Abstract
Sigma B (σB) is a stress-responsive alternative sigma factor that has been identified in various gram-positive bacteria. Seven different regulators of sigma B (Rsbs) are located in the sigB operons of both Bacillus subtilis and Listeria monocytogenes. In B. subtilis, these proteins contribute to regulation of σB activity by conveying environmental and energy stress signals through two well-established branches of a signal transduction pathway. RsbT contributes to regulation of σB activity in response to environmental stresses, while RsbV contributes to σB activation under both environmental and energy stresses in B. subtilis. To probe L. monocytogenes Rsb roles in σB-mediated responses to various stresses, in-frame deletions were created in rsbT and rsbV. Phenotypic characterization of the L. monocytogenes rsbT and rsbV null mutants revealed that both mutants were similar to the ΔsigB strain in their abilities to survive under environmental stress conditions (exposure to synthetic gastric fluid, pH 2.5, acidified brain heart infusion broth [BHI], or oxidative stress [13 mM cumene hydroperoxide]). Under energy stress conditions (carbon starvation in defined media, entry into stationary phase, or reduced intracellular ATP), both ΔrsbT and ΔrsbV showed survival reductions similar to that of the ΔsigB strain. These observations suggest that the pathways for Rsb-dependent regulation of σB activity differ between L. monocytogenes and B. subtilis. As σB also activates transcription of the L. monocytogenes prfAP2 promoter, we evaluated virulence-associated characteristics of ΔprfAP1rsbT and ΔprfAP1rsbV double mutants in hemolysis and tissue culture assays. Both double mutants showed identical phenotypes to ΔprfAP1P2 and ΔprfAP1sigB double mutants, i.e., reduced hemolysis activity and reduced plaque size in mouse fibroblast cells. These findings indicate that RsbT and RsbV both contribute to σB activation in L. monocytogenes during exposure to environmental and energy stresses as well as during tissue culture infection.
Listeria monocytogenes, a gram-positive, non-spore-forming rod-shaped bacterium, is recognized as a foodborne pathogen. This organism is capable of surviving in a broad range of ecological niches (e.g., in farm environments and food processing plants) and in a wide range of hosts, including humans and many species of animals. In L. monocytogenes, the alternative sigma factor B (σB) contributes to survival under stressful environmental conditions, such as exposure to low pH, oxidizing conditions, and starvation (17, 18). Loss of σB also reduces L. monocytogenes virulence in a murine model (32, 44).
The sigB gene, which encodes σB, lies seventh in the sigB operon. This operon also includes seven additional genes, which encode the following regulator of sigma B proteins: RsbR, RsbS, RsbT, RsbU, RsbV, RsbW, and RsbX (4, 19, 25, 44, 45). In B. subtilis, activation of σB by the Rsb proteins is achieved through a complex phosphorylation/dephosphorylation cascade in response to various cellular stimuli, which have been categorized into two general types: environmental or metabolic (1, 6, 7, 9, 15, 21, 26, 27, 29, 42, 43, 46). In Bacillus subtilis, these two types of cellular stimuli are conveyed to σB through two interconnected but separate pathways. The environmental stimulus pathway is transmitted by regulatory proteins encoded in the sigB operon. The “metabolic” or energy stimulus pathway is signaled by proteins encoded in a two-gene operon (rsbQ-rsbP) that is physically distant from the sigB operon (9, 41). The presence of this rsbQ-rsbP operon is not evident in L. monocytogenes. The two primary regulators of B. subtilis σB activity are RsbV and RsbW. Under exponential growth conditions, RsbW, an anti-σ factor, binds directly to σB and blocks association between σB and RNA polymerase. RsbV is inactive as an anti-anti-σ factor when it has been phosphorylated on a conserved serine residue by the kinase activity of RsbW. However, B. subtilis RsbV is dephosphorylated by the phosphatase activity of RsbU or of RsbP under conditions of environmental (43) or energy (41) stress, respectively. The phosphatase activity of RsbU is activated upon protein-protein interaction with serine kinase RsbT (27) and that of RsbP by α/β hydrolase RsbQ (9). After dephosphorylation, RsbV binds to RsbW, thus freeing σB, which then becomes available to bind RNA polymerase core enzyme. In summary, the phosphorylation status of RsbV determines whether σB is bound to RsbW or is free to interact with core polymerase (16).
Expression of the majority of recognized L. monocytogenes virulence genes is regulated by positive regulatory factor A (PrfA). PrfA regulates expression of a set of virulence factors, including listeriolysin O (LLO), actin polymerization protein ActA, phospholipases (PlcA and PlcB), and internalins (30, 40). Transcription of prfA is initiated from three promoters: prfAP1, prfAP2, and a promoter upstream of plcA. In vivo, loss of either prfAP1 or -P2 appears to be compensated for by the remaining promoter; loss of both prfA promoters results in virulence attenuation (20). Our group has shown that the prfAP2 promoter is σB dependent and that a combined loss of prfAP1 and σB also results in reduced virulence-associated characteristics (32). From this evidence, we conclude that, in addition to contributing to survival under environmental stress, σB also plays a role in L. monocytogenes virulence.
As the organization and components of the B. subtilis and L. monocytogenes sigB operons are identical (19), we hypothesized that the regulatory network that determines σB activity in B. subtilis could be used as a model for stress signal transduction by Rsb proteins in L. monocytogenes. To investigate the role of these Rsbs in stress resistance and virulence-associated characteristics in the bacterial pathogen L. monocytogenes: (i) in-frame deletion mutations were created in rsbT and rsbV and (ii) stress survival and PrfA-mediated virulence-associated characteristics of the ΔrsbT and ΔrsbV strains were compared with those of the ΔsigB and wild-type strains.
MATERIALS AND METHODS
Bacterial strains.
L. monocytogenes 10403S and its derivatives were used throughout this study (Table 1). In comparison to the wild-type strain, all mutant strains had identical culture characteristics, including growth at 37°C with shaking (250 rpm) for at least 120 h in brain heart infusion (BHI) broth (Difco, Sparks, Md.) (data not shown). Stock cultures were stored at −80°C in BHI broth with 15% glycerol and streaked onto BHI agar plates prior to each experiment.
TABLE 1.
L. monocytogenes strains used in this studya
| Strain | Characteristics | Source or reference |
|---|---|---|
| DP-L1956 | ΔprfAP1 (−10 promoter deletion) | 20 |
| DP-L1957 | ΔprfAP2 (−10 promoter deletion) | 20 |
| DP-L1964 | ΔprfAP1P2 (−10 deletion each in P1 and P2) | 20 |
| DP-L2161 | Δhly | 24 |
| FSL A1-254 | ΔsigB | 44 |
| FSL B2-002 | ΔprfAP1sigB | 32 |
| FSL C3-015 | ΔrsbT | This study |
| FSL C3-047 | ΔprfAP1rsbT | This study |
| FSL C3-049 | ΔprfAP2rsbT | This study |
| FSL C3-053 | ΔprfAP2sigB | This study |
| FSL C3-057 | ΔrsbV | This study |
| FSL C3-091 | ΔprfAP1rsbV | This study |
| FSL C3-095 | ΔprfAP2rsbV | This study |
| 10403S | Serotype 1/2a wild-type strain | 5 |
All strains are derivatives of 10403S.
Mutant construction.
rsbT and rsbV alleles with in-frame internal deletions were created in the Escherichia coli-L. monocytogenes shuttle vector pKSV7 (10) by SOE (splicing by overlap extension) PCR (23) and introduced into L. monocytogenes 10403S by allelic exchange mutagenesis (44). For the rsbT mutation, SOE PCR primers were designed to amplify two ∼400-bp DNA fragments, one comprising the 5′ end of rsbT (amplified by primers SOE-rsbTA and SOE-rsbTB [Table 2]) and one comprising the 3′ end of rsbT (amplified by primers SOE-rsbTC and SOE-rsbTD [Table 2]). Subsequent PCR amplification of the two PCR products with SOE-rsbTA and SOE-rsbTD created an in-frame 267-bp deletion within the rsbT open reading frame. The resulting fragment was purified with the QIAquick PCR Purification kit (QIAGEN Inc., Valencia, Calif.) and subsequently digested with KpnI and XbaI. The resulting fragment was cloned into pKSV7 and transformed into E. coli DH5α. The resulting plasmid, pSC1, was electroporated into L. monocytogenes 10403S. Transformants were selected on BHI agar plates containing 10-μg/ml chloramphenicol. A transformant was serially passaged in BHI-chloramphenicol at 41°C to direct chromosomal integration of the plasmid by homologous recombination. Confirmation of chromosomal integration was done by PCR and sequencing. A single colony with a chromosomal integration was serially passaged in BHI at 30°C and screened for loss of chloramphenicol resistance. Allelic exchange mutagenesis was confirmed by PCR amplification and direct sequencing of the PCR product with primers CHK-ΔrsbTF and CHK-ΔrsbTR. Similar procedures were performed to create an in-frame 303-bp deletion in the rsbV gene by using the primers shown in Table 2.
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| SOE-rsbTAa | GGG GTA CCG TGA ATA CTG TGG GGA TAC |
| SOE-rsbTBb | GGA TTC AAT ATC AAA ACT GTC GTT CAT TCC CCC AAT TCC TG |
| SOE-rsbTC | GAC AGT TTT GAT ATT GAA TCC |
| SOE-rsbTDc | GCT CTA GAT CAC TAC GTA ATT CTT TTT GC |
| CHK-ΔrsbTFe | GAG ATA GTA GAA AAA GAG GG |
| CHK-ΔrsbTRe | GCA TAT CTT CTG CTT GAG C |
| SOE-rsbVAa | GGG GTA CCT ATG TCA GAT GGT GTA ACA G |
| SOE-rsbVBd | CAT TCA TTT CAC CCT CTA CAA TAT TCA TCA CTT CAC CCC |
| SOE-rsbVC | GTA GAG GGT GAA ATG AAT G |
| SOE-rsbVDc | GCT CTA GAC AAT TAA AAA TAA ACC TAG ACC |
| CHK-ΔrsbVFf | GAT AAA AAT GCT GTC TAC CG |
| CHK-ΔrsbVRf | TAA TAA AGT TTC ACG TCA TCC |
The KpnI restriction site incorporated into this primer to facilitate cloning is underlined.
The overhang complementary to SOE-rsbTC is underlined.
The XbaI restriction site incorporated into this primer to facilitate cloning is underlined.
The overhang complementary to SOE-rsbVC is underlined.
Forward (F) and reverse (R) primers for in-frame rsbT deletion confirmation.
Forward (F) and reverse (R) primers for in-frame rsbV deletion confirmation.
Growth and stress conditions.
For environmental stress survival, L. monocytogenes 10403S, FSL A1-254 (ΔsigB mutant), FSL C3-015 (ΔrsbT mutant), and FSL C3-057 (ΔrsbV mutant) were tested under three different conditions. Acid survival assays were performed by using synthetic gastric fluid (pH 2.5) (12) or acidified BHI (pH 2.5). Overnight cultures were inoculated into 10 ml of BHI broth (1:100 dilution). The cultures were grown at 37°C with shaking (250 rpm) to an optical density at 600 nm (OD600) of 0.4, and then 0.1 ml of each culture was reinoculated into 10 ml of BHI broth. From this point, to prepare mid-log cells, 1 ml of each culture was collected when these cultures reached an OD600 of 0.4. Samples were centrifuged and resuspended in 1 ml of synthetic gastric fluid (pH 2.5) or BHI-HCl (pH 2.5). The assay tubes were incubated at 37°C with shaking (250 rpm). Immediately after acid challenge (t = 0) and 10 min after acid challenge (t = 10), 100-μl aliquots were removed, serially diluted, and plated on BHI agar plates for enumeration. To prepare stationary-phase cells, overnight cultures were inoculated into 10 ml of BHI broth (1:100). Following 12 h of incubation at 37°C with shaking (250 rpm), 1 ml of each culture was centrifuged and resuspended in 1 ml of synthetic gastric fluid (pH 2.5) or BHI-HCl (pH 2.5). Immediately after acid challenge (t = 0) and 60 min after acid challenge (t = 60), 100-μl aliquots were removed, serially diluted, and plated. Plates were incubated at 37°C for 48 h prior to enumeration.
Oxidative stress survival was assessed with the oxidative agent cumene hydroperoxide (CHP) (Sigma, St. Louis, Mo.). CHP survival assays were performed as described by Antelmann et al. (3). Briefly, overnight cultures were inoculated into 10 ml of BHI broth (1:100). Following 12 h of incubation at 37°C, 1 ml of each culture was centrifuged and resuspended in 0.9 ml of dimethyl sulfoxide (Fisher Scientific, Fair Lawn, N.J.). A 100-μl aliquot of 130 mM CHP was added to yield a final CHP concentration of 13 mM. Assay tubes were incubated for 15 min at 37°C with shaking (250 rpm). Aliquots were removed for standard plate counts on BHI agar plates at t = 15 min. A 100-μl portion of a 12-h culture was serially diluted and plated to represent viable cells at t = 0 min (prior to CHP exposure).
For energy stresses, L. monocytogenes 10403S, FSL A1-254, FSL C3-015, and FSL C3-057 were tested under three different conditions. For the first condition, carbon starvation was induced with defined medium (DM) (36) with a growth-limiting concentration of glucose (0.04% [wt/vol]). Briefly, overnight cultures in BHI broth were inoculated into 10 ml of DM (1:100 dilution) supplemented with glucose (0.4% [wt/vol]). After 12 h of incubation with shaking (250 rpm) at 37°C, cultures were reinoculated into DM plus 0.04% glucose (1:100 dilution). A preliminary experiment showed that the ODs of cultures grown under these conditions reflected the viable counts of bacteria in the cultures. Thereafter, culture densities were monitored by measuring absorbance (OD600) for up to 30 h. For the second energy stress condition, wild-type and mutant strains were monitored for survival during entry into stationary phase. Aliquots (100 μl) of overnight cultures from each strain were transferred into 10 ml of BHI (∼1:100 dilution). All strains were incubated statically at 37°C. Bacterial numbers were measured by standard plate count procedures using 100-μl aliquots removed at various time points up to 72 h. Entry into stationary phase for these experiments was defined as the period during and after which bacterial numbers reached maximal levels (between 6 and 36 h postinoculation). For the third energy stress condition, the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) was used to induce energy stress (2, 42). The MIC of CCCP was determined for L. monocytogenes 10403S according to the procedures described by the National Committee for Clinical Laboratory Standards (33). A final CCCP concentration of 32 μg/ml (16 times the MIC) was added to exponentially growing cultures (OD600 = 0.4). In a preliminary experiment, the ODs of the cultures grown under these conditions were found to reflect the viable counts of bacteria in the cultures. Thereafter, culture densities were monitored by measuring absorbance (OD600) over time.
Hemolysin assay.
The enzymatic activity of LLO from each strain was measured as described previously (35), except that 0.5 mM dithiothreitol was used as a reducing agent instead of cysteine. Lysis of sheep red blood cells was measured as hemoglobin release at 420 nm in a Fusion Universal microplate analyzer (Packard, Meriden, Conn.). A hemolytic unit was defined as the reciprocal of the supernatant dilution at which 50% of the sheep red blood cells were lysed. The wild-type level, represented by L. monocytogenes 10403S, was set at 100% hemolysis.
Tissue culture plaque assay.
Cytopathogenicities of different mutants were evaluated with a plaque assay performed with mouse L929 fibroblast cells as previously described (38). Briefly, overnight cultures (∼12 h) grown statically at 30°C in BHI broth were centrifuged, resuspended, and serially diluted in phosphate-buffered saline (pH 7.4). Five microliters of a 102 dilution and 15 μl of a 103 dilution were used as inocula for two wells of a six-well plate containing monolayers of mouse L cells. Inocula were enumerated by plating serial dilutions onto BHI agar and incubating at 37°C overnight. Inocula were equivalent for all strains. For plaque visualization at 3 days postinfection, infected mouse L cells were overlaid with 2× Dulbecco's modified Eagle's medium (Difco) containing 1.4% Bacto agar (Difco) and neutral red solution (Sigma). For each assay, Sigmascan Pro 5.0 software (SPSS, Inc., Chicago, Ill.) was used to measure the areas of at least 25 plaques. Plaque area for each strain was expressed as a percentage of the L. monocytogenes 10403S plaque size, which was included as an internal standard for each assay and assigned a value of 100%.
Statistical analyses.
All comparisons were evaluated by one-way analysis of variance. Statistical significance was established at P < 0.05. Statistical analyses were performed in Minitab (Statistical Software, State College, Pa.).
RESULTS
Mutant survival under acid stress conditions.
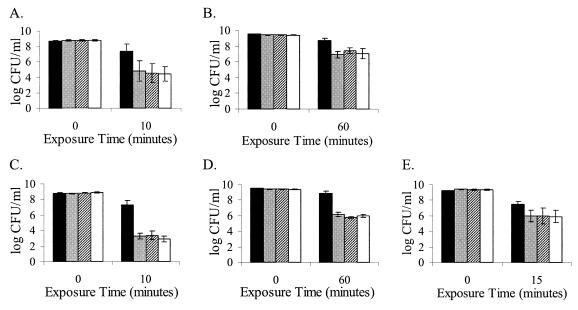
Two different acid stress conditions were used to study the roles of RsbT and RsbV in activation of σB in L. monocytogenes. First, wild-type 10403S and the ΔsigB, ΔrsbT, and ΔrsbV strains were exposed to synthetic gastric fluid (pH 2.5) (Fig. 1A and B). Within 10 min of exposure, numbers of exponential wild-type 10403S cells (OD600 = 0.4) were reduced by 1 log (relative to t = 0 counts) while all mutant strain numbers were reduced by approximately 4 logs. Numbers of wild-type survivors were significantly different from those of all mutant strains after 10 min in synthetic gastric fluid (P = 0.007). Following a 60-min exposure to synthetic gastric fluid, stationary-phase numbers of the ΔsigB, ΔrsbT, and ΔrsbV strains were reduced by an average of 1.6 log more than those of the wild-type strain (P = 0.007). When mid-log wild-type and mutant strains were challenged with acidified BHI broth (pH 2.5) for 10 min (Fig. 1C and D), wild-type strain numbers were reduced by 1.5 log relative to t = 0, while all mutant strains were reduced by about 6 to 6.5 logs. Numbers of wild-type survivors were significantly different from those of the mutant strains (P < 0.0001). Bacterial numbers for wild-type stationary-phase cultures were approximately 2.9 logs higher than those for all mutant strains (P < 0.0001) following exposure to BHI-HCl (pH 2.5) for 60 min. Results from these experiments suggest that RsbT and RsbV contribute to acid stress survival of both exponential- and stationary-phase L. monocytogenes cultures, likely through σB activation.
FIG. 1.
Viabilities of L. monocytogenes wild-type 10403S (black), ΔsigB (gray), ΔrsbT (hatched), and ΔrsbV (white) strains following exposure of mid-log (A and C) or stationary-phase (B, D, and E) cultures to synthetic gastric fluid at pH 2.5 (A and B), acid (pH 2.5) (C and D), and CHP (13 mM) (E). The results shown are mean values from three independent experiments; error bars indicate standard deviations.
Mutant survival under oxidative stress conditions.
CHP (13 mM) was used to induce oxidative stress in this study. As shown in Fig. 1E, after 15 min of exposure, wild-type numbers were reduced by 2 logs; all mutant strains were reduced by 3 logs. Numbers of wild-type survivors were significantly different from those of the mutant strains (P = 0.017). These results show that wild-type L. monocytogenes 10403S is more resistant to oxidative stress imposed by 13 mM CHP than the ΔsigB strain or the strains lacking predicted σB regulatory proteins (ΔrsbT and ΔrsbV). Thus, RsbT and RsbV appear to contribute to σB-mediated survival following exposure to CHP stress.
Mutant survival under energy stress conditions.
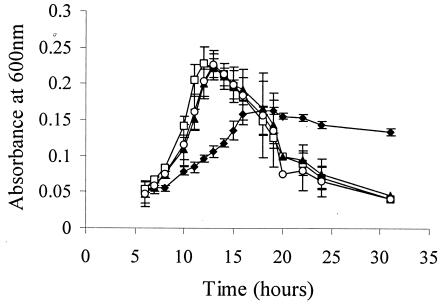
Energy stresses usually result from limitations in key metabolites such as carbon, phosphate, or nitrogen. For this study, we used an L. monocytogenes DM (36) supplemented with limiting glucose (0.04% [wt/vol]) or prolonged static incubation into the stationary phase to induce carbon starvation. As a third condition, the protonophore CCCP was added as a chemical agent to limit ATP synthesis, thereby resulting in an intrinsic energy stress. Figure 2 shows that the ΔsigB, ΔrsbT, and ΔrsbV strains entered exponential growth after 6 h, as compared to 10 h for the wild-type strain when all were grown under limiting glucose conditions. The mutant strains also grew more rapidly (average doubling times of approximately 180 min, between 8 and 13 h) than the wild type (doubling time = 360 min between 10 and 16 h). ODs of all three mutant strains reached higher maxima than the wild type; however, ODs declined rapidly for all three mutant strains, possibly due to cell lysis resulting from depletion of glucose (18). The wild-type strain, which grew more slowly and reached a lower maximum OD than those of the mutant strains, also declined more slowly than the mutant strains. From these data, we hypothesize that activated σB negatively affects growth in exponentially growing cells but positively contributes to maintenance of viability after glucose is depleted in defined media.
FIG. 2.
Growth of L. monocytogenes 10403S (⧫), ΔsigB (□), ΔrsbT (▴), and ΔrsbV (○) in a defined medium with a limiting amount of glucose (0.04% [wt/vol]). OD600 values were recorded. The results shown are mean values from three independent experiments; error bars indicate standard deviations.
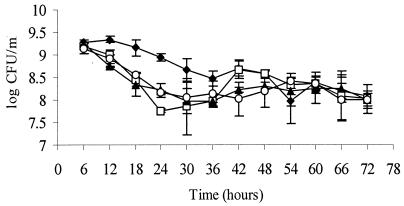
Survival following entry into stationary phase was tested by growing each of the four strains in BHI with static incubation at 37°C for at least 72 h. Cultures were sampled for enumeration every 6 h. As shown in Fig. 3, numbers of ΔsigB, ΔrsbT, and ΔrsbV declined more rapidly than those of the wild-type control following entry into stationary phase (between 6 and 36 h postinoculation). After 54 h, however, all cultures were present at similar numbers. σB appears to contribute to survival between 6 and 36 h under static growth in BHI.
FIG. 3.
Viabilities of L. monocytogenes 10403S (⧫), ΔsigB (□), ΔrsbT (▴), and ΔrsbV (○) cultures that had been grown statically in BHI at 37°C. Bacteria were harvested at indicated times, serially diluted, and plated for enumeration on BHI agar plates. The results shown are mean values from three independent experiments; error bars indicate standard deviations.
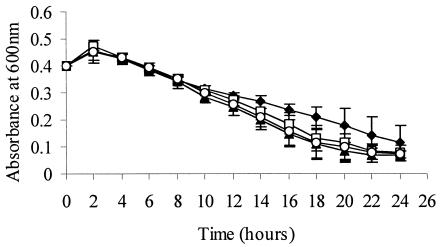
The protonophore, CCCP, was added to exponentially growing cells (OD600 = 0.4) in BHI, and OD was monitored every 2 h for 24 h. OD600 slightly increased above 0.4 for all strains but then declined for all strains. ODs between the wild-type and mutant strains (ΔsigB, ΔrsbT, and ΔrsbV) were significantly different (P = 0.01) after 14 h, presumably following consumption of accumulated intracellular ATP (Fig. 4). σB appears to contribute to maintaining cell viability after ATP depletion.
FIG. 4.
OD600 values for L. monocytogenes 10403S (⧫), ΔsigB (□), ΔrsbT (▴), and ΔrsbV (○) strains after exposure of mid-log cells (OD600 = 0.4) to 32-μg/ml CCCP. The results shown are mean values from three independent experiments; error bars indicate standard deviations.
Tissue culture virulence and hemolysis phenotypes.
Contributions of σB and putative σB activators to virulence-associated phenotypes were characterized in vitro by performing hemolysis assays and tissue culture plaque assays. Strains used in these assays included the ΔsigB, ΔrsbT, ΔrsbV, ΔprfAP1, and ΔprfAP2 strains and selected double mutants (Table 1). The rationale for our research hypothesis regarding the interplay between σB activation and PrfA-mediated virulence arose from the finding that the prfAP2 promoter is σB dependent. A combined loss of prfAP1 and σB results in reduced virulence (32). Therefore, we predicted that loss of prfAP1 and σB activity (through ΔprfAP1rsbT or ΔprfAP1rsbV) would also affect virulence-associated phenotypes.
A hemolysis assay was used to measure the hemolytic activity of LLO in the culture supernatant from each strain. The ability to lyse sheep red blood cells was compared among wild-type and mutant strains. Results are shown in Table 3. The ΔprfAP1sigB, ΔprfAP1rsbT, or ΔprfAP1rsbV strain showed reduced hemolytic activities. These results suggest that the activated σB has a negative effect on LLO activity or perhaps on the prfAP1 promoter.
TABLE 3.
Virulence-associated phenotypes of L. monocytogenes strains used in this study
| Genotype | % Hemolysis (mean ± SD)a | Plaque size relative to 10403S (mean ± SD)a |
|---|---|---|
| Wild type | 100 ± 0 | 100 ± 0 |
| ΔsigB | 250 ± 122b | 103 ± 6 |
| ΔrsbT | 250 ± 122b | 105 ± 8 |
| ΔrsbV | 233 ± 82b | 101 ± 9 |
| ΔprfAP1 | 21 ± 6b | 79 ± 12 |
| ΔprfAP2 | 75 ± 27 | 92 ± 9 |
| ΔprfAP1P2 | 5 ± 3b | 45 ± 1b |
| ΔprfAP1sigB | 6 ± 0b | 62 ± 8b |
| ΔprfAP1rsbT | 9 ± 3b | 58 ± 8b |
| ΔprfAP1rsbV | 10 ± 4b | 63 ± 5b |
| ΔprfAP2sigB | 175 ± 125 | 93 ± 4 |
| ΔprfAP2rsbT | 175 ± 125 | 90 ± 4 |
| ΔprfAP2rsbV | 133 ± 58 | 91 ± 4 |
| Δhly | 0 ± 0b | NTc |
Data report triplicate experiments.
P < 0.05 when data are compared with those from the wild type.
NT, not tested.
A plaque assay was conducted in mouse fibroblast cells (L929). Cells were infected with L. monocytogenes 10403S, ΔsigB, ΔrsbT, or ΔrsbV mutants. The cytopathogenicity of each strain was inferred from bacterial ability to infect and disseminate among host cells, as measured by plaque sizes (Table 3). A combined loss of prfAP1 and sigB, rsbT, or rsbV led to a range in plaque sizes from wild type to very small. The ΔsigB, ΔrsbT, ΔrsbV, Δ prfAP2sigB, ΔprfAP2rsbT, and Δ prfAP2rsbV strains yielded similar plaque sizes as the wild-type strains, in concurrence with previous findings (20, 32).
DISCUSSION
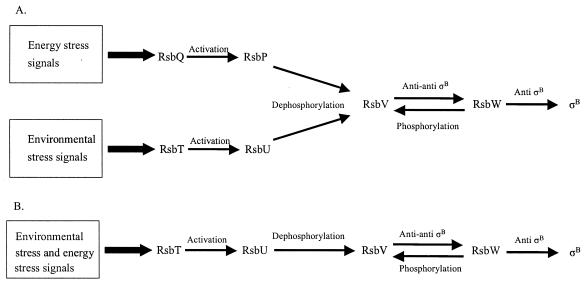
Our overarching research hypothesis is that L. monocytogenes σB promotes bacterial survival under exposure to environmental stresses both outside and inside a host, thus contributing to bacterial pathogenicity. To begin to dissect σB-activating stress signaling pathways in L. monocytogenes, we created two rsb null mutants: one in rsbT and the other in rsbV. We hypothesized that, as with the L. monocytogenes ΔsigB strain, the ΔrsbT and ΔrsbV strains also would be more susceptible than the wild type to environmental stresses such as exposure to synthetic gastric fluid, reduced pH, and oxidative stresses. Based on the B. subtilis model (Fig. 5A), we predicted that only the ΔrsbV and ΔsigB strains would survive less well than the wild type under energy stress conditions such as carbon starvation or ATP depletion. However, we found that the ΔsigB, ΔrsbT, and ΔrsbV strains had identical phenotypes under all stress conditions, suggesting that stress signaling pathways that activate σB differ between L. monocytogenes and B. subtilis.
FIG. 5.
Proposed model of Rsb-mediated σB activation in L. monocytogenes (B) in comparison to Rsb-mediated σB activation in B. subtilis (A).
σB, RsbT, and RsbV enhance environmental and energy stress survivals.
As predicted, the ΔrsbT, ΔrsbV, and ΔsigB strains survived at significantly lower levels than the wild-type strain after exposure to environmental stresses. Specifically, σB-mediated survival of the wild-type strain in synthetic gastric fluid (pH 2.5), acid (pH 2.5), and CHP (13 mM) appears to require σB activation. This activation requires both RsbT and RsbV, since loss of either resulted in the same phenotype as loss of σB. We further hypothesize that RsbT and RsbV contribute to σB activation and L. monocytogenes survival under environmental stresses such as those encountered during gastric passage (bile salt in the host gut) (28), or during acid and reactive oxygen intermediate exposure inside the host cell vacuole.
We defined energy stress as a condition in which bacteria are deprived of a carbon source or after depletion of intracellular ATP. Our results show that σB plays a positive role in maintaining cellular viability in glucose-limiting defined media after glucose is depleted. However, shortly after inoculation into fresh glucose-limiting media, the ΔrsbT, ΔrsbV, and ΔsigB strains grew more rapidly than the wild-type strain (Fig. 2), possibly by utilizing the carbon source more efficiently than the wild type. All three mutant cultures achieved higher numbers of cells more rapidly than the wild-type culture. Similar observations also have been reported with B. subtilis and E. coli strains that bear mutations in their respective general stress response sigma factors, σB or σS (34, 37). Schweder et al. (37) and Notley-McRobb et al. (34) have hypothesized that both B. subtilis and E. coli respond primarily to “hunger” rather than “general stress” during glucose-limited growth and suggest that a hunger signal may be perceived independently from a general stress signal. We hypothesize that at least some genes that are responsible for glucose transport or utilization are regulated by a sigma factor(s) other than σB in L. monocytogenes. For example, we identified putative σA-dependent promoters upstream of lmo0169 and lmo0176, which encode putative glucose uptake proteins, in a preliminary examination of the L. monocytogenes genome (22) with the ListiList Web Server (http://genolist.pasteur.fr/ListiList) (31). It is possible that the loss of σB reduces overall competition among other sigma factors for core RNA polymerase. If this hypothesis is true and if glucose transport is not σB regulated, an L. monocytogenes strain lacking σB (or an activated σB) may initially respond more efficiently to nutrient-limiting conditions than the wild-type strain. Another hypothesis is that σB and genes in its regulon may negatively regulate genes or proteins responsible for glucose transport (C. P. O'Byrne, personal communication) such that loss of activated σB results in very rapid uptake of glucose, leading to rapid growth followed by a dramatic dropoff after the glucose is completely consumed.
We also tested survival of the wild-type strain in comparison to the ΔrsbT, ΔrsbV, and ΔsigB mutants during prolonged static incubation at 37°C. The wild-type strain survived better following entry into stationary phase (6 to 36 h postinoculation), concomitantly with predicted activation of σB, which occurs in a growth phase-dependent manner (18). σB, RsbT, and RsbV are clearly important for survival in nutrient-limiting batch culture, particularly in early stationary phase.
CCCP is a protonophore that carries protons across the plasma membrane, which destroys the electron motive force, inhibits electron transport systems, and prevents proton flow back through ATPase, thereby limiting ATP synthesis. CCCP treatment has been shown to result in increased σB protein levels, while simultaneously reducing intracellular ATP in Bacillus cereus (39). As shown in Fig. 4, the ODs of all four L. monocytogenes strains increased slightly for approximately 2 h after addition of CCCP, possibly resulting from completion of cell division that had been initiated prior to CCCP exposure. After 2 h, the ODs of all four strains decreased at a similar rate until ∼14 h postexposure. The ODs of the three mutant strains were significantly lower than that of the wild-type strain after 14 h. We hypothesize that accumulated intracellular ATP was depleted in all strains after 14 h. Thus, while insufficient for complete viability maintenance, the presence of active σB appears to reduce the rate of death in the presence of CCCP following ATP depletion.
Under the conditions selected for these studies, we did not distinguish phenotypic differences predicted by the B. subtilis model between the L. monocytogenes ΔrsbT and ΔrsbV mutants following energy stress (i.e., the B. subtilis model predicts that the ΔrsbT strain would be affected by environmental, but not energy, stresses) (Fig. 5A). Based on the evidence presented, we propose that, in contrast with B. subtilis, in L. monocytogenes, Rsb activation of σB by energy and environmental stresses is achieved through a single pathway (Fig. 5B). Both types of stresses appear to be conveyed to σB by RsbT, RsbU, RsbV, and RsbW. Our model does not rule out the existence of additional contributors to energy stress signaling pathways in L. monocytogenes. Additional σB-activating pathways have recently been proposed in B. subtilis. For example, Zhang et al. (48) suggest that, in addition to Rsb proteins, RelA also is associated with activation of σB under energy stresses in B. subtilis. Moreover, Brigulla et al. (8) put forward the possibility of the existence of yet another environmental stress signaling pathway responsible for σB-dependent contributions to low-temperature growth of B. subtilis. Given that L. monocytogenes is capable of growth at temperatures as low as 0°C (47), it is possible that a low-temperature σB activation pathway exists in this organism as well.
σB, RsbT, and RsbV contribute to virulence-associated phenotypes.
In contrast with the B. subtilis system, the L. monocytogenes system also can be used to investigate the role of σB in bacterial virulence. Plaque assays and relative comparisons of hemolytic activities were used to examine the role of σB activation in L. monocytogenes virulence-associated characteristics. In the hemolysis assays, we observed that the loss of σB resulted in more than a twofold increase in apparent LLO activity relative to that measured in the wild-type strain. This finding is consistent with previous studies in which increased hemolysin activity was observed in a Staphylococcus aureus sigB mutant (11). We used sodium dodecyl sulfate-polyacrylamide gel electrophoresis to examine protein samples collected from wild-type, ΔsigB, ΔrsbT, and ΔrsbV culture supernatants. We found no gross differences in protein secretion patterns between the wild type and mutants (data not shown), suggesting that the observed differences in hemolytic activities are not a consequence of vastly different quantities of hemolysin in the culture supernatants. As predicted, the ΔprfAP1rsbT and ΔprfAP1rsbV double mutants had dramatically reduced LLO activity, similar to the ΔprfAP1P2 and ΔprfAP1sigB double mutant phenotypes (32). We propose that the reduced LLO production phenotypes are likely a consequence of reduced prfA expression. We conclude that RsbT and RsbV contribute to σB activation required to reach the wild-type level of PrfA expression from the prfAP2 promoter, which leads to the wild-type level of LLO expression.
We also examined the role of σB in virulence-associated characteristics in a tissue culture plaque assay. Although the ΔsigB, ΔrsbT, or ΔrsbV and the ΔprfAP2sigB, ΔprfAP2rsbT, or ΔprfAP2rsbV strain had higher levels of LLO activity than the wild-type strain, these strains all yielded wild-type plaque sizes. Earlier studies have shown that LLO activity levels do not directly correlate with virulence in vivo (14). The L. monocytogenes requirement for LLO during intracellular infection appears to be minute and temporal (13). The wild-type plaque sizes among the ΔsigB-equivalent strains listed above suggest that while the increased LLO activity that we observed might possibly assist the bacterium in phagosomal escape, it does not appear to enhance bacterial ability to spread from cell to cell. We also show that σB, RsbT, and RsbV are not absolutely required for L. monocytogenes infection and dissemination in mouse fibroblast cells, as previously suggested for σB (32). However, the apparent contributions of these proteins to PrfA expression, which is important for cell-to-cell spread and overall virulence-associated phenotypes, are illustrated by the significantly reduced plaque sizes of the ΔprfAP1P2, ΔprfAP1sigB, ΔprfAP1rsbT, and ΔprfAP1rsbV strains in comparison to those of the wild-type strain.
In summary, we used a genetic approach to analyze the σB-activating stress signaling pathway in L. monocytogenes. We disproved our initial hypothesis that the σB activation pathways in L. monocytogenes are identical to those in B. subtilis. σB activation, which requires both RsbT and RsbV, contributes to L. monocytogenes survival under both environmental and energy stresses outside of the host as well as to virulence-associated characteristics in in vitro systems.
Acknowledgments
We thank H. Marquis for assistance with the protein work and for intellectual discussions. We are grateful for the generosity of D. Portnoy for providing bacterial strains and for the valuable ideas of C. P. O'Byrne and M. Wiedmann. We truly appreciate the technical assistance of B. Bowen and T. Arvik with mutant strain creation.
This work was supported in part by National Institutes of Health award no. RO1-AI052151-01A1 (to K.J.B.). S.C. was supported by the Office of the Civil Service Commission (Thailand).
REFERENCES
- 1.Akbar, S., C. M. Kang, T. A. Gaidenko, and C. W. Price. 1997. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24:567-578. [DOI] [PubMed] [Google Scholar]
- 2.Alper, S., L. Duncan, and R. Losick. 1994. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell 77:195-205. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann, H., S. Engelmann, R. Schmid, and M. Hecker. 1996. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 178:6571-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, L. A., M. S. Çetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 6.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor σB by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brigulla, M., T. Hoffmann, A. Krisp, A. Völker, E. Bremer, and U. Völker. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brody, M. S., K. Vijay, and C. W. Price. 2001. Catalytic function of an α/β hydrolase is required for energy stress activation of the σB transcription factor in Bacillus subtilis. J. Bacteriol. 183:6422-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of PlcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., Y.-T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 13.Dancz, C. E., A. Haraga, D. A. Portnoy, and D. E. Higgins. 2002. Inducible control of virulence gene expression in Listeria monocytogenes: temporal requirement of listeriolysin O during intracellular infection. J. Bacteriol. 184:5935-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decatur, A. L., and D. A. Portnoy. 2000. A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science 290:992-995. [DOI] [PubMed] [Google Scholar]
- 15.Delumeau, O., R. J. Lewis, and M. D. Yudkin. 2002. Protein-protein interactions that regulate the energy stress activation of σB in Bacillus subtilis. J. Bacteriol. 184:5583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira, A., M. Gray, M. Wiedmann, and K. J. Boor. 2004. Comparative genomic analysis of the sigB operon in Listeria monocytogenes and in other Gram-positive bacteria. Curr. Microbiol. 48:39-46. [DOI] [PubMed] [Google Scholar]
- 20.Freitag, N. E., and D. A. Portnoy. 1994. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol. Microbiol. 12:845-853. [DOI] [PubMed] [Google Scholar]
- 21.Gaidenko, T. A., X. Yang, Y. M. Lee, and C. W. Price. 1999. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis. J. Mol. Biol. 288:29-39. [DOI] [PubMed] [Google Scholar]
- 22.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couv, A. D. Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K.-D. Entian, H. Fsihi, F. G.-D. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueo, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. D. Pablos, J.-C. Prez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J.-A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 23.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-534. [PubMed] [Google Scholar]
- 24.Jones, S., and D. A. Portnoy. 1994. Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect. Immun. 62:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalman, S., M. L. Duncan, S. M. Thomas, and C. W. Price. 1990. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J. Bacteriol. 172:5575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang, C. M., K. Vijay, and C. W. Price. 1998. Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol. Microbiol. 30:189-196. [DOI] [PubMed] [Google Scholar]
- 28.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovacs, T., A. Hargitai, K. L. Kovacs, and I. Mecs. 1998. pH-dependent activation of the alternative transcriptional factor σB in Bacillus subtilis. FEMS Microbiol. Lett. 165:323-328. [DOI] [PubMed] [Google Scholar]
- 30.Milohanic, E., P. Glaser, J. Y. Coppée, L. Frangeul, Y. Vega, J. A. Vázquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 31.Moszer, I., P. Glaser, and A. Danchin. 1995. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology 141:261-268. [DOI] [PubMed] [Google Scholar]
- 32.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 34.Notley-McRobb, L., T. King, and T. Ferenci. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Portnoy, D. A., P. S. Jacks, and D. J. Hinrichs. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167:1459-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Premaratne, R. J., W. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schweder, T., A. Kolyschkow, U. Völker, and M. Hecker. 1999. Analysis of the expression and function of the σB-dependent general stress regulon of Bacillus subtilis during slow growth. Arch. Microbiol. 171:439-443. [DOI] [PubMed] [Google Scholar]
- 38.Sun, A. N., A. Camilli, and D. A. Portnoy. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 58:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. de Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vázquez-Boland, J. A., G. Domínguez-Bernal, B. González-Zorn, J. Kreft, and W. Goebel. 2001. Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 3:571-584. [DOI] [PubMed] [Google Scholar]
- 41.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 42.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voelker, U., A. Voelker, and W. G. Haldenwang. 1996. Reactivation of the Bacillus subtilis anti-σB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J. Bacteriol. 178:5456-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wise, A. A., and C. W. Price. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J. Bacteriol. 177:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 47.Yuqian, L., and A. E. Yousef. 1999. Characteristics of Listeria monocytogenes important to food processors, p. 131-224. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 48.Zhang, S., and W. G. Haldenwang. 2003. RelA is a component of the nutritional stress activation pathway of the Bacillus subtilis transcription factor σB. J. Bacteriol. 185:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]