Abstract
A highly enriched culture that reductively dechlorinates trichloroethene (TCE), cis-1,2-dichloroethene (cDCE), and vinyl chloride (VC) to ethene without methanogenesis is described. The Dehalococcoides strain in this enrichment culture had a yield of (5.6 ± 1.4) × 108 16S rRNA gene copies/μmol of Cl− when grown on VC and hydrogen. Unlike the other VC-degrading cultures described in the literature, strains VS and BAV1, this culture maintained the ability to grow on TCE with a yield of (3.6 ± 1.3) × 108 16S rRNA gene copies/μmol of Cl−. The yields on an electron-equivalent basis measured for the culture grown on TCE and on VC were not significantly different, indicating that both substrates supported growth equally well. PCR followed by denaturing gradient gel electrophoresis, cloning, and phylogenetic analyses revealed that this culture contained one Dehalococcoides 16S rRNA gene sequence, designated KB-1/VC, that was identical (over 1,386 bp) to the sequences of previously described organisms FL2 and CBDB1. A second Dehalococcoides sequence found in separate KB-1 enrichment cultures maintained on cDCE, TCE, and tetrachloroethene was no longer present in the VC-H2 enrichment culture. This second Dehalococcoides sequence was identical to that of BAV1. As neither FL2 nor CBDB1 can dechlorinate VC to ethene in a growth-related fashion, it is clear that current 16S rRNA gene-based analyses do not provide sufficient information to distinguish between metabolically diverse members of the Dehalococcoides group.
Vinyl chloride (VC) is a known human carcinogen (ToxFAQs: vinyl chloride, Agency for Toxic Substances and Disease Registry, Centers for Disease Control and Prevention; http://www.atsdr.cdc.gov/toxfaq.html) and a common groundwater pollutant present at over 384 of 1,232 Final National Priority List sites identified by the U.S. Environmental Protection Agency (http://cfpub.epa.gov/supercpad/cursites/srchsites.cfm. It is the highest ranked organic compound on the 2001 Comprehensive Environmental Response, Compensation, and Liability Act Priority List of Hazardous Substances (http://www.atsdr.cdc.gov/01list.html). VC causes angiosarcoma, a rare form of liver cancer, as well as nervous system disorders and immune reactions. The U.S. Environmental Protection Agency has set a maximum contaminant level of 2 μg/liter of drinking water for this chemical (http://www.atsdr.cdc.gov/toxfaq.html). VC is found in the subsurface, due to incomplete degradation of other priority pollutants such as perchloroethene (PCE), trichloroethene (TCE), and 1,1,2-trichloroethane (11, 17; http://www.atsdr.cdc.gov/toxfaq.html). Other sources of VC in groundwater include waste from polyvinyl chloride plastic production. Although VC volatilizes rapidly and is subject to aerobic oxidation both abiotically and biotically (21), VC is persistent in the subsurface at many contaminated sites.
Complete biological reductive dechlorination to ethene is gaining acceptance as a viable remediation method for some chlorinated ethene-contaminated sites (5, 15). However, the organisms that carry out this degradation to completion are, so far, restricted to the Dehalococcoides group of bacteria (10). Two isolates from this group, strains 195 (16) and FL2 (12, 13), degrade VC to ethene cometabolically while metabolically degrading higher chlorinated ethenes. Another isolate from this group, CBDB1, cannot degrade chlorinated ethenes but instead dechlorinates tetrachlorobenzenes and trichlorobenzenes to trichlorobenzenes and dichlorobenzenes (1). Recently, He et al. (9) described BAV1, the first isolated organism proven to gain energy for growth from VC dechlorination to ethene. Growth of Dehalococcoides has been linked to VC dechlorination in only two mixed cultures (2, 8). In both cases, the cultures lost their ability to dechlorinate TCE and PCE as a growth-related process once they were enriched on VC.
The original purpose of the present work was to isolate a VC-dechlorinating organism from the parent KB-1 mixed culture (3). Although not 100% pure, a highly enriched culture was obtained and designated KB-1/VC-H2 because it was enriched on VC and hydrogen. KB-1/VC-H2 was capable of growth-related anaerobic reductive dechlorination of VC and maintained its ability to grow on TCE as well. The Dehalococcoides sequence in the KB-1/VC-H2 enrichment culture, designated KB-1/VC, had a 16S rRNA gene sequence (of over 1,386 bp) identical to those of strains FL2 and CBDB1, despite important metabolic differences. Moreover, the parent TCE-dechlorinating enrichment culture (KB-1) contained two distinct strains of Dehalococcoides, while only one of these strains remained in the VC-H2 enrichment culture.
MATERIALS AND METHODS
Chemicals and analytical procedures.
Chlorinated ethenes, methane, and ethene were analyzed by injecting a 300-μl headspace sample onto a Hewlett-Packard 5890 Series II gas chromatograph fitted with a GSQ column (30-m by 0.53-mm [inner diameter] PLOT column; J&W Scientific) and a flame ionization detector. The oven temperature was programmed to hold at 50°C for 1 min and then to increase to 190°C at 60°C/min and hold at 190°C for 4.6 min. Calibration of cis-1,2-dichloroethene (cDCE), TCE, and PCE was performed with aqueous external standards prepared gravimetrically from a concentrated methanolic stock solution. VC was added to these external standards via a gastight syringe. Chlorinated ethenes were of >97% purity (Sigma-Aldrich). Ethene and methane were calibrated with a 1% gas mixture (Scotty II; Alltech Associates, Inc.). Acetate was measured on a Dionex 300 series ion chromatograph with an IonPac AS14 4-mm column. The eluent was 3.5 mM sodium carbonate and 1 mM sodium bicarbonate at a flow rate of 1.2 ml/min run isocratically.
Culture maintenance.
Four separate enrichment cultures derived from a common PCE- and TCE-dechlorinating parent culture were each maintained with a different chlorinated ethene as described previously (3). The KB-1/PCE, KB-1/TCE, KB-1/cDCE, and KB-1/VC cultures were maintained and analyzed as previously described (3), except that cultures in 250-ml bottles with 200 ml of liquid were amended with 240 to 320 microelectron equivalents (μeeq) of chlorinated ethenes and 1,100 to 1,700 μeeq (0.9 to 1.4 mM) of methanol every 2 weeks. For TCE, this is equal to an aqueous concentration of 225 μM, obtained by adding 4.6 μl of undiluted TCE to the bottle. Electron equivalents are a unit of measure describing the number of electrons accepted or donated by a compound during a given redox reaction. One mole of PCE accepts eight electron equivalents of hydrogen during dechlorination to ethene; during each dechlorination step, two electron equivalents are accepted by each mole of chlorinated ethene. Hence, 1 mol of PCE is the same on an electron equivalent basis as 1.33 mol of TCE, 2 mol of cDCE, and 4 mol of VC. It is convenient to compare data on the basis of moles of chloride (Cl−) released because 1 mol of Cl− corresponds to 2 eeq of substrate degraded, regardless of which chlorinated ethene is used.
Enrichment of KB-1/VC-H2.
One bottle of the KB-1/VC culture was further enriched by 2% (vol/vol) transfers into anaerobic mineral medium after each feeding. These transfer cultures were prepared in 110-ml glass bottles sealed with Mininert caps (VICI Precision Sampling) filled with 90 ml of medium as previously described (4). The amount of VC added to the culture at each feeding increased gradually, while the electron donor amounts were decreased to favor dechlorination over methanogenesis (18). Amounts of VC added increased from 41 to 410 μeeq, and electron donor amounts decreased from 1,500 μeeq of methanol to 410 μeeq of hydrogen as an 80% H2-20% CO2 gas mixture (Praxair). Gases were added from stock bottles via an anaerobic, disposable syringe fitted with a stopcock (Cole-Parmer Instrumentation Co.) and a 0.2-μm Supor Acrodisc syringe filter (Pall Corporation) to prevent contamination.
After the third transfer, the culture was amended with 0.5 mM sodium acetate (BDH) to provide a carbon source for the dechlorinators, 0.25 mM 2-bromoethanesulfonic acid (Sigma) to inhibit methanogens, and 3 g of ampicillin (Sigma)/liter to attempt to inhibit other nondechlorinating species, including some acetogens. The culture was transferred into medium without inhibitors after 2 weeks and then transferred once more into inhibitor-amended medium 2 weeks later. KB-1/VC-H2 has since been maintained by 50% transfer into acetate-amended (but not inhibitor-amended) medium every 3 to 4 months (10 transfers in total) and is amended with 410 μeeq of VC (5 ml of undiluted VC gas) and 330 μeeq of hydrogen (5 ml of 80% H2-20% CO2 gas mixture) at the start of each feeding cycle. As dechlorination proceeds, additional hydrogen is periodically added to the culture (in 330-μeeq doses, usually three or four doses per single VC feeding) to sustain rapid and complete conversion of VC to ethene.
Microscopy.
Five microliters of KB-1/VC-H2 was pipetted into one well of a 12-well Teflon-coated slide (Erie Scientific) and allowed to dry prior to flame fixing. Seven microliters of a DAPI (4′,6′-diamidino-2-phenylindole) (Sigma) solution (2 μg/ml) was added to the well, and the slide was kept in the dark while being air dried for 10 min. Excess DAPI was rinsed off three times with filtered water. The slide was kept refrigerated prior to being viewed with a Leica DMRA2 microscope at a magnification of ×1,000. Three images at slightly different focal lengths were captured with a Qimaging Retiga EX CCD camera and superimposed with Improvision Openlab version 3.1.7 software.
DNA extraction.
DNA was extracted with the UltraClean Soil DNA kit (Mo Bio Laboratories, Inc.), with the following modifications to the manufacturer's instructions. Aliquots of culture (each, 5 to 50 ml) were centrifuged for 40 min at 4°C and 1,900 × g. The supernatant was decanted, leaving approximately 1 ml of liquid in which the pelleted cells were resuspended and then transferred to the bead tubes. At the end of the extraction process, DNA was eluted from the spin filters with 50 μl of sterile water instead of the kit's elution buffer. The reproducibility and linearity of this DNA extraction method was evaluated by extracting replicate samples and different sample volumes ranging from 0.5 to 50 ml from the same culture.
PCR-denaturing gradient gel electrophoresis (PCR-DGGE).
Dehalococcoides-specific 16S rRNA gene fragments were PCR amplified with the 1f-GC and 259r primer sets listed in Table 1. Each 100 μl of PCR mixture contained the following: 1× PCR buffer, 200 nmol of MgCl2, 30 nmol of deoxynucleoside triphosphate, 25 pmol of each primer, 2.5 U of Taq polymerase, and approximately 100 pg of template DNA. The touchdown thermocycling program was as follows: initial denaturation for 5 min at 94°C; 20 cycles of 94°C for 1 min, annealing for 1 min, and 72°C for 2 min (with the annealing temperature decreasing from 69 to 59°C at −0.5°C/cycle); an additional 15 cycles with annealing at 59°C; and a 5-min final extension at 72°C. The success of the PCR was verified with agarose gel electrophoresis prior to DGGE. DGGE was performed with the DCode mutation detection system (Bio-Rad). PCR-amplified fragments were electrophoresed on an 8% polyacrylamide gel with a 30 to 60% urea-formamide gradient for 6 h at 160 V and 60°C.
TABLE 1.
Primer sequences used in this study
| Name | Sequence | Specificitya | Reference or source |
|---|---|---|---|
| Fp DHC 1b | 5′-GAT GAA CGC TAG CGG CG-3′ | Dehalococcoides, Flexibacter, Microscilla | 10 |
| 1f-GCb | 5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GGA TGA ACG CTA GCG GCG-3′ | Same as Fp DHC 1 | This study |
| 259rb | 5′-CAG ACC AGC TAC CGA TCG AA-3′ | Dehalococcoides, Rubritepida, Defluvicoccus | Hendrickson, personal communication |
| 341f-GC | 5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG-3′ | Bacteria | 19 |
| 534r | 5′-ATT ACC GCG GCT GCT GG-3′ | Bacteria | 19 |
| 582f | 5′-CTG TTG GAC TAG AGT ACA GC-3′ | Dehalococcoides | This study |
| 582r | 5′-GCT GTA CTC TAG TCC AAC AG-3′ | Same as 582f | This study |
| 728rc | 5′-GTG ACA ACC TAG AAA ACC GCC TT-3′ | Dehalococcoides | 13 |
| 962r | 5′-TCC TGA CTT AAG AGG TCG TT-3′ | Dehalococcoides | This study |
| 1386rb | 5′-CCT CCT TGC GGT TGG CAC ATC-3′ | Dehalococcoides | Hendrickson |
| T7 | 5′-TAA TAC GAC CTA TAG GG-3′ | Cloning vectors, viruses | Invitrogend |
| M13 Reverse | 5′-CAG GAA ACA GCT ATG AC-3′ | Cloning vectors, viruses | Invitrogend |
Specificity indicates a 100% match in GenBank (not including unidentified clones) as of April 28, 2004.
Sequences subject to E. I. Du Pont de Nemours & Co.'s patent application with exclusive licence to GeoSyntec Consultants, Ltd.
728r is the reverse complement of 728f as listed in reference 13.
Invitrogen, TOPO TA cloning manual.
General bacterial PCR-DGGE was conducted similarly to the Dehalococcoides-specific PCR-DGGE procedure described above, except that the primers 341f-GC and 534r (19) were used. The annealing temperature for the PCR decreased from 65 to 55°C at −0.5°C/cycle and was kept constant at 55°C for the last 15 cycles.
Cloning and sequencing.
Dehalococcoides-specific 16S rRNA genes were PCR amplified with the primers Fp DHC 1 and 1386r (Table 1). PCR conditions were similar to those described above but with a constant annealing temperature of 52°C and a total of 30 cycles. Products were cloned with the TOPO TA cloning kit (TOPO TA cloning manual, version K 25-0184; Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Both forward and reverse sequences were determined to obtain at least triple coverage in all regions with the primers Fp DHC 1, 1386r, 582f, 582r, 728r, 962r, T7, and M13 Reverse (Table 1).
TCE degradation experiment.
Three treatments were examined in this experiment: the KB-1/VC-H2 culture amended with VC (positive control), KB-1/VC-H2 amended with TCE, and an uninoculated treatment (negative control). Treatments were set up in triplicate in 110-ml Mininert-capped bottles containing 40 ml of medium and 10 ml of culture. Inoculated bottles were amended with 68 μeeq of chlorinated ethene (0.83 ml of undiluted VC gas or 1 μl of undiluted TCE, both of >98% purity [Sigma-Aldrich]) and five times the electron equivalents of hydrogen (340 μeeq or 5.2 ml of gas mixture). Negative controls were amended with 68 μeeq each of VC and TCE, as well as 340 μeeq of hydrogen. Medium, chlorinated ethenes, and hydrogen were added 3 days prior to the first gas chromatography sampling and 5 days prior to inoculation (20% [vol/vol] inoculum) to ensure the precision of initial measurements. Chlorinated ethenes were respiked when degradation was complete.
VC yield determination.
Four bottles were set up with 0.5 mM acetate-amended medium with 20 to 30 μeeq of VC and 100 μeeq of H2. After being shaken vigorously and after 1 h of equilibration time, a 5% (vol/vol) inoculum was added and time zero gas chromatography measurements were taken. Ten milliliters of culture was removed for DNA extraction as described above. Samples were monitored by gas chromatography every 3 to 4 days. After the VC was consumed, another 10 to 50 ml of culture was removed for DNA extraction. Quantitative PCR was conducted with an Opticon 2 (MJ Research) and the DyNAmo SYBR Green kit (MJ Research) with the primers Fp DHC 1 and 259r. Each 50-μl reaction mixture contained 25 μl of the DyNAmo SYBR Green mixture, 19 μl of sterile water, 25 pmol of each primer, and 4 μl of DNA template. The thermocycling program was as follows: initial denaturation for 10 min at 94°C; 45 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 30 s; and a final melting curve analysis from 72 to 95°C, measuring fluorescence every 0.5°C. Calibration was performed with serial dilutions of a known quantity of one of the Dehalococcoides 16S rRNA gene-containing plasmids generated in the cloning study described above.
TCE yield determination.
Three treatments were examined in this experiment: KB-1/VC-H2 amended with VC, KB-1/VC-H2 amended with TCE, and uninioculated negative controls containing both VC and TCE. Treatments were set up in triplicate in 42-ml screwcap vials with Mininert caps. Nineteen milliliters of 0.5 mM acetate-amended medium was added to each vial before the headspace was purged for 5 min with 80% H2-20% CO2. After purging was complete, 30 μeeq of chlorinated ethenes was added to each vial. After vigorous shaking and overnight equilibration were carried out, initial gas chromatography measurements were taken and 1 ml of inoculum (5% [vol/vol]) was added to each of the VC and TCE vials. Five milliliters of the inoculum was used for DNA extraction. Gas chromatography measurements were taken every 3 to 4 days thereafter until more than 85% of the initial electron equivalents of chlorinated ethene had been converted to ethene. At this time, 15 ml of culture was removed from each vial for DNA extraction as described above. A second sample of 1.2 ml was used for cell counts with microscopy. Cell counts were performed with serial dilutions of culture stained with DAPI and filtered onto 0.2-μm-pore-size Nuclepore membranes (Whatman). Membranes were rinsed with water to remove excess DAPI and dried prior to being viewed under the microscope at a magnification of ×1,000 as described earlier. Quantitative PCR on both the inoculum and final DNA templates was conducted as described above but with the SYBR Green JumpStart kit (Sigma) instead of the DyNAmo kit. Each 40 μl of reaction mixture contained 20 μl of the kit's SYBR Green JumpStart Taq ReadyMix, 15.6 μl of sterile water, 20 pmol of each primer, and 3 μl of DNA template.
Nucleotide sequence accession numbers.
The sequences for KB-1/VC (AY146779) and KB-1/PCE (AY146780) were submitted to GenBank and compared with those available (www.ncbi.nlm.nih.gov) as of November 2003.
RESULTS
Characteristics of the highly enriched KB-1/VC-H2 culture.
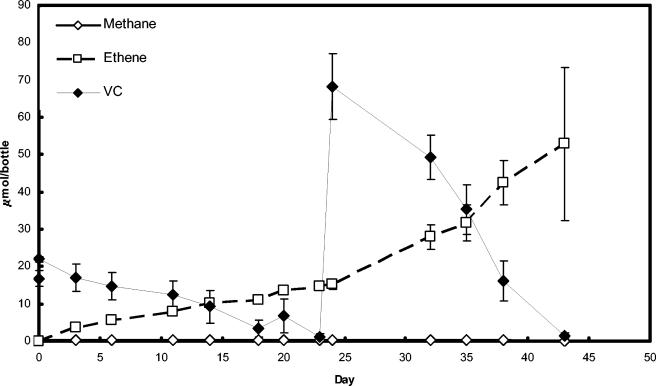
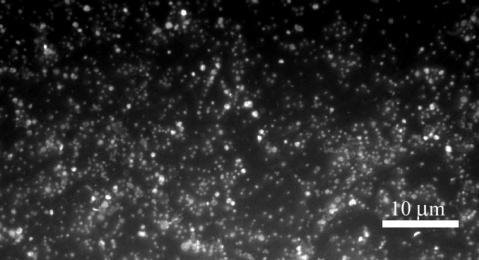
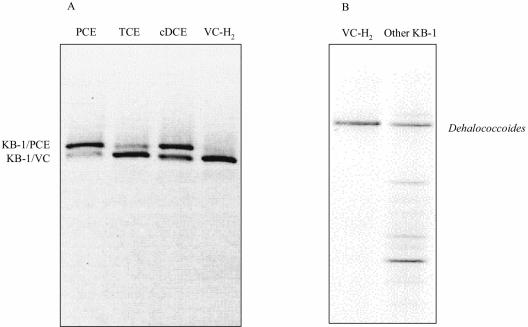
VC dechlorination proceeded in the absence of methanogenesis and at increasing rates in a 20% (vol/vol) transfer of KB-1/VC-H2 (Fig. 1). Methanogenesis present in the parent KB-1 cultures was eliminated through a combination of 2% (vol/vol) transfers, amendment with 2-bromoethanesulfonic acid, higher VC concentrations, and lower hydrogen concentrations. While the highly enriched culture still produced acetate (data not shown) and therefore comprised some acetogens, over 97% of the cells viewed under the microscope were small, coccoid bacteria of a size similar to other published Dehalococcoides spp. (Fig. 2). Further evidence of culture purity was provided by general bacterial DGGE, in which the only band visible on the gel was that of Dehalococcoides species (Fig. 3B).
FIG. 1.
VC dechlorination in 20% (vol/vol) transfer cultures of KB-1/VC-H2. Note that 20 μmol of VC/bottle corresponded to 190 μM (aqueous) VC. Error bars are standard deviations of results for three replicate cultures.
FIG.2.
The KB-1/VC-H2 culture, DAPI stained and viewed at an original magnification of ×1,000.
FIG. 3.
(A) PCR-DGGE targeting Dehalococcoides in four KB-1 enrichment cultures. (B) General bacterial PCR-DGGE, illustrating that the KB-1/VC-H2 culture (left lane) had lost all but the Dehalococcoides band compared to the mixed KB-1 culture (right lane).
Dehalococcoides-specific PCR-DGGE.
A PCR-DGGE method was developed that distinguished different Dehalococcoides sequences in a mixed culture based on sequence differences in the region amplified (positions 1 to 259). Up to two distinct bands corresponding to Dehalococcoides were detected in the four KB-1 enrichment cultures maintained on different chlorinated ethenes (Fig. 3A). Lanes corresponding to the PCE, TCE, and cDCE enrichment cultures each showed two Dehalococcoides bands, while the lane corresponding to the VC-H2 culture showed only the lower band. This result was consistently observed in at least four separate DGGE analyses of over 20 different KB-1 enrichment cultures. Because band separation is based on sequence differences, each band should theoretically correspond to a distinct sequence. To determine if the DGGE patterns observed corresponded to true sequence differences or to PCR or DGGE artifacts, the near-full-length Dehalococcoides 16S rRNA gene sequences (positions 1 to 1386) in the various enrichment cultures were amplified, cloned, and sequenced. Sequencing of clones and subsequent analysis of the cloned DNA by the same DGGE method revealed that the upper band, designated KB-1/PCE (GenBank accession number AY146780), corresponded to a 16S rRNA gene sequence identical to that of BAV1 (9). The lower band, designated KB-1/VC (GenBank accession number AY146779), corresponded to a sequence identical to FL2 (13) and CBDB1 (1). The difference between these two sequences was just one base pair substitution (G/A) at position 148. Of 25 clones analyzed from the KB-1/VC-H2 enrichment culture, none matched the upper band. In contrast, of more than 10 clones analyzed from each of the cDCE, TCE, and PCE enrichment cultures, at least 25% of the clones displayed the upper band, with the remaining corresponding to the lower band (data not shown).
TCE dechlorination by KB-1/VC-H2.
When the DGGE results showed that the KB-1/VC-H2 enrichment culture had lost a Dehalococcoides phylotype, we sought to discover if this culture had also lost its capacity to degrade TCE. Previous experiments had shown that the KB-1/VC enrichment culture had lost the ability to degrade PCE (3). However, the KB-1/VC-H2 enrichment culture maintained its ability to degrade TCE over several feedings. Hence, the loss of the upper band seemed to correspond to the loss of an organism capable of PCE degradation, while the remaining lower-band organism degraded TCE to ethene. TCE was degraded to ethene rapidly without accumulation of intermediates and without methanogenesis. Initial rates of VC and TCE degradation by KB-1/VC-H2 were similar on an electron equivalent basis (Table 2). Corresponding data for a typical KB-1/TCE enrichment culture (containing two Dehalococcoides phylotypes) are shown for comparison. In the KB-1/TCE culture, the rate of TCE degradation was faster than the rate of VC degradation (Table 2); as a result, unlike in the KB-1/VC culture, cDCE and VC accumulated before being reduced to ethene. No degradation in uninoculated controls was observed.
TABLE 2.
Daily rates of TCE and VC dechlorination by KB-1/VC-H2 and KB-1/TCE cultures
| Substrate | Rate of degradation by:
|
|||
|---|---|---|---|---|
| KB-1/VC-H2a | KB-1/TCEa | KB-1/VC-H2b | KB-1/TCEb | |
| VC | 86 ± 16 | 74 ± 8 | 43 ± 8 | 37 ± 4 |
| TCE | 78 ± 10 | 120 ± 12 | 13 ± 2 | 60 ± 6 |
Values are in microelectron equivalents of substrate per liter of culture.
Values are in micromoles of substrate per liter of culture.
Yield on VC.
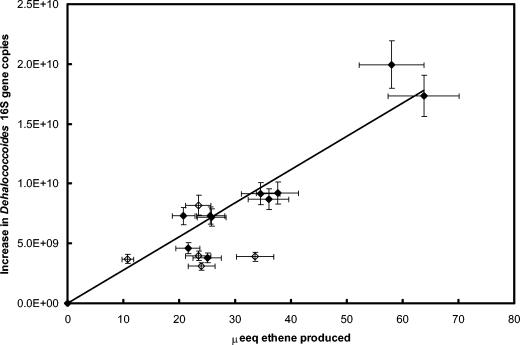
The growth yield on VC of the Dehalococcoides strain present in the KB-1/VC-H2 culture was (5.6 ± 1.4) × 108 16S rRNA gene copies/μmol of ethene produced (10 experiments), calculated from the slope of the line shown in Fig. 4. As shown in Table 3, this yield agreed closely with the value obtained by Cupples et al. (2), although it was an order of magnitude higher than that obtained by He et al. (9). To try to reconcile these different yield values, bioenergetic calculations were performed to obtain a theoretical maximum yield (20). Theoretical maximum cell yields can be estimated from the free energy released from H2 oxidation coupled to VC reduction to ethene. These calculations resulted in an estimate of the maximum yield equal to 0.25 eeq of cells/eeq of H2. This theoretical yield corresponds to 2.8 g of dry cells/mol of H2, assuming 113 g of cells/20 eeq of cells, and 2 eeq/mol of H2 (20).
FIG. 4.
Increase in Dehalococcoides 16S rRNA gene copy number during VC and TCE degradation. Solid diamonds correspond to VC degradation, and open diamonds correspond to TCE degradation. The x axis error bars represent the 10% uncertainty in gas chromatography measurements. The y axis error bars represent the error in quantitative PCR measurements, estimated to be 10%, based on analysis of replicate samples.
TABLE 3.
Comparison of yields of Dehalococcoides grown on VC
| Strain | Yield
|
Reference or source | ||
|---|---|---|---|---|
| 16S gene copies/ μmol Cl−a | g (dry wt)/ mol of Cl− | |||
| VS | (5.2 ± 1.5) × 108 | 2.2 ± 0.6c | 2 | |
| BAV1 | (6.3 ± 0.3) × 107 | 0.27 ± 0.01c; 0.48 ± 0.05d | 9 | |
| KB-1/VC | (5.6 ± 1.4) × 108 | 2.4 ± 0.6c | This study | |
| Theoreticalb | 2.8 | This study | ||
For VC dechlorination, moles of ethene (C2H4) produced are equal to the moles of Cl− produced and the moles of VC (C2H3Cl) degraded because of the following stoichiometry: C2H3Cl + H2→C2H4 + H+ + Cl−.
See text for bioenergetics assumptions and calculations.
Calculated from quantitative PCR data with conversion factor (see text).
Measured as protein; converted to dry weight of cells by assuming 0.5 g of protein/g (dry weight) of cells.
To compare yields reported in grams of protein or grams of cells per mole of substrate to yields measured by quantitative PCR (16S rRNA gene copies/mole of substrate), a conversion factor of 4.2 × 10−15 g of cells/copy was developed, based on the characteristics of Dehalococcoides strains 195 and BAV1. We chose not to use the factor of 1.6 × 10−14 g of cells/copy calculated by Cupples (2) because electron micrographs of strains 195 and BAV1 show that Dehalococcoides is more disk shaped than spherical (9, 16), with a diameter of approximately 0.5 μm and a thickness of approximately 0.1 μm. Assuming that this shape is present, that cell density is equal to that of water, and that a cell is 80% water, a conversion factor of 4.2 × 10−15 g (dry weight) per cell was calculated. This is equivalent to 4.2 × 10−15 g of cells/16S rRNA gene copy, assuming that there was only one copy of the 16S rRNA gene per genome, as was shown for strain 195 (www.tigr.org). The yield for BAV1 calculated from quantitative PCR data with the conversion factor of 4.2 × 10−15 g of cells/copy, assuming 0.5 g of protein/g of cells, was relatively close to the measured protein yield previously reported (9), supporting the use of these conversion factors for Dehalococcoides cells (Table 3).
Yield on TCE and DNA extraction efficiency.
The growth yield on TCE of the Dehalococcoides strain present in the KB-1/VC-H2 culture was determined from quantitative PCR data to be (3.6 ± 1.3) × 108 16S rRNA gene copies/μmol Cl− produced (five experiments). Therefore, on an electron equivalent or Cl− basis, the yield on TCE was not significantly different from the yield on VC, indicating that this organism gained energy for growth from all dechlorination steps from TCE to ethene. Cell counts were also measured to corroborate the increase in cell number measured by quantitative PCR. In addition, cell counts were used to estimate the DNA extraction efficiency to validate the quantitative PCR approach for determining cell yield. Cell count data were much more variable than the quantitative PCR data because of the difficulty in distinguishing and detecting such small cells under the microscope and because of the tedious nature of the measurements. Nevertheless, an increase in cell counts was generally observed in proportion to chlorinated ethene degradation (data not shown). Given that there are 1.486 × 106 bp per Dehalococcoides genome (www.tigr.org) and 1.1 × 10−12 ng of DNA/bp ([660 g of bp/mol]/6.02 × 1023), the maximum DNA yield is 1.6 × 10−6 ng of DNA/cell (note that this DNA mass per cell is a significant 38% of the estimated total dry weight per cell of 4.2 × 10−6 ng calculated above; see Discussion). In our experiments with KB-1/VC-H2, the ratio of the amount of DNA extracted (nanograms per milliliter of culture) to measured cell counts (cells per milliliter of culture) averaged 78% and varied considerably from 43 to 120% of the maximum yield of 1.6 × 10−6 ng of DNA per cell. However, the variability in these estimates of DNA extraction efficiency was largely due to variability in cell count measurements and not to the amount of DNA extracted. The DNA extraction procedure itself was highly reproducible (<10% variability in nanograms of DNA per milliliter of culture for replicate samples), and the total amount of DNA recovered was proportional to the volume of culture extracted for a given culture. Thus, the true DNA extraction efficiency was relatively constant between samples and, based on the comparisons with cell counts, likely high enough (∼78%) not to be a significant source of error in this study. Moreover, cell yields were more easily and reliably obtained from quantitative PCR data than from cell count data, because the former were more reproducible, accurate, and faster to obtain.
DISCUSSION
The KB-1/VC-H2 culture has thrived for over 3 years in defined mineral medium. This enrichment culture dechlorinates VC to ethene stoichiometrically and without methanogenesis in the presence of hydrogen as an electron donor and with acetate as a carbon source. As expected, neither 2-bromoethanesulfonic acid nor ampicillin affected dechlorination rates at the concentrations used in this study. 2-Bromoethanesulfonic acid is a structural analog to coenzyme M, the cofactor required for the terminal step in methanogenesis, and thereby inhibits this process (7). Ampicillin inhibits peptidoglycan cross-linking in many bacteria (14), but perhaps due to Dehalococcoides' unique cell wall structure (16), ampicillin had no impact on dechlorination rates.
The >97%-pure Dehalococcoides KB-1/VC-H2 enrichment culture maintained its ability to degrade TCE and VC at similar rates (on an electron equivalent basis) over many feedings (Table 2). The KB-1/TCE rate data (Table 2) were obtained from a mixed culture containing two Dehalococcoides phylotypes and had higher cell numbers than KB-1/VC-H2, so rates cannot be directly compared between cultures. In the KB-1/TCE culture, the higher relative rates of TCE compared to those of VC dechlorination on an electron equivalent basis may reflect a greater contribution to TCE dechlorination from the second phylotype in the culture. Comparing relative rates of VC and TCE degradation on a molar basis within each culture showed that the KB-1/VC-H2 enrichment degraded VC more rapidly than TCE; therefore, during TCE degradation there was no accumulation of intermediates. In contrast, the KB-1/TCE culture degraded TCE more rapidly than VC, and hence cDCE and VC accumulated transiently during dechlorination. KB-1/VC-H2 was thus more specialized towards VC degradation than KB-1/TCE.
To establish that the single Dehalococcoides phylotype in KB-1/VC-H2 was capable of growth on TCE, Dehalococcoides copy numbers were measured over a single feeding. The cell yield per mole of chloride released (i.e., on an electron equivalent basis) was similar regardless of whether TCE or VC was the electron acceptor, indicating that both substrates supported growth equally well. The ability of this VC enrichment culture to grow on TCE distinguishes it from other anaerobic VC-dechlorinating cultures in the literature (2, 8). The measured yield on VC of (5.6 ± 1.4) × 108 16S rRNA gene copies/μmol of ethene was estimated to correspond to a yield of 2.4 ± 0.6 g (dry weight) of cells/mol of Cl− produced, which is very similar to the value of 2.2 ± 0.3 g of cells/mol of Cl− obtained by Cupples et al. (2) for strain VS and the theoretical maximum yield of 2.8 g of cells/mol of Cl− obtained with thermodynamic calculations. However, these yields are nearly an order of magnitude higher than the value obtained by He et al. (9) for BAV1. This disparity in yield may reflect intrinsic differences in culture or may be the result of systematic biases in different measurement methods.
Relatively simple calculations of mass per unit of cells estimated from observed cell dimensions and DNA content per cell based on genome size revealed that DNA may comprise roughly 38% of the mass of Dehalococcoides cells. This is significantly higher than 3%, the approximate proportion of DNA in a typical bacterium (14), but not surprising because in larger cells the genome is only two to four times larger than that of Dehalococcoides, while the cell volume is over 100 times larger. The implication for Dehalococcoides cells is that mass proportions of proteins, ribosomes, and other major macromolecules in these tiny cells may be significantly different from those reported for larger cells, such as Escherichia coli.
The KB-1/VC-H2 culture lost its ability to degrade PCE, unlike the other KB-1 enrichment cultures. This metabolic difference appears to have arisen from differences on a community structure level. DGGE with Dehalococcoides-specific primers showed that KB-1 mixed cultures maintained on PCE, TCE, and cDCE all contained two Dehalococcoides sequences, while KB-1/VC-H2 contained only one (Fig. 3A). While many factors prevent the theoretical association of one band per species or strain from holding true (e.g., polymerase transcription errors, heteroduplex formation, multiple 16S rRNA genes per genome, different organisms with identical 16S rRNA genes, etc.), our results strongly suggest that KB-1/PCE, KB-1/TCE, and KB-1/cDCE all contained at least two Dehalococcoides strains. First, whole-genome sequencing of Dehalococcoides ethenogenes strain 195 revealed only one copy of the 16S rRNA gene in the genome (www.tigr.org). Second, the pattern observed in the results shown in Fig. 3A has been observed in DNA extracted from over 20 separate bottles of KB-1. Third, the sequence difference between the upper and lower bands of Fig. 3A—a single base pair substitution at position 148 of the 16S rRNA gene sequence—has been identified in clones from both the KB-1/PCE and KB-1/TCE cultures. No clones from the VC enrichment culture showed the upper-band sequence (KB-1/PCE), while at least 25% of the clones from each of the other KB-1 enrichment cultures did contain this sequence. The organism corresponding to the KB-1/PCE sequence was likely responsible for PCE degradation to TCE and possibly beyond, while the KB-1/VC sequence seems to correspond to an organism capable of TCE-to-ethene degradation but incapable of PCE degradation.
Hendrickson et al. (10) classified Dehalococcoides into three groups: Cornell, Pinellas and Victoria. As part of their study, Hendrickson et al. analyzed KB-1 DNA extracted in 2000 and found two distinct sequences, one in the Cornell group (strain DHC-kb1C; GenBank accession number AF388539) and one in the Pinellas group (strain DHC-kb1P; GenBank accession number AF388540). In 2002, DGGE evidence supporting the presence of up to three Dehalococcoides sequences in the KB-1 enrichment cultures was reported (3). Here, we report two sequences, both from the Pinellas group: one which corresponds to Hendrickson's DHC-kb1P (KB-1/VC) (Fig. 3A, lower band), and the other which differs from DHC-kb1P by one base pair (KB-1/PCE) (Fig. 3A, upper band). We attribute the loss over time of the DHC-kb1C sequence identified by Hendrickson et al. to culture enrichment. Transfers, encouragement of complete VC degradation in all cultures prior to refeeding, and amendment with lower levels of electron donor seem to have selected for strains in the Pinellas group as opposed to the Cornell group of Dehalococcoides.
Phylogenetic analysis of the near-full-length (1,386 bp) KB-1/VC 16S rRNA gene sequence revealed a 100% match with the published sequences for strains CBDB1 and FL2 and several uncultured Dehalococcoides strains, and a 1-bp difference in sequence from strain BAV1. However, important metabolic differences exist between these organisms: chlorobenzene-degrading strain CBDB1 did not dechlorinate chlorinated ethenes (1), strain FL2 degraded VC only cometabolically (12), and strain BAV1 is reported to grow on all dichloroethenes and VC, but not on TCE (9). Our strain of Dehalococcoides grew on both TCE and VC.
The diversity exhibited by such closely related organisms presents challenges for bioremediation. 16S rRNA gene-based PCR techniques for tracking Dehalococcoides at contaminated sites are gaining acceptance (6, 10, 15), but these techniques may not offer enough resolution to show whether or not the organism(s) detected at a site is capable of degrading the contaminants present. The ability to track specific Dehalococcoides species by molecular approaches offering greater strain resolution would be particularly useful. The PCR-DGGE method described herein may be one such approach, although its resolution remained somewhat limited because it still targeted the 16S rRNA gene. Sequences with single-base-pair differences electrophoresed in reproducible, distinct positions; such differences might otherwise have been dismissed as PCR errors if analyzed by sequencing alone. In this study, the PCR-DGGE technique clearly revealed differences between cultures that could or could not dechlorinate PCE. The resolution of the technique depends entirely on the choice of primers; obviously, differences outside of the region amplified by the primers would not be detected by this technique. To improve resolution, future application of this method should focus on areas of the Dehalococcoides genome offering greater resolution than the 16S rRNA gene, such as dehalogenase genes or other conserved genes that could be used as phylogenetic or functional markers.
Acknowledgments
We thank Edwin Hendrickson (E. I. Du Pont de Nemours & Co.) for designing and sharing the Dehalococcoides primers used in this work, David Major and Sandra Dworatzek (GeoSyntec Consultants, Ltd.) for invaluable comments and discussion, and Cheryl Washer and Sasha Necakov (University of Toronto) for their microscopy expertise. Sequencing was performed by Ivana Miljanic at the Ontario Cancer Institute.
Funding for this research was provided by the Natural Sciences and Engineering Research Council of Canada and GeoSyntec Consultants, Ltd.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Gorlsch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 2.Cupples, A. M., A. M. Spormann, and P. L. McCarty. 2003. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl. Environ. Microbiol. 69:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duhamel, M., S. Wehr, L. Y. Yu, H. Rizvi, D. Seepersad, S. Dworatzek, E. E. Cox, and E. A. Edwards. 2002. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-1,2-dichloroethene and vinyl chloride. Water Res. 36:4193-4202. [DOI] [PubMed] [Google Scholar]
- 4.Edwards, E. A., and D. Grbić-Galić. 1994. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl. Environ. Microbiol. 60:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis, D. E., E. J. Lutz, J. M. Odom, R. J. J. Buchanan, C. L. Bartlett, M. D. Lee, M. R. Harkness, and K. A. DeWeerd. 2000. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ. Sci. Technol. 34:2254-2260. [Google Scholar]
- 6.Fennell, D. E., A. B. Carroll, J. M. Gossett, and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 7.Gunsalus, R. P., J. A. Romesser, and R. S. Wolfe. 1978. Preparation of coenzyme M analogues and their activity in the methyl coenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochemistry 17:2374-2377. [DOI] [PubMed] [Google Scholar]
- 8.He, J., K. M. Ritalahti, M. R. Aiello, and F. E. Loffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, J., K. M. Ritalahti, K.-L. Yang, S. S. Koenigsberg, and F. E. Loffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 10.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunkeler, D., R. Aravena, and E. Cox. 2002. Carbon isotopes as a tool to evaluate the origin and fate of vinyl chloride: laboratory experiments and modeling of isotope evolution. Environ. Sci. Technol. 36:3378-3384. [DOI] [PubMed] [Google Scholar]
- 12.Loffler, F. E., J. R. Cole, K. M. Ritalahti, and J. M. Tiedje. 2003. Diversity of dechlorinating bacteria. In M. M. Haggblom and I. D. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Academic Publishers, Boston, Mass.
- 13.Loffler, F. E., Q. Sun, J. R. Li, and J. M. Tiedje. 2000. 16S rRNA gene-based detection of tetrachloroethene dechlorinating Desulfuromonas and Dehalococcoides species. Appl. Environ. Microbiol. 66:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. Brock biology of microorganisms, 9th ed. Prentice-Hall, Inc., Upper Saddle River, N.J.
- 15.Major, D. W., M. L. McMaster, E. E. Cox, E. A. Edwards, S. M. Dworatzek, E. R. Hendrickson, M. G. Starr, J. A. Payne, and L. W. Buonamici. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 36:5106-5116. [DOI] [PubMed] [Google Scholar]
- 16.Maymo-Gatell, X., Y.-T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 17.McCarty, P. L. 1994. An overview of anaerobic transformation of chlorinated solvents, p. 135-142. In EPA Symposium on Intrinsic Bioremediation of Ground Water. Office of Research and Development, U.S. Environmental Protection Agency, Washington, D.C.
- 18.McCarty, P. L., and Y. Yang. 1998. Competition for hydrogen within a chlorinated solvent dehalogenating anaerobic mixed culture. Environ. Sci. Technol. 32:3591-3597. [Google Scholar]
- 19.Muyzer, G., E. C. D. Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rittmann, B. E., and P. L. McCarty. 2001. Environmental biotechnology: principles and applications. McGraw-Hill, Inc., New York, N.Y.
- 21.Verce, M. F., R. L. Ulrich, and D. L. Freedman. 2000. Characterization of an isolate that uses vinyl chloride as a growth substrate under aerobic conditions. Appl. Environ. Microbiol. 66:3535-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]