Abstract
Here we developed the new expression system PZn zitR, based on the regulatory signals (PZn promoter and zitR repressor) of the Lactococcus lactis zit operon, involved in Zn2+ high-affinity uptake and regulation. A PZn zitR-controlled expression vector was constructed, and expression regulation was studied with two reporter genes, uspnuc and lacLM; these genes encode, respectively, a protein derived from Staphylococcus aureus secreted nuclease and Leuconostoc mesenteroides cytoplasmic β-galactosidase. Nuclease and β-galactosidase activities of L. lactis MG1363 cells expressing either uspnuc or lacLM under the control of PZn zitR were evaluated on plates and quantified from liquid cultures as a function of divalent metal ion, particularly Zn2+, availability in the environment. Our results demonstrate that PZn zitR is highly inducible upon divalent cation starvation, obtained either through EDTA addition or during growth in chemically defined medium, and is strongly repressed in the presence of excess Zn2+. The efficiency of the PZn zitR expression system was compared to that of the well-known nisin-controlled expression (NICE) system with the same reporter genes cloned under either PZn zitR or PnisA nisRK control. lacLM induction levels reached with both systems were on the same order of magnitude, even though the NICE system is fivefold more efficient than the PZn zitR system. An even smaller difference or no difference was observed after 3 h of induction when nuclease was used as a reporter for Western blotting detection. PZn zitR proved to be a powerful expression system for L. lactis, as it is tightly controlled by the zinc concentration in the medium.
The model lactic acid bacterium species Lactococcus lactis has been extensively engineered for the production and secretion of heterologous proteins, and several genetic tools are available for these purposes (10, 12, 31, 32). Systems that allow the controlled expression of foreign genes permit the choice of the time and rate of production, which are essential for toxic proteins. Several lactococcal promoters regulated by environmental conditions have been characterized; these include the P170 promoter, which is upregulated at a low pH during the transition to stationary phase (23, 34), the dnaJ promoter, which can be induced by heat shock (50), and sugar-regulated promoters or bacteriophage promoters (for a review, see reference 11).
The most commonly used inducible expression system in L. lactis is the nisin-controlled expression (NICE) system, which is controlled by the antimicrobial peptide nisin (30). It is based on the promoter (PnisA) of the nisin biosynthesis gene cluster (including the structural gene nisA) and on the two-component regulatory system gene nisRK, which is triggered by nisin, resulting in autoregulation. Subinhibitory amounts of nisin highly induce the expression of genes cloned under PnisA control in nisRK+ strains (28, 30). Several reports have demonstrated the versatility and efficient use of the NICE system for heterologous protein overproduction in lactic acid bacteria, with a reported induction factor that exceeds 1,000 (2, 5, 10, 16, 27, 46).
Although the usefulness of all of the above-mentioned systems has been documented, most of them have some disadvantages, such as low induction levels, undesirable basal expression, and a need for regulatory genes (provided on plasmids or cloned on the chromosome), which restrict the suitable choice of production conditions in biotechnological and laboratory applications (for a critical review, see reference 29).
Here we developed a new tool for controlled expression in L. lactis that relies on a zinc uptake regulation system. Although they have until now received little attention in L. lactis, high-affinity uptake ABC transporters specific for metals, including zinc, have been described for several microorganisms (14, 20, 22, 24, 25, 48). As they are necessary to ensure homeostasis, they are finely regulated systems. In some instances, the gene encoding the metalloregulatory protein is located in the same operon as the uptake genes (14, 15, 22, 48). In other instances, genes for specific metalloregulators are independently transcribed (24, 25). Some uptake transporters for different metal ions have been demonstrated to respond to general regulators, such as Fur or Zur proteins (17, 18, 20, 21, 40).
Our interest in the zinc regulation system began with the identification of an L. lactis MG1363 gene, encoding the exported protein Nlp3 (new lipoprotein 3), as an active translational fusion to the ΔSPNuc reporter open reading frame (ORF) (ΔSPNuc is derived from staphylococcal nuclease by signal peptide deletion and is active exclusively in an extracytoplasmic location) (42). Nlp3 is similar to streptococcal adhesins that appeared to be metal binding lipoproteins (42) and is identical to the N-terminal part of L. lactis IL1403 ZitS lipoprotein (3). ZitS seems to form with ZitQ ATPase and ZitP permease a typical ABC transporter putatively involved in high-affinity Zn2+ uptake (3). The ABC transporter-encoding genes zitSQP are organized with an upstream repressor gene, zitR, as a putative zitRSQP operon (3). Sequence and homology data suggested that zit expression could be regulated in response to environmental zinc concentrations, as for other zinc transport operons in gram-positive bacteria (14, 17, 20; J. P. Claverys, personal communication).
As regulation of the zit operon could depend on ZitR, we explored the use of the PZn zitR promoter-regulator region to control the expression of heterologous genes. Here we constructed an expression vector based on this promoter-regulator system. PZn zitR proved to be a tightly regulated system and a useful tool for controlling protein production in L. lactis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are listed in Table 1. Lactococcal strains were routinely grown in M17 medium (49) supplemented with 1% glucose (GM17 medium) or, when needed, in chemically defined SA medium (26) at 30°C without shaking. Escherichia coli strains were grown in Luria-Bertani (LB) broth at 37°C with shaking. Antibiotics were used for plasmid maintenance at the following concentrations: erythromycin (5 μg/ml for L. lactis and 75 μg/ml for E. coli) and chloramphenicol (10 μg/ml for L. lactis and 12 μg/ml for E. coli).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | 47 | |
| TG1 pcnB | TG1 pcnB::kan Kmr | 33 |
| L. lactis | ||
| MG1363 | Laboratory strain; plasmid free | 19 |
| NZ9000 | MG1363 pepN::nisRK | 30 |
| IL1403 | Laboratory strain; plasmid free | 6 |
| Plasmids | ||
| pFUN | Apr ColE1 Emr pAMβ1 Δnuc | 42 |
| pVE8020 | PZnzitRS′-Δnuc (derived from pFUN by PZnzitRS′ cloning) | 42 |
| pVE8061 | PZnzitR (derived from pFUN) | This work |
| pVE8062 | PZnzitR (derived from pVE8061) | This work |
| pSEC:Nuc | Cmr pWV01 PnisA uspnuc | 1, 5 |
| pVE5239 | Apr T1T2 terminator | 12 |
| pAMJ769 | Emr pCT1138 lacLM | 34 |
| pVE8063 | T1T2 (derived from pSEC:Nuc) | This work |
| pVE8064 | PZnzitR uspnuc T1T2 (derived from pVE8062) | This work |
| pVE8065 | PZnzitR lacLM T1T2 (derived from pVE8064) | This work |
| pVE8066 | PnisA uspnuc T1T2 (derived from pVE8064) | This work |
| pVE8067 | PnisA lacLM T1T2 (derived from pVE8066) | This work |
ColE1, pAMβ1, pWV01, and pCT1138 refer to the replicon or vector. Kmr, Cmr, Apr, and Emr, resistance to kanamycin, chloramphenicol, ampicillin, and erythromycin, respectively.
Oligonucleotides.
Oligonucleotide 9 (5′-CTAATGAGCGGGCTTTTT-3′) and oligonucleotide MUT (5′-GCTCTAGAGCGGGATCCTTCATCGAAACTCTTCAG-3′, where BamHI and XbaI restriction sites are underlined and two single silent mutations are in bold type) were used to amplify an ∼700-bp MG1363 DNA fragment containing PZn zitR from plasmid pVE8020 (pVE8020 harbors the original fusion between a PZn zitRS′ fragment and a Δnuc reporter; it encodes both ZitR and Nlp3-ΔSPNuc and is expressed under the control of native zit expression signals [GenBank accession number U95834]) (42) (Table 1). Oligonucleotides 9 and MUT, respectively, hybridize with the 5′ end of the insert in pVE8020 and with the region covering the zitR stop and the zitS start (Fig. 1A). In the MUT sequence, the two mutations (bold type) are expected to alter the zitS potential ribosome binding site (RBS) (ACGGAGGAG is converted to ACTGAAGAG) without modifying the ZitR sequence (ACT, like ACG, and GAA, like GAG, are, respectively, Thr and Glu codons), although the possibility of the presence of a low-efficiency RBS cannot be excluded.
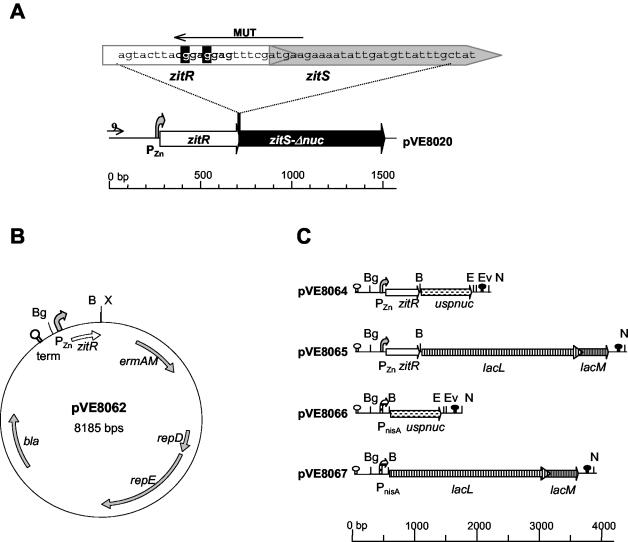
FIG. 1.
PZn zitR-regulated expression plasmids. (A) Detail of the wild-type PZn zitRS region from MG1363 cloned into pVE8020 (42). To amplify PZn zitR, oligonucleotide MUT was designed on the basis of the zitRS-overlapping region (top sequence); mismatches (wild-type base pairs highlighted in black) were introduced in MUT to alter the zitS putative RBS (bold type). (B) Schematic representation of expression vector pVE8062, which is an E. coli and gram-positive bacterium shuttle vector allowing ORF cloning under PZn zitR control. (C) Reporters and expression systems used. uspnuc and lacLM reporter ORFs were placed under the control of the PZn zitR or PnisA expression system. Hairpin symbols represent the ρ-independent trpA (white) and T1T2 (black) terminators. Curved arrows represent PZn (stippled) and PnisA (hatched) promoters. Unique restriction sites pertinent for reporter or promoter exchange are denoted as Bg (BglII), B (BamHI), E (EcoRI), Ev (EcoRV), N (NotI), and X (XbaI).
Oligonucleotides Lac5 (5′-CGCGGATCCTTTGAAAGGATATTCCTC-3′, where the lacL putative RBS is in bold type and the BamHI site is underlined) and Lac3 (5′-CCTACGTATTAGAAATGAATGTTAAAGC-3′, where the lacM stop codon is in bold type and the SnaBI site is underlined) were designed to amplify from pAMJ769 (Table 1) the lacLM genes, encoding β-galactosidase of Leuconostoc mesenteroides subsp. cremoris (34).
Plasmid construction.
Parental plasmids and the plasmids constructed in this study are listed in Table 1. To construct plasmid pVE8061, the PCR-amplified fragment containing PZn zitR (see above) was treated with the E. coli DNA polymerase Klenow fragment (Pol1k), digested with XbaI, and then ligated to vector pFUN (broad host range within gram-positive bacteria) (42) that had been previously digested with EcoRV and XbaI. The ligation mixture was used to electrotransform strain MG1363, and erythromycin-resistant clones were selected. One of them was confirmed by both digestion and sequence analysis to contain the correct sequence for pVE8061. This plasmid was digested with endonucleases EcoRI and EcoRV, made blunt with Pol1k, and religated, giving plasmid pVE8062 (Fig. 1B). Electrotransformation and selection processes were performed as described for pVE8061.
The T1T2 terminator (41) resulting from SacI digestion of pVE5239 (12) followed by filling in with T4 DNA polymerase and by ClaI digestion was cloned 3′ of the uspnuc reporter gene (encoding SPUsp45Nuc, hereafter referred to as UspNuc) into plasmid pSEC:Nuc (1, 5) that had been previously treated with XhoI, filled in with T4 DNA polymerase, and digested with ClaI. Ligation was used to transform E. coli strain TG1 with chloramphenicol resistance selection, and plasmid pVE8063 was selected as the correct construct. The uspnuc reporter gene-T1T2 terminator cassette was excised by digestion with SacII, filling in with T4 DNA polymerase, and BamHI digestion of pVE8063 and cloned into pVE8062 that had been previously treated with XbaI, filled in with T4 DNA polymerase, and digested with BamHI to generate plasmid pVE8064 (Fig. 1C).
L. mesenteroides subsp. cremoris lacLM genes were placed under PZn zitR control on plasmid pVE8065 (Fig. 1C). The lacLM PCR-amplified fragment (see above) was digested with BamHI and SnaBI and ligated to previously BamHI/EcoRV-digested pVE8064 to replace the uspnuc reporter gene. Transformation and selection procedures similar to those described above were used to obtain pVE8065.
PZn zitR was replaced in pVE8064 and pVE8065 by the PnisA promoter, giving rise to pVE8066 and pVE8067 (Fig. 1C). For pVE8066, the BglII/EcoRI fragment of pSEC:Nuc (Table 1), containing the uspnuc reporter gene under the control of PnisA, was cloned into BglII/EcoRI-digested pVE8064. pVE8067 was constructed like pVE8065 by cloning of lacLM into pVE8066. Both pVE8066 and pVE8067 were transferred into strain NZ9000 (nisRK+) (30) and selected on erythromycin-containing solid GM17 medium supplemented with nisin (0.1 ng/ml) and with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Euromedex; 160 μg/ml) only for pVE8067. The Nuc+ or LacLM+ phenotype was used for screening, and regulation by nisin was confirmed on plates by using a streak of concentrated nisin (data not shown).
DNA manipulations.
Plasmid DNA was extracted from L. lactis by a modified alkaline lysis method (47). Restriction and modification enzymes (Fermentas, New England Biolabs, and Amersham Biosciences) were used according to the manufacturers' instructions. PCR amplifications were performed with a Perkin-Elmer GeneAmp PCR system 2400 thermocycler according to the protocol provided with the amplification kit (DyNAzyme EXT Finnzymes). Electroporation of L. lactis was performed as described previously (31). DNA sequencing was performed by the BigDye Terminator method according to the protocol recommended by Applied Biosystems.
Reporter activity tests.
The nuclease activity of colonies on plates was detected on a toluidine blue-DNA-agar (TBDAgar) overlay by pink halos (31, 42). For liquid cultures, aliquots were spotted on 10 ml of TBDAgar (which does not allow bacterial growth) poured into petri dishes. The nuclease production level was estimated by comparing the intensity and the size of the pink halos of tested samples to those of purified Staphylococcus aureus nuclease protein dilutions.
The β-galactosidase activity of colonies was detected by their blue coloration in medium containing X-Gal at final concentrations of 160 μg/ml for L. lactis and 80 μg/ml for E. coli. Quantification assays were performed basically as described by Miller (35). Growing cells were harvested from different culture volumes and suspended in 500 μl of buffer Z (35). Permeabilization was carried out by the addition of 0.1% sodium dodecyl sulfate (SDS) (25 μl) and chloroform (25 μl). After vortexing, the reaction mixture was prewarmed at 30°C for 5 min, and o-nitrophenyl-β-d-galactopyranoside (Sigma) was added at 0.66 mg/ml to start the reaction. The reaction was stopped by the addition of 250 μl of 1 M CaCO3. Samples then were centrifuged at room temperature for 10 min at 10,000 × g, and the A420 in the supernatants was measured. β-Galactosidase activity in Miller units was calculated as (1,000 × A420)/(t × V × OD600), where t is the reaction time (in minutes), V is the volume (in milliliters) of the culture analyzed, and OD600 is the optical density of the culture at 600 nm.
Protein extracts, SDS-polyacrylamide gel electrophoresis, and Western blotting.
Cell and supernatant protein extracts were obtained as described previously (42). For each sample, equivalent amounts of protein were loaded on SDS-12% polyacrylamide gels, and polyacrylamide gel electrophoresis was performed at 50 mA for ∼2 h. After electroblotting on polyvinylidene difluoride membranes (Millipore), immunoblotting was performed with a commercial rabbit antinuclease antibody (Eurogentec) as described previously (47). Immunodetection was carried out with Western Lightning chemiluminescence reagent (Perkin-Elmer) according to the manufacturer's recommendations.
RESULTS
Description of PZn zitR expression vectors.
A new expression system was developed with the PZn promoter and the zitR regulator gene from the L. lactis MG1363 zit (zitRSQP) operon (3, 42). As there is translational coupling in wild-type zitRS genes (Fig. 1A) (42) (GenBank accession number U95834), expression under PZn zitR control might result in undesirable translation from the zitS RBS. To avoid this phenomenon, two silent mutations were introduced at the 3′ end of PZn zitR to alter the zitS RBS without modifying the ZitR C-terminal sequence (Fig. 1A).
Expression vector pVE8062 (Fig. 1B) was constructed with an E. coli and L. lactis shuttle vector derived from pFUN (42) by cloning PZn zitR instead of the reporter region. Two different reporter genes were placed under PZn zitR control on plasmids pVE8064 and pVE8065 (Fig. 1C)—uspnuc and lacLM, which encode, respectively, an exported protein (UspNuc) and a cytoplasmic protein (LacLM). UspNuc is derived from the staphylococcal nuclease which has been extensively used as a secretion reporter in gram-positive bacteria (2, 12, 31, 36, 37, 44), and is endowed with the signal peptide of the L. lactis Usp45 protein (51) to ensure efficient secretion (32). LacLM is the β-galactosidase of L. mesenteroides subsp. cremoris (9) and has been used as a reporter for transcriptional fusions in L. lactis (23, 34). Plasmids pVE8064 and pVE8065 carry two ρ-independent terminators; the trpA terminator (7), upstream of the PZn promoter, ensures that transcription is exclusively directed by PZn zitR, while the T1T2 terminator (41), downstream of the reporter ORF, controls the final lengths of transcripts. Unique restriction sites were designed to facilitate reporter exchange.
Reporter expression controlled by PZn zitR is constitutive in E. coli.
As PZn zitR-controlled expression plasmids (pVE8064 and pVE8065) are replicative in E. coli via ColE1, we tested the eventual regulation of uspnuc and lacLM expression in this species. As some instability of high-copy-number pFUN derivative vectors (including pVE8020) was observed in some E. coli strains, such as TG1 or DH5α (data not shown), both pVE8064 and pVE8065 were transferred into strain TG1 pcnB, known to reduce the ColE1 replicons copy number (33). TG1 pcnB(pVE8064) presented a Nuc+ phenotype on LB agar plates, and TG1 pcnB(pVE8065) developed a blue color on LB agar plates supplemented with X-Gal (data not shown), albeit only after several days of incubation at 25°C, probably because of the previously observed LacLM instability in E. coli (S. Madsen, personal communication). In both instances, the addition of concentrated Zn2+ (up to 10 μM) on plates had no effect on expression (data not shown), indicating that the PZn zitR expression system is not regulated in E. coli.
PZn zitR expression depends on environmental Zn2+ concentrations in L. lactis.
We tested whether the expression of reporter genes cloned into pVE8064 and pVE8065 under PZn zitR control might be regulated by metallic cations, particularly Zn2+, in L. lactis. When MG1363(pVE8064) or MG1363(pVE8065) was cultivated on solid GM17 medium, nuclease or LacLM activities were undetectable, suggesting that on this rich medium, PZn promoter activity is completely repressed (data not shown).
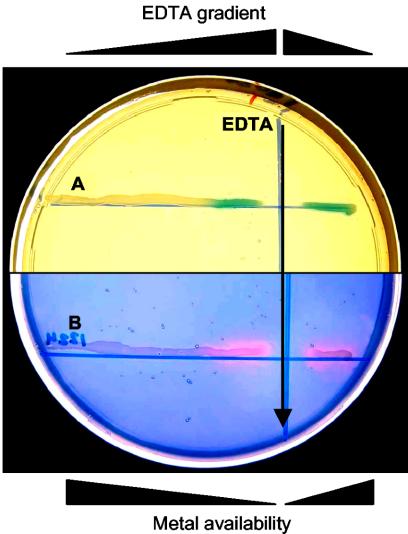
We used the general divalent cation chelator EDTA to mimic metal ion starvation. In accordance with the technique described by Patzer and Hantke (39), concentrated EDTA and cells of MG1363(pVE8064) or MG1363(pVE8065) were perpendicularly streaked on GM17 agar plates and incubated overnight at 30°C. Simple diffusion into the agar created a decreasing concentration gradient of EDTA from its deposition line, where the highest metal ion starvation was obtained. MG1363(pVE8065) on X-Gal-containing plates was detected as blue β-galactosidase-producing colonies only near the EDTA streak (Fig. 2). Similarly, MG1363(pVE8064) showed a typical pink halo of nuclease activity on a TBDAgar overlay exclusively near the EDTA streak (Fig. 2). These results revealed that EDTA is a useful inducer of the system. In both instances, reporter activity was not detectable or was hardly detectable at the lowest EDTA concentration even several hours after the tests, indicating that GM17 medium under these conditions allows strong or total repression.
FIG. 2.
EDTA induction of PZn zitR-controlled expression. Twenty microliters of 0.5 M EDTA (black arrow) and cultures of MG1363 carrying plasmid pVE8065 (A) or pVE8064 (B) were cross-streaked on a GM17 agar plate containing erythromycin (5 μg/ml) and X-Gal (160 μg/ml). EDTA diffusion produced a concentration gradient (decreasing from the deposition line) resulting in an inverse metal availability gradient. Enzymatic activity for β-galactosidase was detected directly (blue color in panel A); enzymatic activity for nuclease was detected after the plate was covered with a 10-ml TBDAgar overlay and incubated at 37°C for 1 h (pink halos in panel B).
Similar results were also obtained with MG1363(pVE8020), producing the original Nlp3-ΔSPNuc fusion protein (data not shown). However, its nuclease activity was slightly higher than that of MG1363(pVE8064) under the same conditions (data not shown), indicating that translational coupling of zitR and zitS in the natural system may be beneficial for expression efficiency. With strain MG1363(pVE8020), additional experiments were carried out to define the nature of the metallic cation involved in zit regulation. Detection of nuclease activity of MG1363(pVE8020) cells on solid chemically defined SA medium after cross-streaking of metal solutions showed that repression was essentially mediated by Zn2+ and, to a lesser extent, by Cu2+ (data not shown). Finally, all of our findings showed that PZn zitR is a powerful tool for regulating the production of heterologous proteins in L. lactis, whatever their final localization.
PZn zitR can be induced by Zn2+ depletion during growth in chemically defined medium.
We anticipated that the initial Zn2+ concentration in the culture medium would affect the induction level of the system. To examine this point, we evaluated uspnuc reporter expression in chemically defined SA medium with different starting Zn2+ or EDTA concentrations. After overnight growth in 1 μM ZnSO4-supplemented SA medium (to ensure repression of the system), MG1363(pVE8064) cells were washed and diluted in SA medium initially devoid of any added Zn2+ and then supplemented either with ZnSO4 at 0.01 (the concentration of normal SA medium), 0.1, or 1 μM or with EDTA at 10 or 100 μM.
Growth was not significantly affected, except with the addition of 100 μM EDTA, which led to considerable growth inhibition, probably because of drastic chelation of divalent cations (Fig. 3). However, the addition of 10 μM EDTA or the absence of any addition (neither EDTA nor Zn2+) (data not shown) was harmless, indicating that divalent cations are present in excess in SA medium initially devoid of Zn2+ and that sufficient trace amounts of Zn2+ may be provided either by water or by other medium components, including metals, to allow growth. Similarly, no toxic effect of Zn2+, even at the highest concentration tested (1 μM), was observed.
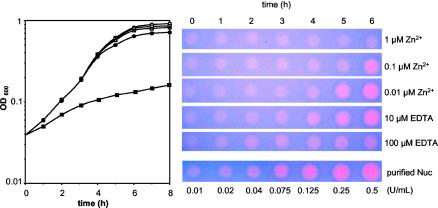
FIG. 3.
Induction of nuclease production in liquid cultures as a function of initial zinc concentrations. (Left panel) Growth of MG1363(pVE8064) in SA medium with different Zn2+ concentrations. SA medium devoid of Zn2+ was supplemented either with Zn2+ at 1 μM (open circles), 0.1 μM (open squares), or 0.01 μM (open triangles; normal SA medium) or with EDTA at 10 μM (filled circles) or 100 μM (filled squares). (Right panel) Nuclease activity of aliquots (5 μl) collected at different times during growth. Various concentrations of purified S. aureus nuclease protein (purified Nuc) were also spotted at 5 μl (bottom row) as a quantification control. The growth and nuclease detection experiments were reproduced at least five times, and the same results were obtained without or with 0.01 μM Zn2+ added to the initial medium (SA medium devoid of Zn2+ or normal SA medium).
Culture samples taken during growth were tested for nuclease activity (Fig. 3). Pink halos of nuclease activity increased in size and intensity as a function of the age of the culture and initial Zn2+ content; there was a sequential induction from Zn2+- to EDTA-supplemented cultures, reaching a maximal level of nuclease activity with 10 μM EDTA after 6 h of growth (estimated amounts, 0.25 to 0.5 U/ml of culture; i.e., 1.14 to 2.27 μg/ml). The addition of 10 μM EDTA also allowed the earliest induction, as clear induction was detectable after only 4 h of growth. Growth in normal SA medium (0.01 μM Zn2+) led to a natural (achieved without any addition) induction that was slightly weaker and delayed compared to that provided by the addition of 10 μM EDTA; although a comparable maximal induction level was obtained after 6 h of growth, induction was hardly detectable after 4 h (Fig. 3). The addition of 1 μM Zn2+ efficiently blocked expression throughout growth, despite the basal level (∼0.01 U/ml) observed under these conditions (it remained constant and probably corresponds to a drawback of the nuclease reporter). The level of induction after 6 h of exposure to 10 μM EDTA or 0.01 μM Zn2+ could be estimated as approximately >25- to 50-fold (by comparison to the standard) (Fig. 3), although the basal level of nuclease activity could result in underestimation.
These results indicate that PZn zitR allows tight control of gene expression. In chemically defined SA medium, strong repression can be achieved by the addition of Zn2+, while a comparable induction level can be obtained either by the addition of EDTA or by Zn2+ consumption during growth. The latter represents a convenient means of induction, as there is no need for any addition that might perturb cell physiology; a low production level that is maintained during exponential growth might be beneficial for many proteins, and induction is obtained at the end of exponential growth, when the biomass is maximal.
Quantification of PZn zitR induction levels to define optimal conditions for use.
To precisely quantify expression levels obtained in SA medium with different EDTA concentrations, cytoplasmic β-galactosidase LacLM was chosen as a reporter. Strain MG1363(pVE8065) was grown overnight on 1 μM Zn2+-supplemented SA medium, washed, diluted with normal SA medium, grown to mid-exponential phase (OD600, 0.25), and then divided into subcultures which received different amounts of EDTA (final concentrations of 0, 10, 30, 50, 300, and 500 μM) or Zn2+ (1 μM) as a control. β-Galactosidase activity of cell samples collected at different times was determined (Fig. 4).
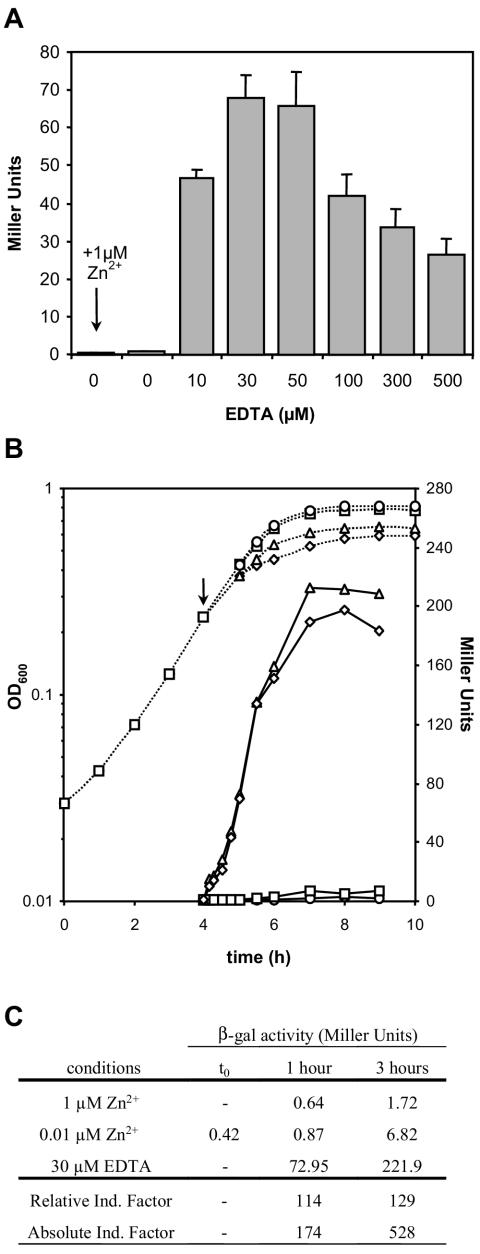
FIG.4.
Induction of β-galactosidase production as a function of EDTA concentration and time. An exponential-phase culture (OD600, 0.2) of MG1363(pVE8065) grown in SA medium was divided into aliquots that were exposed either to the addition of various EDTA concentrations (0, 10, 30, 50, 100, 300, and 500 μM) or, as a control, to the addition of Zn2+ (1 μM). (A) Histogram showing β-galactosidase activity of LacLM produced after 1 h of induction. (B) Growth (dotted lines) and β-galactosidase activity (solid lines) drawn as a function oftime in the presence of 1 μM Zn2+ (circles), normal SA medium (squares), EDTA at 30 μM (triangles), or EDTA at 50 μM (diamonds). The arrow indicates the moment of induction (time zero [t0]). (C) Induction factors for PZn zitR-controlled expression calculated as the ratio of induced (30 μM EDTA) β-galactosidase (β-gal) activity at the indicated times to repressed (1 μM Zn2+) β-galactosidase activity at the indicated times (Relative Induction [Ind.] Factor) or to β-galactosidase activity at t0 (Absolute Induction Factor). The values represent the means of at least three independent measures; standard deviations are shown only in panel A.
At 1 h after addition, cells that had not been exposed to EDTA showed little or no enzymatic activity (Fig. 4A), regardless of Zn2+ addition; this result suggests that the Zn2+ concentration, even in normal SA medium, is high enough to maintain repression. This result is in agreement with that observed for the natural induction of UspNuc, which was hardly detectable after 4 h, in cultures whose OD600 had reached 0.4 (Fig. 3). A clear induction of β-galactosidase activity was observed in all cultures treated with EDTA. Increasing amounts were observed for 10 and 30 μM EDTA, while 50 μM EDTA had essentially the same effect as 30 μM EDTA; concentrations of 100 μM or higher reduced both LacLM production and growth, indicating toxicity (Fig. 4A). Maximal induction, obtained in cultures treated with 30 μM EDTA, reached >100-fold compared to the basal level detected under repression conditions (Fig. 4C). Similar results were estimated for UspNuc production by use of a quantification method for nuclease activity (8, 45), despite its elevated basal level (data not shown).
β-Galactosidase assays were also performed at different times up to 5 h after the addition of the inducer or the control (Fig. 4B). In contrast to the situation observed with Zn2+-supplemented SA medium, in normal SA medium, a slight increase in the β-galactosidase activity level was observed as a function of the age of the culture (Fig. 4B); this result suggests consumption of zinc in the medium and the beginning of a natural induction event. The induction factor under these conditions (∼64-fold after 5 h) is higher (although of the same order) than the previously estimated one (Fig. 3), confirming that the β-galactosidase assay is more accurate than the nuclease activity test. Finally, a very strong increase in enzymatic activity was observed in cultures treated with 30 and 50 μM EDTA after 3 to 4 h of exposure (Fig. 4B), reaching an induction factor higher than 500-fold compared to the basal level (Fig. 4C). These results define 3 h of induction with 30 μM EDTA as the optimal conditions for use of the PZn zitR system to induce maximal gene expression.
Comparison with the standard NICE protein expression system.
To evaluate the usefulness of the PZn zitR expression system, we compared its efficiency to that of the NICE system (PnisA nisRK), the most commonly used expression system in L. lactis (29). The PZn zitR promoter-regulator system was replaced in pVE8064 (PZn zitR uspnuc) and pVE8065 (PZn zitR lacLM) by the PnisA promoter, giving rise to plasmids pVE8066 (PnisA uspnuc) and pVE8067 (PnisA lacLM) (Fig. 1C). This plasmid design, with an identical backbone vector, allowed direct comparison of the PZn zitR and PnisA expression systems, once pVE8066 and pVE8067 had been transferred into strain NZ9000 (MG1363 nisRK+) to allow PnisA system regulation (30).
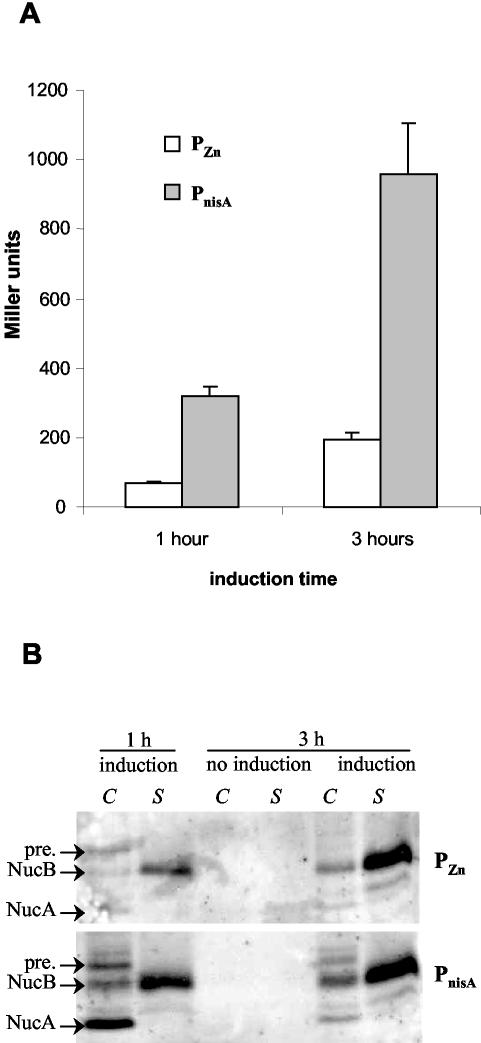
For comparing LacLM production levels, strains MG1363(pVE8065) and NZ9000(pVE8067) were induced with 30 μM EDTA and with 1 ng of nisin/ml, respectively (i.e., the recommended nisin concentration for optimal induction under laboratory conditions) (1; P. Langella, personal communication), for 1 or 3 h, and cells were analyzed for β-galactosidase activity. PnisA gave fivefold higher levels of enzymatic activity than did PZn in the first 3 h of induction (Fig. 5A) (322 and 68 units of β-galactosidase, respectively, after 1 h of induction; 959 and 196, respectively, after 3 h of induction).
FIG. 5.
Comparison of the PZn zitR and PnisA nisRK induction systems. (A) β-Galactosidase LacLM production driven by PZn zitR or by PnisA after 1 or 3 h of induction. Activity was measured from exponential-phase cultures of MG1363(pVE8065) (PZn) and MG1363(pVE8067) (PnisA) induced with 30 μM EDTA and with 1 ng of nisin/ml, respectively. Noninduced cultures produced no significant β-galactosidase activity in either system (data not shown). (B) Western blot analysis of cellular (C) and supernatant (S) fractions of 1- and 3-h-induced or noninduced cultures of MG1363(pVE8064) (PZn, upper panel) or MG1363(pVE8066) (PnisA, lower panel). NucA, NucB, and precursor (pre.) forms of UspNuc are shown.
We also examined the production of UspNuc by Western blotting of both cellular and supernatant fractions with an antiserum specific for S. aureus nuclease. Three different forms were observed: the precursor, exclusively in the cell fraction; the NucB form (after signal peptide processing), which is mainly secreted into the medium; and NucA, a form resulting from HtrA surface protease processing (43). After 1 h of induction, PnisA produced considerably larger amounts of UspNuc forms than did PZn, with an accumulation of the NucA processed form in the cellular fraction (Fig. 5B). NucA is believed to be entirely translocated and to remain cell surface associated (by electrostatic interactions) after the secretion process has been completed (43). Both systems gave essentially the same yields after 3 h of induction, with the cell-associated NucA form being hardly detectable in both instances (Fig. 5B). These results suggested that while there might be a slowdown of induction for PnisA, production driven by PZn might be continuous, resulting in similar final yields of protein.
Taken together, our results indicate that PZn zitR allows induction factors comparable to those obtained with PnisA under classical laboratory conditions.
DISCUSSION
A new inducible gene expression system was developed by use of the L. lactis zit operon, encoding an emergency Zn2+ uptake ABC transporter and its regulator (3, 42; D. Llull et al., unpublished data). Cloning of the promoter-regulator region of the system (PZn zitR) on a shuttle vector is sufficient to obtain controlled expression of reporter genes in L. lactis; divalent cation starvation leads to upregulation, whereas concentrated Zn2+ maintains repression. Chromosomal zit mRNA detection (Northern blotting) confirmed that zit is regulated by environmental zinc concentrations: it is repressed by an excess of Zn2+ and induced under conditions of extreme zinc depletion (at a Zn2+ concentration of <10 nM) (Llull et al., unpublished). Repression depends on the binding of Zn2+-ZitR to PZn (Llull et al., unpublished).
The absence of PZn zitR regulation in E. coli is not due to problems with ZitR expression (ZitR can be overproduced in E. coli and is stable) (Llull et al., unpublished) and may reflect a lower Zn2+ cytoplasmic content. It has been demonstrated that free Zn2+ does not persist in the E. coli cytoplasm (the concentration is 6 orders of magnitude less than one atom per cell), probably due to a very high zinc binding capacity (38) and/or the presence of three efflux systems (20). It is therefore possible that the L. lactis cytoplasmic environment is different from that of E. coli and/or that the affinity of ZitR for zinc is not high enough to compete with that of E. coli zinc binding proteins. Alternatively, as DNA fragments from AT-rich organisms, such as Streptococcus pneumoniae, can function as low-efficiency promoters in E. coli (13), the L. lactis PZn zitR sequence (62.5% AT) may lead to constitutive reporter expression.
The efficiency of the developed PZn zitR expression system was demonstrated in this study by the conditional production of two different reporter proteins, one secreted and one cytoplasmic. The system can be strongly repressed in SA medium by the addition of 1 μM Zn2+ without any growth defect, representing an obvious advantage for the overproduction of heterologous proteins. Otherwise, Zn2+ depletion is the necessary condition for induction. It was obtained either (i) by the addition of EDTA, a divalent cation chelator agent that removes zinc from the medium, or (ii) by gradual zinc starvation of the culture due to bacterial growth. In the latter situation, limited amounts of zinc in chemically defined SA medium are consumed or become unavailable, promoting a progressive increase in expression in mid-exponential phase of growth. This phenomenon can also be observed in SA medium supplemented with Zn2+ at a moderate concentration (0.1 μM), although in that situation it is delayed to the beginning of stationary phase (Fig. 3). Both induction conditions are interesting for practical applications. The use of EDTA allows accurate control of the time and rate of induction, leading to maximal induction factors (100- to 500-fold). On the other hand, natural PZn zitR induction may prevent problems of protein quality or stability due to overly rapid folding under maximal induction conditions and may also be useful when the required level of protein overproduction is moderate (∼50-fold) (Fig. 3 and 4).
EDTA is an efficient inducer of PZn zitR. A 100-fold induction factor for PZn zitR-controlled expression was reached when LacLM was induced by the addition of subinhibitory amounts of EDTA (30 μM) for 1 h, and a 500-fold induction factor was reached 2 h later (Fig. 4C). EDTA is simple to use, inexpensive, and readily available in molecular biology laboratories. Furthermore, EDTA is a food additive (for information, please visit website http://www.chem.ox.ac.uk/mom/EDTA/EDTA.html), suggesting the possible adaptation of the PZn zitR system to food-grade conditions.
Induction factors obtained with the PZn zitR system are much higher than those obtained with other reported expression systems (11), except for the NICE system, where the induction factor reaches 1,000-fold (10). After cloning in the presence of identical genetic backbone vectors, PnisA directed the synthesis of fivefold more β-galactosidase than did PZn zitR (Fig. 5A). One drawback of the original NICE system is that protein overproduction with the inducible PnisA promoter requires the presence of the nisRK genes in the strains (30); an improved vector containing both PnisA and the nisRK genes has been developed and should prove useful (4). Like the NICE system (16), the PZn zitR system should also be useful in different gram-positive bacteria. Our expression system is functional in two lactococcal strains, MG1363 and IL1403 (data not shown). Furthermore, our results concerning the nuclease reporter (Fig. 5B) suggest that the continuous slower production rate of PZn could be advantageous for secretion compared to the faster production rate of PnisA.
Finally, the availability of multiple possibilities for expression control allows one to choose the most suitable expression system for a particular gene of interest and to coordinate the efficient expression of two distinct genes in a single bacterial system.
Acknowledgments
We thank B. Cesselin for DNA sequencing and P. Regent for photography. We are indebted to P. Langella, J.-C. Piard, S. Madsen, and B. Michel for kindly providing, respectively, pSEC:Nuc, pVE5239, pAMJ769, and TG1pcnB. We thank our colleagues from Unité de Recherches Laitières et Génétique Appliquée for helpful discussions during the course of this work and A. Gruss and M. van de Guchte for critical reading of the manuscript. Thanks are also due to S. Madsen, who generously gave useful advice about the LacLM reporter.
This work was partially funded by the Fondation pour la Recherche Médicale. D. Llull was the recipient of a Marie Curie Individual Fellowship from the European Commission.
REFERENCES
- 1.Bermúdez-Humarán, L., P. Langella, J. Commissaire, S. Gilbert, Y. Le Loir, R. L'Haridon, and G. Corthier. 2003. Controlled intra- or extracellular production of staphylococcal nuclease and ovine omega interferon in Lactococcus lactis. FEMS Microbiol. Lett. 224:307-313. [DOI] [PubMed] [Google Scholar]
- 2.Bermúdez-Humarán, L., P. Langella, A. Miyoshi, A. Gruss, R. T. Guerra, R. Montes de Oca-Luna, and Y. Le Loir. 2002. Production of human papillomavirus type 16 E7 protein in Lactococcus lactis. Appl. Environ. Microbiol. 68:917-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 5.Chatel, J. M., P. Langella, K. Adel-Patient, J. Commissaire, J. M. Wal, and G. Corthier. 2001. Induction of mucosal immune response after intranasal or oral inoculation of mice with Lactococcus lactis producing bovine betalactoglobulin. Clin. Diagn. Lab. Immunol. 8:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 7.Christie, G. E., P. J. Farnham, and T. Platt. 1981. Synthetic sites for transcription termination and a functional comparison with tryptophan operon termination sites in vitro. Proc. Natl. Acad. Sci. USA 78:4180-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuatrecasas, P., S. Fuchs, and C. B. Anfinsen. 1967. Catalytic properties and specificity of the extracellular nuclease of Staphylococcus aureus. J. Biol. Chem. 242:1541-1547. [PubMed] [Google Scholar]
- 9.David, S., S. Herman, M. van Riel, G. Simons, and W. M. de Vos. 1992. Leuconostoc lactis β-galactosidase is encoded by two overlapping genes. J. Bacteriol. 174:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Ruyter, P. G. G. A., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vos, W. M. 1999. Gene expression systems for lactic acid bacteria. Curr. Opin. Microbiol. 2:289-295. [DOI] [PubMed] [Google Scholar]
- 12.Dieye, Y., S. Usai, F. Clier, A. Gruss, and J. C. Piard. 2001. Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol. 183:4157-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillard, J. P., and J. Yother. 1991. Analysis of Streptococcus pneumoniae sequences cloned into Escherichia coli: effect of promoter strength and transcription terminators. J. Bacteriol. 173:5105-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dintilhac, A., G. Alloing, C. Granadel, and J.-P. Claverys. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25:727-739. [DOI] [PubMed] [Google Scholar]
- 15.Dintilhac, A., and J.-P. Claverys. 1997. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res. Microbiol. 148:119-131. [DOI] [PubMed] [Google Scholar]
- 16.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garrido, M. E., M. Bosch, R. Medina, M. Llagostera, A. M. Pérez de Rosas, I. Badiola, and J. Barbé. 2003. The high-affinity zinc-uptake system znuACB is under control of the iron-uptake regulator (fur) gene in the animal pathogen Pasteurella multocida. FEMS Microbiol. Lett. 221:31-37. [DOI] [PubMed] [Google Scholar]
- 19.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hantke, K. 2001. Bacterial zinc transporters and regulators. Biometals 14:239-249. [DOI] [PubMed] [Google Scholar]
- 21.Hantke, K. 2002. Members of the Fur protein family regulate iron and zinc transport in E. coli and characteristics of the Fur-regulated fhuF protein. J. Mol. Microbiol. Biotechnol. 4:217-222. [PubMed] [Google Scholar]
- 22.Hazlett, K. R. O., F. Rusnak, D. G. Kehres, S. W. Bearden, C. J. La Vake, M. E. La Vake, M. E. Maguire, R. D. Perry, and J. D. Radolf. 2003. The Treponema pallidum tro operon encodes a multiple metal transporter, a zinc-dependent transcriptional repressor, and a semi-autonomously expressed phosphoglycerate mutase. J. Biol. Chem. 278:20687-20694. [DOI] [PubMed] [Google Scholar]
- 23.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca Mn2+ permease in Streptococcus gordonii is regulated by diphteria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 25.Janulczyk, R., J. Pallon, and L. Bjorck. 1999. Identification and characterization of a Streptococcus pyogenes ABC transporter with multiple specificity for metal cations. Mol. Microbiol. 34:596-606. [DOI] [PubMed] [Google Scholar]
- 26.Jensen, P. R., and K. Hammer. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleerebezem, M., M. M. Beerthuyzen, E. E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuipers, O. P., M. M. Beerthuyzen, P. G. G. A. de Ruyter, E. J. Luesnik, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 29.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1997. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 15:135-140. [DOI] [PubMed] [Google Scholar]
- 30.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 31.Le Loir, Y., A. Gruss, S. D. Ehrlich, and P. Langella. 1994. Direct screening of recombinants in gram-positive bacteria using the secreted staphylococcal nuclease as a reporter. J. Bacteriol. 176:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Loir, Y., S. Nouaille, J. Commissaire, L. Bretigny, A. Gruss, and P. Langella. 2001. Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl. Environ. Microbiol. 67:4119-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopilato, J., S. Bortner, and J. Beckwith. 1986. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol. Gen. Genet. 205:285-290. [DOI] [PubMed] [Google Scholar]
- 34.Madsen, S. M., J. Arnau, A. Vrang, M. Givskov, and H. Israelsen. 1999. Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis. Mol. Microbiol. 32:75-87. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Miyoshi, A., I. Poquet, V. Azevedo, J. Commissaire, L. Bermúdez Humarán, E. Domakova, Y. Le Loir, S. C. Oliveira, A. Gruss, and P. Langella. 2002. Controlled production of stable heterologous proteins in Lactococcus lactis. Appl. Environ. Microbiol. 68:3141-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okabayashi, K., and D. Mizuno. 1974. Surface-bound nuclease of Staphylococcus aureus: localization of the enzyme. J. Bacteriol. 117:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488-2492. [DOI] [PubMed] [Google Scholar]
- 39.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199-1210. [DOI] [PubMed] [Google Scholar]
- 40.Patzer, S. I., and K. Hantke. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321-24332. [DOI] [PubMed] [Google Scholar]
- 41.Peschke, U., V. Beuck, H. Bujard, R. Gentz, and S. Le Grice. 1985. Efficient utilization of Escherichia coli transcriptional signals in Bacillus subtilis. J. Mol. Biol. 186:547-555. [DOI] [PubMed] [Google Scholar]
- 42.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poquet, I., V. Saint, E. Seznec, N. Simoes, A. Bolotin, and A. Gruss. 2000. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042-1051. [DOI] [PubMed] [Google Scholar]
- 44.Ravn, P., J. Arnau, S. M. Madsen, A. Vrang, and H. Israelsen. 2000. The development of TnNuc and its use for the isolation of novel secretion signals in Lactococcus lactis. Gene 242:347-356. [DOI] [PubMed] [Google Scholar]
- 45.Ravn, P., J. Arnau, S. M. Madsen, A. Vrang, and H. Israelsen. 2003. Optimization of signal peptide SP310 for heterologous protein production in Lactococcus lactis. Microbiology 149:2193-2201. [DOI] [PubMed] [Google Scholar]
- 46.Ribeiro, L. A., V. Azevedo, Y. Le Loir, S. C. Oliveira, Y. Dieye, J. C. Piard, A. Gruss, and P. Langella. 2002. Production and targeting of the Brucella abortus antigen L7/L12 in Lactococcus lactis: a first step toward food-grade live vaccines against brucellosis. Appl. Environ. Microbiol. 68:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Solioz, M., and J. V. Stoyanov. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27:183-195. [DOI] [PubMed] [Google Scholar]
- 49.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Asseldonk, M., W. M. de Vos, and G. Simons. 1993. Cloning, nucleotide sequence, and regulatory analysis of the Lactococcus lactis dnaJ gene. J. Bacteriol. 175:1637-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Asseldonk, M., G. Rutten, M. Oteman, R. J. Siezen, W. M. de Vos, and G. Simons. 1990. Cloning of usp45, a gene encoding secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 95:155-160. [DOI] [PubMed] [Google Scholar]