RNA template transcribed from plasmid pAW109 (pAW109 RNA), purchased from Applera France and used as an internal amplification quality marker. We designed the primers and probe with the software Primer Express (Applera) in order to use this RNA in a PCR duplex format to monitor the amplification reaction quality. The software defined the primers pAW1 (5′-TCCCCAGGAACAGTTGAAAGA-3′) and pAW2 (5′-AACAGGGAACCCAGGCTCC-3′) and pAW-probe (5′-TET-CAGTGCCTGCCCATTCGGAGGA-TAMRA-phosphor-3′; TET is 6-tetrachlorofluorescein). The compatibility of these primers and probe with enterovirus amplification was tested with the software Oligo 4 (National Biosciences, Inc.) in order to determine the multiplex conditions.
Reaction mixture.
The reaction mixture (final volume, 25 μl) was prepared in a single tube as follows: 1× TaqMan buffer (Eurogentec), 6 mM MgCl2 (Applera), 700 μM deoxynucleoside triphosphates (Eurogentec), 120 nM each of primers Ev1 and Ev2 (Genosys, Pampisford, England), 100 nM Ev-probe (Eurogentec), 60 nM each of primers pAW1 and pAW2 (Genosys), 80 nM pAW-probe (Eurogentec), 1.5% PVP-25 (Coger), 0.6 μg bovine serum albumin (Roche Diagnostic), 2 μg of T4 gene 32 protein (Roche Diagnostic), 1.5 U of murine leukemia virus reverse transcriptase (Applera), 1.5 U of HotStart Gold (Eurogentec), 20 U of RNasin (Promega), and 1 μl of internal positive control RNA (1,000 copies/μl); 20 μl of the reaction mixture was added to PCR tubes containing 5 μl of RNA extract from sludge samples or RNA standard in serial dilution. Enterovirus RNA and the internal positive control RNA were reverse transcribed into cDNA (40 min at 50.1°C), followed by a murine leukemia virus Taq Gold denaturation-activation step (10 min at 94°C). The cDNA fragments were amplified by PCR (15 s at 94°C and 1 min at 60°C) for 45 cycles on an ABI Prism 7700 (Applera).
Controls.
The BGM cell culture microplates inoculated with samples were compared to the microplate inoculated with the assay medium only for cytopathic effect. The negative control for reverse transcription-PCR was the RNA diluent.
Analysis of fluorescence signals with the ABI Prism 7700.
Real-time fluorescence measurements were obtained, and the threshold cycle (CT) value for each sample was calculated by determining the point at which fluorescence exceeded a threshold limit (10 times the baseline standard deviation). A standard graph of the CT values obtained with a serially diluted external RNA standard was prepared. CT values obtained from the sludge samples were plotted on the standard curve, and the number of copies was calculated automatically by the software Sequence Detector v1.7 (Applera).
Validation of results.
Results were validated, in essence, by evaluating the quality of amplification in the reaction tube. The quality of amplification was evaluated by the CT of the internal positive control (CT-ipc). In practice, we chose to validate those results for which CT-ipc was <39 cycles. When amplification is inhibited, no fluorescence signal can be detected, and CT-ipc = 45 cycles. The analysis cannot therefore be validated, and the RNA extract has to be diluted (twofold or fourfold) for a new amplification. A result was considered negative (true) if its CT for enterovirus (CT-ev) = 45 cycles and its CT-ipc = <39 cycles.
RESULTS
Fluorogenic RT-PCR with internal control.
During the initial optimizing steps of the multiplex fluorogenic RT-PCR, the optimal simultaneous amplification conditions were determined. The RNA standard constructed was used at concentrations of 50 to 500,000 copies/reaction, and the internal control was used at 1,000 copies/reaction. The first results showed that the internal positive control was not amplified in the presence of the RNA standard at quantities over 50,000 copies/reaction and that the enterovirus detection limit was 5,000 copies/reaction. Different concentrations of each primer (100 to 400 nM for enterovirus and 50 to 100 nM for the internal positive control) were therefore systematically tested. The best results were obtained with 120 nM enterovirus primers and 60 nM internal positive control primers. These primer concentrations made possible a constant amplification level for the internal positive control in the presence of RNA standard at 50 to 500,000 copies/reaction.
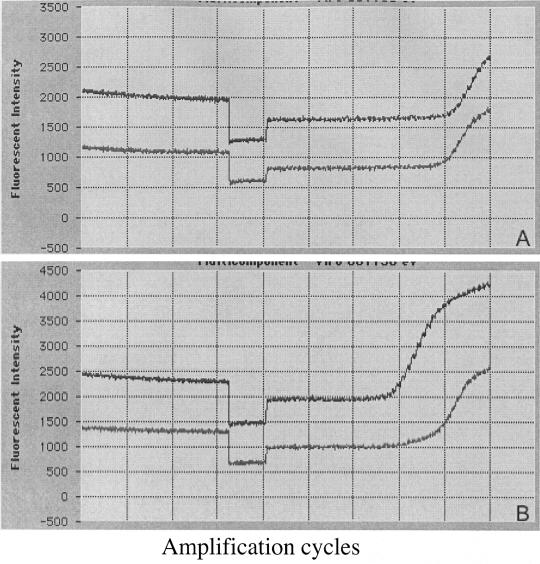
Figure 1 shows the amplification plots (multicomponent curves) of two points (50 and 5,000,000 copies) of the enterovirus RNA standard (upper curve) with the internal control (lower curve). The detection limit and intra-assay and interassay variability were determined in 10 replicates of 10-fold dilutions of the standard from 5 to 50,000 RNA copies amplified in eight independent assays. The intra-assay coefficient of variation varied between 2.2 and 1.4%, and the interassay coefficient of variation varied between 5.5 and 1.95%. The analytical sensitivity of our method, taking all extraction steps into account and with the RNA standard serial dilutions, is 250 copies of RNA per gram of sludge (5 copies of enterovirus RNA standard per reaction). Smaller quantities, however, can be measured because the linearity of the standard curve allows values below the lower limit of the dynamic range to be interpolated (14).
FIG. 1.
Multiplex amplification plots (multicomponent curves) of two points, 50 copies (A) and 5 × 106 copies (B), of the enterovirus RNA standard (upper curve) with the internal control (lower curve).
Liming.
Results from the 12-month period over which this process was studied are shown in Table 1. Enteroviruses were consistently present all year round in the dehydrated biological sludge entering the process. The infectious particle count was between 3.4 and 167 MPNCU/g (dry matter) of dehydrated sludge (mean, 48 MPNCU/g). In terms of genome titers, quantification values were between 7.28E + 02 and 4.50E + 04 copies/g (dry matter) of dehydrated sludge (mean, 1.03E + 04 copies/g). Low viral loads, in terms of both infectious particles and genome titers, were found during the summer period (July, August, and September), while in spring (April, May, and June) viral loads were high. At the end of the process, no viable enterovirus or genome was detected in sludge treated by liming.
TABLE 1.
Quantification of enteroviruses in sludge before and after liming
| Date (mo and yr) | Plant | Virus enumeration on BGM cells (MPNCU/g)
|
Enterovirus RT-PCR (copies/g)
|
||
|---|---|---|---|---|---|
| Dehydrated sludges | Limed sludges | Dehydrated sludges | Limed sludges | ||
| Jan 00 | 1 | 11 | <1.7a | 3.4E + 03 | <50a |
| 2 | 16 | <1.7 | 2.7E + 03 | <50 | |
| Feb 00 | 1 | 37 | <1.7 | 3.4E + 03 | <50 |
| 2 | 18 | <1.7 | 6.0E + 03 | <50 | |
| Mar 00 | 1 | 17 | <1.7 | 1.9E + 04 | <50 |
| 2 | 34 | <1.7 | 1.2E + 04 | <50 | |
| Apr 00 | 1 | 39 | <1.7 | 1.4E + 03 | <50 |
| 2 | 130 | <1.7 | 4.5E + 04 | <50 | |
| May 00 | 1 | 48 | <1.7 | 3.2E + 03 | <50 |
| 2 | 152 | <1.7 | 1.3E + 04 | <50 | |
| Jun 00 | 1 | 78 | <1.7 | 5.5E + 03 | <50 |
| 2 | 112 | <1.7 | 1.5E + 04 | <50 | |
| Jul 00 | 1 | 3.4 | <1.7 | 1.2E + 03 | <50 |
| 2 | 25 | <1.7 | 7.1E + 03 | <50 | |
| Aug 00 | 1 | 6.8 | <1.7 | 1.1E + 03 | <50 |
| 2 | 3.4 | <1.7 | 2.1E + 03 | <50 | |
| Sep 00 | 1 | 6.8 | <1.7 | 1.6E + 04 | <50 |
| 2 | 17 | <1.7 | 7.3E + 02 | <50 | |
| Oct 00 | 1 | 73 | <1.7 | 1.6E + 03 | <50 |
| 2 | 167 | <1.7 | 9.9E + 03 | <50 | |
| Nov 00 | 1 | 6.8 | <1.7 | 1.1E + 03 | <50 |
| 2 | 19 | <1.7 | 1.1E + 04 | <50 | |
| Dec 00 | 1 | 68 | <1.7 | 4.2E + 04 | <50 |
| 2 | 73 | <1.7 | 2.4E + 04 | <50 | |
Limit of detection.
Composting.
The results for this process are shown in Table 2. In sludge entering the process, infectious particles were not detected over the course of the year in a regular pattern (6 of 10). Infectious titers were between <1.7 and 112 MPNCU/g (mean, 37 MPNCU/g). The mean infectious titer of 37 MPNCU was calculated without taking negative results into account. Viral genomes were detected consistently throughout the study period. Genome titers were between 2.19E + 03 and 4.20E + 04 copies/g (mean, 9.70E + 03 copies/g).
TABLE 2.
Quantification of enteroviruses in sludge samples from five stages of the composting process
| Date (mo and yr) | Virus enumeration on BGM cells (MPNCU/g)
|
Enterovirus RT-PCR (copies/g)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dehydrated sludges | Sludge + coproduct | Before sifting | After sifting | End product | Dehydrated sludges | Sludge + coproduct | Before sifting | After sifting | End product | |
| Apr 00 | 3.4 | 6.8 | <1.7 | <1.7 | <1.7 | 7.9E + 3 | 700 | <200a | <50 | <50 |
| May 00 | 14 | <1.7 | <1.7 | <1.7 | <1.7 | 4.6E + 3 | 1.4E + 3 | <200 | <200 | <200 |
| Jun 00 | 6.8 | 6.8 | <1.7 | <1.7 | <1.7 | 3.2E + 3 | 5.5E + 2 | <100a | <50 | <100 |
| Jul 00 | <1.7 | 3.4 | <1.7 | <1.7 | <1.7 | 5.2E + 3 | 4.7E + 2 | <50 | <50 | <50 |
| Aug 00 | <1.7 | 3.4 | <1.7 | <1.7 | <1.7 | 1.1E + 4 | 2.4E + 3 | <50 | <50 | <100 |
| Oct 00 | <1.7 | <1.7 | <1.7 | <1.7 | <1.7 | 2.2E + 3 | 1.1E + 3 | <100 | <100 | <100 |
| Nov 00 | Not taken | <1.7 | <1.7 | <1.7 | <1.7 | Not taken | 1.0E + 3 | <100 | <50 | <100 |
| Dec 00 | 6.8 | 3.4 | <1.7 | <1.7 | <1.7 | 4.2E + 4 | 1.0E + 4 | <50 | <100 | <50 |
| Jan 01 | 80 | 74 | <1.7 | <1.7 | <1.7 | 7.5E + 3 | 1.6E + 3 | <100 | 490 | <100 |
| Feb 01 | 112 | 93 | <1.7 | <1.7 | <1.7 | 3.7E + 3 | <200 | <100 | <100 | <100 |
detection threshold of RT-PCR as a function of RNA extract dilution (dilution 1:2 = <100 and dilution 1:4 = <200).
Many of the samples taken at the mixture stage (19 of 49) showed PCR inhibition phenomena (no fluorescence signal for the internal control). This made further manipulation necessary (extraction and RT-PCR) after diluting the RNA extract. The consequences, as seen in the tables, are higher detection thresholds: <100 corresponds to a 1:2 dilution and <200 to a 1:4 dilution.
Enteroviruses were found in the samples (sludge plus coproduct) of this first stage of composting in which sludge is mixed with bark. Counts in this mixture were between <1.7 and 92.6 MPNCU/g (mean, 27.1 MPNCU/g) and between 4.70E + 02 and 1.04E + 04 copies/g (mean, 2.37E + 03 copies/g). A decrease in the presence of virus was observed from this stage onwards and was more marked in terms of genomes than in terms of infectious particles.
After the compost had been aerated for 3 to 4 weeks, all samples taken before sifting and after sifting (with one exception) and of the mature compost were negative, both in culture and by TaqMan RT-PCR.
Mesophile anaerobic digestion.
The results of the virological study of the mesophile anaerobic digestion process are shown in Table 3. Enterovirus was consistently detected in samples of sludge entering the process. Counts in the thickened sludge were between 28.7 and 901.4 MPNCU/g (mean, 287.6 MPNCU/g) and between 8.9E + 03 and 2.95E + 05 copies/g (dry matter) (mean, 1.04E + 05 copies/g). At the end of the process, infectious virus particles were detected on six occasions in the digested sludge at loads of between <1.7 and 219.8 MPNCU/g (mean, 38.2 MPNCU/g) and of between 9.1E + 02 and 3.2E + 04 copies/g (mean, 1.2E + 04 copies/g). Over the study period, the level of enterovirus reduction (about 1 log), which varied according to month, was also a function of the viral quantification method, as seen in Table 3.
TABLE 3.
Quantification of enteroviruses in sludges before and after anaerobic digestion
| Date (mo and yr) | Virus enumeration on BGM cells (MPNCU/g)
|
Enterovirus RT-PCR (copies/g)
|
Enterovirus reduction (%)
|
|||
|---|---|---|---|---|---|---|
| Thickened sludges | Digested sludges | Thickened sludges | Digested sludges | Culture | RT- PCR | |
| Mar 00 | 83 | 10 | 5.6E + 04 | 5.7E + 03 | 87 | 90 |
| May 00 | 84 | 6.1 | 3.8E + 04 | 5.8E + 03 | 93 | 85 |
| Jun 00 | 900 | <3.5a | 1.3E + 05 | 9.1E + 02 | 100 | 99 |
| Jul 00 | 29 | <3.1 | 8.2E + 04 | 2.4E + 04 | 100 | 71 |
| Sep 00 | 43 | <3.5 | 2.3E + 04 | 7.1E + 03 | 100 | 69 |
| Oct 00 | 280 | 12 | 8.9E + 03 | 2.9E + 03 | 96 | 67 |
| Nov 00 | 45 | <3.9 | 1.5E + 05 | 1.6E + 03 | 100 | 99 |
| Dec 00 | 93 | 53 | 3.0E + 05 | 3.2E + 04 | 43 | 89 |
| Jan 01 | 570 | 81 | 1.3E + 05 | 8.8E + 03 | 86 | 93 |
| Feb 01 | 740 | 220 | 1.3E + 05 | 1.6E + 04 | 70 | 88 |
Detection threshold varied as a function of sludge siccity, taking account a constant test sample of 50 ml for thickened sludge and 100 ml for digested sludge.
Heat treatment.
The results for the study of the heat treatment process are shown in Table 4. In contrast to infectious virus particles, genomes were consistently measurable in samples of sludge entering the process. They were found at levels of between 1.1E + 04 and 1.5E + 05 copies/g (dry matter) of thickened sludge (mean, 4.5E + 04 copies/g). At the end of the process, neither genomes nor infectious particles were observed during the period studied.
TABLE 4.
Quantification of enteroviruses in sludges before and after heat treatment
| Date (mo and yr) | Virus enumeration on BGM cells (MPNCU/g)
|
Enterovirus RT-PCR (copies/g)
|
||
|---|---|---|---|---|
| Thickened sludges | Treated sludges | Thickened sludges | Treated sludges | |
| Mar 00 | <3.8a | <2.1a | 3.8E + 04 | <63.8a |
| May 00 | <3.5 | <2.1 | 1.1E + 04 | <62.5 |
| Jun 00 | <4.6 | <2.7 | 3.5E + 04 | <79.1 |
| Jul 00 | <4.3 | <2.3 | 6.6E + 03 | <69.6 |
| Sep 00 | <4.8 | <2.5 | 4.2E + 04 | <73.5 |
| Oct 00 | <5.3 | <3.1 | 3.0E + 04 | <90.9 |
| Nov 00 | <5.2 | <2.9 | 1.5E + 05 | <86.2 |
Detection threshold varied as a function of sludge siccity, taking account a constant test sample of 50 ml for thickened sludge and 100 ml for digested sludge.
DISCUSSION
Quantification of viruses in sludge samples by RT-PCR requires a sensitive technique with an internal control, in view of the low levels of virus often present (in treated sludge) and of problems due to PCR inhibitors. During the optimization of our previous fluorogenic RT-PCR (16, 17), we overcame problems of sensitivity related to the secondary structures present in the 5′ noncoding region of the enterovirus genome and problems due to PCR inhibitors by using PCR adjuvants such as T4 gene 32 protein and polyvinylpyrrolidone (16). In order to detect PCR inhibitors without a second run, we decided to work on the incorporation of an internal control. We chose to develop a multiplex amplification technique so as to spare ourselves the dilutions required to bring a competitive RT-PCR within the equivalence zone. This internal control is completely different from those used in competitive PCR. It is simply an internal quality control of the amplification, as described by some authors for real-time quantification of DNA (1, 4, 21). The sole requirement is that the two amplification reactions be independent. The fact that the internal control is amplified at the same level (CT = 36 cycles) over a range of 50 to 500,000 copies of enterovirus RNA standard makes it a marker of good quality and improves its usefulness. The intra- and interassay variability values of 2.2 and 5.5%, respectively, obtained with low numbers of RNA standard copies are consistent with values observed in work on other TaqMan RT-PCRs (3.4 and 6.2%, respectively) (9, 14).
After optimizing our RT-PCR protocol, we chose to investigate four processes used for stabilizing wastewater sludge. These processes are employed in treatment plants located in different regions of France (north, west, and south). These plants receive effluent from towns of various sizes. The degree of viral pollution in the sewage received by any one plant is a function of a number of factors: state of health of the population, level of hygiene, industrial premises connected to the sewerage network, season, and climate (26).
At the start of each process, enteroviruses (infectious particles and genomes) were consistently present in greater or smaller quantities. This consistent presence of enteroviruses confirms their value as indicators of stabilization process efficiency. The differences found in this study between the viral loads measured at the start of each treatment process may be explained by several factors: the capacity of the treatment plant, the pretreatment method (dehydration, filtration) to which sludge is subjected prior to the stabilization process itself, and the nature of any chemical substances entering the sewerage network (bleach, detergents, etc.) Another factor that had to be taken into account during this study was how the sludge samples to be tested were transported. During the summer, this was by post (4 to 5 days), which may have contributed to the loss of virus. From our study, it emerges that sludge prior to treatment can be divided into two groups, and this was particularly shown by enterovirus genome titers. On the one hand were the liquid sludges (raw material of the digestion and heat treatment processes, 3 to 4% dry matter) with 5 log copies/g, and on the other the solid sludges (dehydrated raw material of the liming and composting processes, 18 to 20% dry matter) with 4 log copies/g. This difference in viral load (1 log) may be explained by desiccation (29) and the mechanical process used for dehydration, this author reporting that sludge with a water content of less than 10% contains less than 0.01% enteric viruses.
The different stabilization processes are designed to reduce this viral pollution. In this study, investigating the enterovirus load in sludges after stabilization treatment allowed the efficacy of these processes in eliminating enteric viruses to be evaluated, in terms of both infectious particles and genomes.
For the liming process, no enterovirus (infectious particle or genome) was detected in limed sludge samples. This process was also effective for other viruses, such as adenoviruses and reoviruses (23), which were, in fact, observed in certain samples during the study (data not shown). It has been shown by Hurst (10) that the rate of reduction of polioviruses in limed sludge is 90% in 0.465 days in the presence of 5 kg of lime per m3 of sludge. This decontaminating activity is principally due to the alkaline pH (pH = 12). At this pH, the NH4+ ions in the sludge lose a proton, and ammonium gas is produced. The combination of a high pH and ammonium can diminish enterovirus titers by at least 4 log10 (23, 26).
In practice, good stabilization and decontamination are obtained when the amount of lime present makes up over 40% of the sludge mass as dry matter in a homogenous mixture. The two treatment plants studied have chosen to use a system of mixer-aerator, which allows a homogenous and identical pH (pH > 12) to be obtained at all points of the blend of sludge and lime. Moreover, the use of quicklime tends to raise the temperature, increasing the efficacy of decontamination.
For composting, enterovirus inactivation was appreciable from the first stage when sludge was mixed with the coproduct. The addition of structure-providing carbonaceous matter or of metals (e.g., Cu2+) in the bark may promote the irreversible adsorption or the inactivation of the viruses. When the compost was sifted, i.e., after 3 or 4 weeks of fermentation, the temperature reached 65°C; enterovirus infectious particles were completely eliminated, while genomes remained present at a very low level before disappearing in the mature end product. It should, however, be pointed out that levels of enterovirus infectious particles present in the sludge prior to treatment (9.7E + 03 copies/g dry matter) appeared low, and this perhaps contributed to the good results observed with this process. One reason for such low virus levels may be sludge dehydration (strip filtering) before composting.
Composting is a heat-dependent stabilization process, and the temperatures reached during treatment (55 to 70°C) seem sufficiently high to eliminate enteric microorganisms (26, 30). Such temperatures are capable of promoting a decrease in virus of between 3 and 4 log10. Dehydration also plays an important role in virus inactivation by rupturing the virus capsid and releasing nucleic acid (29).
From the point of view of testing technique, attention should be drawn to two points: the frequent presence of PCR inhibitors in the compost extracts (38.7%), which made it necessary to carry out dilutions of 1:2 or 1:4 for the results obtained to be correctly interpreted; since these inhibitors were not found in the dehydrated sludge, they must have been derived from the coproduct, the tree bark; and the extreme heterogeneousness of the material which, during the composting process, made the taking of test samples difficult to reproduce.
For mesophile anaerobic digestion, the results of our study confirm the low decontaminating power of this process in virological terms. The mean level of virus decrease (87.5%) in terms of infectious particles is comparable to that reported in the literature (80 to 90%) (8, 26). Virus inactivation depends on a number of parameters, of which the temperature of digestion and chemical and biological factors apparently play an important role. Between 30 and 37°C, 50 to 90% of poliovirus I (Sabin) is inactivated (3, 6), while at 50°C the level of decrease can reach 99.99% (22). However, the contribution of the temperature in mesophile anaerobic digestion (35°C) has been estimated to be between 19% (27) and 46% (22). Other factors such as ammonium, detergents, bacterial enzymes, or microorganisms may also play a part (28). Ammonium inactivates viruses at a pH of more than 8 (22) and causes fragmentation of RNA (10). In this study, the pH measured in the vat, which was generally alkaline (mean, 7.85), was sometimes over 8, and a decrease in infectious particles (87.5%) parallel to the loss of genomes (85%) was observed. Partial degradation of genome RNA has previously been shown (32) by a decrease in the sedimentation coefficient of RNA extracted from virus that had spent 10 days in a mesophile digester. The suggested mechanism was limited proteolysis of the viral capsid with infiltration by ammonium or any other substance capable of degrading RNA. Proof of the role of chemical and biological factors has also been contributed by the work of Spillmann et al. (25), who have shown that a virus as heat resistant as parvovirus (1 h at 56°C) is two or three times more sensitive to mesophile digestion than rotaviruses and coxsackieviruses.
In the months of June, July, September, and November, only genomes could be quantified (9,100 to 24,000 copies/g) in sludge after mesophile anaerobic digestion. They were most likely being protected by a virus capsid. Two possibilities can be considered: the genomes detected belonged to enteroviruses that are difficult to grow in BGM cell culture (echovirus and coxsackievirus A), or virus capsids were partially damaged, making culture impossible but leaving the virus RNA intact, either in its entirety or at least in the 5′ noncoding region. Alteration of the virus capsid when viruses are adsorbed onto solid particles in suspension has been shown by some authors (18, 19, 33, 34). But, as a general rule, the phenomenon of adsorption allows the physical integrity of a virus to be protected from chemical or biological factors present in the digester (5, 13, 24) and therefore to conserve its RNA. Virus adsorption in combination with a minimally raised temperature (35°C) could thus explain the relatively low efficacy of the mesophile anaerobic digestion process (22, 31).
For heat treatment, no microorganism proved capable of resisting 100 min at 195°C at a pressure of 21 bars, and thus theoretically, of the four processes studied, this process clearly has by far the highest decontaminating power.
During this study, the absence of genomes corresponded every time to the absence of infectious virus particles, which agrees with other studies in the literature (7, 11). Nevertheless, one discordance should be pointed out in the results of the composting treatment. The February sample of the sludge-coproduct mixture (Table 2) which contained no enterovirus RNA but showed, on BGM cell culture, an enterovirus viral load of 92 MPNCU/g. It is true that the RNA extract contained PCR inhibitors, but the 1:4 dilution (hence the <200 MPNCU/g limit) yielded a negative result which was validated by correct amplification of the internal control. An enterovirus load of infectious particles cannot, in principle, yield a negative result even when diluted 1:4. Finally, typing of the these viruses made it possible to identify type 1 reoviruses. Confusion of the cytopathic effect of enteroviruses with that of other viruses (reovirus) in cell culture has been reported (20).
This study enabled us to verify the virological decontaminating power of four stabilization processes. Three stabilization methods (liming, composting, and heat treatment) showed complete efficacy in the elimination of enteroviruses (in terms of both genomes and infectious particles). Mesophile anaerobic digestion proved to be the least effective process, with a reduction in enteroviruses of about 1 log in both infectious particle titers and genome titers. The advantage of our sludge testing protocol is that it allows all enteroviruses to be quantified with a sensitivity considerably higher than that of cell culture. We have demonstrated that virological follow-up of stabilization processes is entirely feasible with this technology. However, the infectivity of any genomes quantified by RT-PCR needs to be confirmed by cell culture. Otherwise, a model needs to be developed for correlating infectious particle titers with genome titers if the virological risk inherent in the agricultural use of wastewater sludge is to be fully addressed by RT-PCR.
Acknowledgments
This study was based on work supported by the French Ministry of the Environment, A.D.E.M.E (the French Environment and Energy Management Agency), and Anjou Recherche (Vivendi Water).
REFERENCES
- 1.Aberham, C., C. Pendl, P. Gross, G. Zerlauth, and M. Gessner. 2001. A quantitative, internally controlled real-time PCR Assay for the detection of parvovirus B19 DNA. J. Virol. Methods 92:183-191. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, A. U., and D. L. Sorensen. 1995. Kinetics of pathogen destruction during storage of dewatered biosolids. Water Environ. Res. 67:143-150. [Google Scholar]
- 3.Bertucci, J. J., G. Sullivan, and A. D. Venosa. 1988. Low temperature stability of viruses in sludges. Appl. Environ. Microbiol. 54:839-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa, J. M., P. Ernault, E. Gautier, and S. Bretagne. 2001. Prenatal diagnosis of congenital toxoplasmosis by duplex real-time PCR using fluorescence resonance energy transfer hybridization probes. Prenat. Diagn. 21:85-88. [DOI] [PubMed] [Google Scholar]
- 5.De Flora, S., G. De Renzi, and G. Badolati. 1975. Detection of animal viruses in coastal seawater and sediments. Appl. Microbiol. 30:472-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenhardt, A., E. Lund, and B. Nissen. 1977. The effect of sludge digestion on virus infectivity. Water Res. 11:579-581. [Google Scholar]
- 7.Enriquez, C. E., M. Abbaszadegan, I. L. Pepper, K. J. Richardson, and C. P. Gerba. 1993. Poliovirus detection in water by cell culture and nucleic acid hybridization. Water Res. 27:1113-1118. [Google Scholar]
- 8.Goddard, M. R., J. Bates, and M. Butler. 1981. Recovery of indigenous enteroviruses from raw and digested sewage sludges. Appl. Environ. Microbiol. 42:1023-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gut, M., C. M. Leutenegger, J. B. Huder, N. C. Pedersen, and H. Lutz. 1999. One-tube fluorogenic reverse transcription-polymerase chain reaction for the quantitation of feline coronaviruses. J. Virol. Methods. 77:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurst, C. J. 1989. Fate of viruses during wastewater sludge treatment processes. Crit. Rev. Environ. Control 18:317-343. [Google Scholar]
- 11.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis, G. D., and T. G. Metcalf. 1988. polyethylene glycol precipitation for recovery of pathogenic viruses including hepatitis A and human rotavirus from oyster, water, and sediments. Appl. Environ. Microbiol. 54:1983-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liew, P. F., and C. P. Gerba. 1980. Thermostabilization of enteroviruses by estuarine sediment. Appl. Environ. Microbiol. 40:305-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martell, M., J. Gomez, J. I. Esteban, S. Sauleda, J. Quer, B. Cabot, R. Esteban, and J. Guardia. 1999. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J. Clin. Microbiol. 37:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maul, A. 1991. Aspects statistiques des methodes de quantification en virologie, p. 143-171. In G. Lavoisier (ed.), Virologie des milieux hydriques. Schwartzbrod Tec & Doc, Paris, France.
- 16.Monpoeho, S., A. Dehee, B. Mignotte, L. Schwartzbrod, V. Marechal, J. C. Nicolas, S. Billaudel, and V. Ferre. 2000. Quantification of enterovirus RNA in sludge samples using single tube real-time RT-PCR. BioTechniques 29(1):88-93. [DOI] [PubMed] [Google Scholar]
- 17.Monpoeho, S., A. Maul, C. B. Mignotte, L. Schwartzbrod, S. Billaudel, and V. V. Ferre. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore, R. S., D. H. Taylor, M. M. Reddy, and L. S. Sturman. 1982. Adsorption of reovirus by minerals and soils. Appl. Environ. Microbiol. 44:852-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray, J. P., and S. J. Laband. 1979. Degradation of poliovirus by adsorption on inorganic surfaces. Appl. Environ. Microbiol. 37:480-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muscillo, M., G. La Rosa, C. Marianelli, S. Zaniratti, M. R. Capobianchi, L. Cantiani, and A. Carducci. 2001. A new RT-PCR method for identification of reoviruses in seawater samples. Water Res. 35:548-556. [DOI] [PubMed] [Google Scholar]
- 21.Najioullah, F., D. Thouvenot, and B. Lina. 2001. Development of a real-time PCR procedure including an internal control for the measurement of HCMV viral load. J. Virol. Methods 92:55-64. [DOI] [PubMed] [Google Scholar]
- 22.Sanders, D. A., J. F. Malina, B. E. Moore, B. P. Sagik, and C. A. Sorber. 1979. Fate of poliovirus during anaerobic digestion. J. Water Pollut. Control Fed. 51:333-343. [PubMed] [Google Scholar]
- 23.Sattar, S. A., S. Ramia, and J. C. Westwood. 1976. Calcium hydroxide (lime) and the elimination of human pathogenic viruses from sewage: studies with experimentally-contaminated (poliovirus type 1, Sabin) and pilot plant samples. Can. J. Public Health 67:221-225. [PubMed] [Google Scholar]
- 24.Sobsey, M. D., C. H. Dean, M. E. Knuckles, and R. A. Wagner. 1980. Interactions and survival of enteric viruses in soil materials. Appl. Environ. Microbiol. 40:92-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spillmann, S. K., F. Traub, M. Schwyzer, and R. Wyler. 1987. Inactivation of animal viruses during sewage sludge treatment. Appl. Environ. Microbiol. 53:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Straub, T. M., I. L. Pepper, and C. P. Gerba. 1993. Hazards from pathogenic microorganisms in land-disposed sewage sludge. Rev. Environ. Contam. Toxicol. 132:55-91. [DOI] [PubMed] [Google Scholar]
- 27.Traub, F., S. K. Spillmann, and R. Wyler. 1986. Method for determining virus inactivation during sludge treatment processes. Appl. Environ. Microbiol. 52:498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward, R. L. 1982. Evidence that microorganisms cause inactivation of viruses in activated sludge. Appl. Environ. Microbiol. 43:1221-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward, R. L. 1984. Mechanisms of enteric virus inactivation in treatment processes. Monogr. Virol. 15:175-183. [Google Scholar]
- 30.Ward, R. L., and C. S. Ashley. 1978. Heat inactivation of enteric viruses in dewatered wastewater sludge. Appl. Environ. Microbiol. 36:898-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward, R. L., and C. S. Ashley. 1977. Identification of the virucidal agent in wastewater sludge. Appl. Environ. Microbiol. 33:860-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward, R. L., and C. S. Ashley. 1976. Inactivation of poliovirus in digested sludge. Appl. Environ. Microbiol. 31:921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeager, J. G., and R. T. O'Brien. 1979. Enterovirus inactivation in soil. Appl. Environ. Microbiol. 38:694-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeager, J. G., and R. T. O'Brien. 1979. Structural changes associated with poliovirus inactivation in soil. Appl. Environ. Microbiol. 38:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]