Abstract
We developed for Bacteria in environmental samples a sensitive and reliable mRNA fluorescence in situ hybridization (FISH) protocol that allows for simultaneous cell identification by rRNA FISH. Samples were carbethoxylated with diethylpyrocarbonate to inactivate intracellular RNases and pretreated with lysozyme and/or proteinase K at different concentrations. Optimizing the permeabilization of each type of sample proved to be a critical step in avoiding false-negative or false-positive results. The quality of probes as well as a stringent hybridization temperature were determined with expression clones. To increase the sensitivity of mRNA FISH, long ribonucleotide probes were labeled at a high density with cis-platinum-linked digoxigenin (DIG). The hybrid was immunocytochemically detected with an anti-DIG antibody labeled with horseradish peroxidase (HRP). Subsequently, the hybridization signal was amplified by catalyzed reporter deposition with fluorochrome-labeled tyramides. p-Iodophenylboronic acid and high concentrations of NaCl substantially enhanced the deposition of tyramides and thus increased the sensitivity of our approach. After inactivation of the antibody-delivered HRP, rRNA FISH was performed by following routine protocols. To show the broad applicability of our approach, mRNA of a key enzyme of aerobic methane oxidation, particulate methane monooxygenase (subunit A), was hybridized with different types of samples: pure cultures, symbionts of a hydrothermal vent bivalve, and even sediment, one of the most difficult sample types with which to perform successful FISH. By simultaneous mRNA FISH and rRNA FISH, single cells are identified and shown to express a particular gene. Our protocol is transferable to many different types of samples with the need for only minor modifications of fixation and permeabilization procedures.
One of the long-standing goals of environmental microbiology is to simultaneously ascertain the identity, activity, and biogeochemical impact of individual organisms in situ. For identification, fluorescence in situ hybridization (FISH) with rRNA-targeted probes is routinely used. To link a specific metabolic activity to the identity of a microorganism in situ, without introducing alterations due to incubation with tracers or substrates, it is necessary to detect either a key enzyme of a particular process or its mRNA. In situ hybridization (ISH) of mRNA sequences is a popular technique for studying gene expression in eukaryotic cells and tissues. Unfortunately, microbiology lacks stable protocols for in situ detection and quantification of gene expression in environmental samples.
Since the first ISH experiments (18), mainly radioactive nucleotides have been used to synthesize probes (9). The advantage of radiolabeled probes is their ability to detect very low levels of transcripts. The major limitations are poor spatial resolution and long exposure times for microautoradiography, depending on the radioisotope used and the amount of target molecules in the cell. More recently, the application of nonradioactively labeled nucleotides (e.g., biotin-UTP and digoxigenin [DIG]-UTP) considerably improved ISH (8, 21, 27). Of all the nonradioactive labeling methods developed, DIG-based detection has proven to be the most appropriate for rare transcripts. Since DIG is synthesized only in plants of the genus Digitalis, background problems due to nonspecific antibody binding in cells of other organisms are avoided (8).
In many mRNA ISH protocols, precipitating substrates are used for chromogenic detection of probes (5). Recently, however, fluorescence-labeled tyramides proved to be considerably more sensitive for immunochemical detection systems (34, 35). Catalyzed reporter deposition (CARD) has been applied to increase signal intensities in various immunochemical and FISH applications (23, 29, 33, 40). Through the use of horseradish peroxidase (HRP), many tyramide molecules, preconjugated with either haptens or fluorescent reporters, are deposited in close vicinity to the HRP binding site, resulting in superior spatial resolution. In combination with HRP-labeled antibodies, the CARD-FISH method has the potential to detect low-abundance mRNAs, probably even a single copy (28, 29, 33, 40).
Here we report an improved protocol for the simultaneous detection of mRNA and rRNA in environmental microorganisms. As a case study, we used the expression of pmoA, which encodes subunit A of particulate methane monooxygenase (pMMO). This is the first enzyme in the methane oxidation pathway, catalyzing the oxidation of methane into methanol. There are two distinct types of methane monooxygenase (MMO) enzymes: a soluble, cytoplasmic enzyme complex (sMMO) and membrane-bound pMMO. Virtually all methanotrophic Bacteria possess pMMO. pmoA is often used as a phylogenetic marker and as a marker for methanotrophy (22, 31, 32). Copper ions have been shown to play a key role in regulating MMO expression. When the copper/biomass ratio is high, pMMO is expressed and sMMO expression is inhibited (22). For inducible genes, such as the MMO genes, the presence of the respective mRNAs is an indicator for an ongoing metabolic process. With a combination of mRNA FISH and rRNA FISH, key players of biochemical processes can be identified.
MATERIALS AND METHODS
Sample collection and preparation.
Bathymodiolus azoricus specimens were collected in July 2001 at the northern Mid-Atlantic Ridge site Rainbow (36°13′N, 33°54′W; depth, 2,350 m). Gills were fixed in 4% (wt/vol) paraformaldehyde, dehydrated in increasing alcohol series and xylene, and embedded in paraffin. Paraffin sections of 4 μm were cut and mounted on aminosilane-coated slides. To prevent mixing or loss of different solutions during hybridization and antibody reactions, each section was encircled with a thin film of a 1:1 mixture of silicone and toluene. After being air dried, the slides were kept at 4°C in the dark until further processing.
Sediment samples from the Baltic Sea were collected in March 2002 at Eckernförde in the Kiel Bight, at a water depth of 8 m, by using a multicorer. The upper 10 cm of the sediment was mixed with water from the same site (1:1), and the slurry was incubated for 4 weeks at 12°C with an air-methane headspace (80:20 [vol/vol]) on a rotary shaker at 80 rpm. Sediment samples were fixed in 2% (vol/vol) formaldehyde for 30 min at room temperature (RT), centrifuged, and washed once with phosphate-buffered saline (PBS) (pH 7.6) and twice with 50% ethanol in PBS. Cells then were resuspended in absolute ethanol and stored at −80°C until further processing.
Cultures of Methylocystis echinoides 10491 (14) were grown at 37°C on a rotary shaker (100 rpm) in NMS medium (38) supplemented with trace element solution SL10A (39). In cultures used for the repression of pmoA expression, CuCl2 was omitted from the trace element solution. Cultures were fixed in 2% (vol/vol) formaldehyde for 30 min at RT, centrifuged, and washed once with PBS and twice with 50% ethanol in PBS. Cells then were resuspended in absolute ethanol and stored at −80°C until further processing.
PCR amplification.
Almost complete fragments of pmoA from B. azoricus symbionts, M. echinoides, and sediment samples were amplified from DNA extracts as described previously (6). Templates for probe synthesis were PCR amplified with a T7 polymerase promoter on either the forward primer for the control probe or the reverse primer for the mRNA-targeted antisense probe. PCR conditions were the same as those described above. Amplicons were precipitated, and the correct length of 450 nucleotides was checked on a 1.5% (wt/vol) agarose gel. Concentrations of T7 templates were determined photometrically (NanoDrop Technologies, Rockland, Del.).
Cloning and expression of pmoA.
pmoA from B. azoricus symbionts was cloned and expressed by using vector pBAD according to the manufacturer's instructions (Invitrogen, Karlsruhe, Germany). Overnight cultures of Escherichia coli Top10 were diluted 1:100 and grown for 2 h at 37°C. Cells were induced with 0.2% l-arabinose for 3 h and subsequently fixed and stored as described above. pmoA amplicons from sediment samples were cloned (TOPO TA; Invitrogen) and sequenced (ABI Prism 3100 genetic analyzer; Perkin-Elmer, Boston, Mass.) for phylogenetic analysis.
Processing.
All of following steps were performed with either RNase-free plasticware or baked glassware. Water and buffers were treated with 0.1% (vol/vol) diethylpyrocarbonate (DEPC; Sigma, Taufkirchen, Germany) and then autoclaved.
Probe synthesis.
Probes for pmoA from B. azoricus symbionts and M. echinoides and a probe mixture targeting different pmoA mRNAs from sediment bacteria were synthesized under the following conditions. The transcription reaction mixture (30 μl) was mixed in the following order: 8 μl of RNase-free water, 3 μl of 10 × nucleotide mixture (10 mM ATP, 10 mM CTP, 10 mM GTP, 10 mM UTP), 3 μl of dithiothreitol (100 mM), 3 μl of 10× transcription buffer (400 mM Tris-HCl [pH 8.0], 60 mM MgCl2, 20 mM spermidine), 10 μl of T7 template DNA (100 ng μl−1), and 3 μl of T7 polymerase (50 U μl−1; Epicentre, Madison, Wis.). This mixture was incubated at 37°C for 2 h. To remove the template DNA, 1 μl of RNase-free DNase (1 U μl−1; Epicentre) was added, and the mixture was incubated at 37°C for another 15 min and subsequently purified by precipitation. The length of the transcript was checked on a 1.5% (wt/vol) agarose gel. The concentration was determined photometrically.
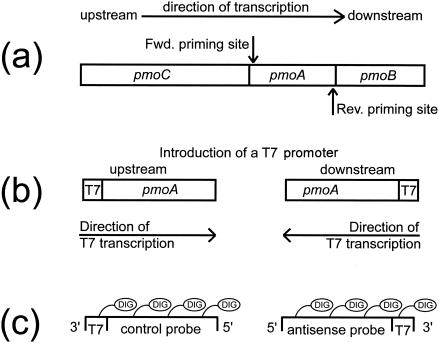
Transcripts were chemically labeled with cis-platinum-linked DIG (DIG-Chem-link; Roche Diagnostics, Mannheim, Germany) by following the manufacturer's recommendations with one modification: the reaction temperature was decreased to 75°C to reduce probe hydrolysis. Alternatively, probes can be labeled with DIG-UTP during the transcription reaction. In that case, the following nucleotide mixture must be used: 10 mM ATP, 10 mM CTP, 10 mM GTP, 3.5 mM DIG-UTP, and 6.5 mM UTP. Purification and determination of the concentrations of the probes were done as described above. Polyribonucleotide probes were diluted with deionized, particle-free water (MilliQ; Millipore, Eschborn, Germany) to a concentration of 10 ng μl−1. Ten-microliter portions were stored at −80°C until use. Figure 1 shows an overview of probe synthesis.
FIG. 1.
Flow diagram of probe synthesis. (a) MMO genes. (b) Subunit A PCR amplified with a T7 polymerase promoter on either the forward (Fwd.) primer (left) or the reverse (Rev.) primer (right). (c) PCR amplicons used as templates for probe synthesis.
Pretreatment of tissue sections.
B. azoricus gill sections were dried at 58°C for 20 min and then deparaffinized through successive baths of xylene (three times for 5 min each time), 96% ethanol (two times for 5 min each time), and 70% ethanol (once for 5 min). For carbethoxylation and inactivation of endogenous RNases, slides were treated with freshly prepared 0.1% (vol/vol) active DEPC in PBS for 12 min at RT with gentle mixing and then rinsed with PBS and MilliQ water for 1 min each. To allow penetration of detection molecules (probes and antibodies), the tissue was digested with proteinase K (7 μg ml−1) (from Tritirachium album; 30 mAnson U mg−1; Merck, Darmstadt, Germany) in 50 mM Tris-HCl-5 mM EDTA (pH 8) (TE) for 15 min at RT. Slides then were washed three times in MilliQ water for 1 min each time, dehydrated in 70 and 96% ethanol for 1 min each, air dried, and processed for FISH.
Pretreatment of sediment and pure cultures.
Quantities of 2 to 10 μl of sediment, M. echinoides cultures, or pmoA clones were spotted on slides (Superfrost Plus; Menzel, Braunschweig, Germany) in fields encircled with a silicone-toluene mixture. Slides were dried at 46°C, covered with 0.1% (wt/vol) agarose (MetaPhor; Bioproducts, Rockland, Maine), dried at 46°C, dehydrated in absolute ethanol for 1 min, and dried at RT.
To reduce background fluorescence, slides with sediment samples were covered with 1 ml of 0.2 M HCl and incubated at RT for 10 min. Slides then were washed in 50 ml of PBS and carbethoxylated by incubation in 50 ml of freshly prepared 0.1% (vol/vol) DEPC in PBS for 12 min at RT; the latter was removed by washing in PBS and MilliQ water for 1 min each. Cells were digested with highly purified lysozyme (5 mg ml−1) (from chicken egg white; nuclease free; Sigma) in TE for 30 min at RT by pipetting 1 ml of lysozyme solution onto the slides. Lysozyme then was washed off with 50 ml of MilliQ water. Sediment samples were treated with proteinase K (1 μg ml−1) in TE for 15 min at RT as described before. Slides then were washed three times in MilliQ water, washed once in absolute ethanol, dried, and processed for FISH.
Cultures were carbethoxylated and treated with lysozyme as described before. Proteinase K digestion was omitted.
FISH of mRNA.
A step-by-step protocol for mRNA FISH is shown in Table 1. Slides were covered with 100 μl of hybridization buffer, containing 50% (vol/vol) formamide, 2× SSC (1× SSC is 15 mM sodium citrate plus 0.15 M sodium chloride), 10% (wt/vol) dextran sulfate, 1% (wt/vol) blocking reagent (Boehringer, Mannheim, Germany), Denhardt's solution (Sigma), yeast RNA (0.2 mg ml−1; Ambion, Huntington, United Kingdom), and sheared salmon sperm DNA (0.2 mg ml−1; Ambion). Slides were placed in a chamber humidified with 50% (vol/vol) formamide-2× SSC and prehybridized for 60 min at 58°C. A total of 100 μl of hybridization buffer was mixed with 50 ng of transcript probe and incubated for 5 min at 80°C in order to denature the probe. The probe mixture was added to the prehybridized preparations to obtain a probe concentration of 250 ng ml−1. Slides were hybridized overnight at 58°C and then washed in 1× SSC-50% (vol/vol) formamide at 58°C for 1 h and in 0.2× SSC-0.01% (wt/vol) sodium dodecyl sulfate (SDS) at 58°C for 30 min. Slides were blocked in PBS-0.5% blocking reagent for 30 min at RT and then incubated with an anti-DIG-HRP antibody (0.75 U ml−1; Fab fragments; Roche) in PBS-1% (wt/vol) blocking reagent for 1 h at 37°C. Unbound antibody was removed by three washes in PBS for 10 min each. The antibody was detected by incubation with fluorescein-labeled tyramide (0.25 to 0.5 μg ml−1) in amplification buffer (PBS [pH 7.6], 0.1% [wt/vol] blocking reagent, 10% [wt/vol] dextran sulfate, 2 M NaCl, freshly added 0.0015% [vol/vol] H2O2) for 5 min at RT. For double or triple hybridizations, the antibody was detected with Alexa488-labeled tyramide (0.25 to 0.5 μg ml−1). p-Iodophenylboronic acid (IPBA) was added to the tyramide solution (20 mg of IPBA per 1 mg of tyramide in dimethylformamide) to further enhance CARD (3). All tyramides were custom labeled as described previously (24). Slides were washed in PBS for 3 min and three times with MilliQ water for 1 min each time. For subsequent microscopy, preparations were dehydrated with 50% ethanol and absolute ethanol and then dried.
TABLE 1.
Protocol for mRNA FISHa
| Step(s) | Procedures |
|---|---|
| Sample fixation | 2% Formaldehyde in PBS, 30 min at RT. |
| Sample preparation and immobilization | |
| Sediment and cultures | Centrifuge, wash two times with 50% ethanol in 2% Formaldehyde in PBS. Resuspend in absolute ethanol and store at −80°C. Immobilize by pipetting onto glass slides, let dry, cover with 0.1% agarose, let dry, wash in absolute ethanol, and let dry. |
| Tissue | Paraffinize, cut sections, and mount onto slides. Store at 4°C. |
| Pretreatment and permeabilization | |
| Cultures | Incubate in 0.1% active DEPC in PBS for 12 min at RT and then wash in PBS and MilliQ water. Incubate in lysozyme (5 mg ml−1 in TE) for 30 min at RT. Wash in MilliQ water, wash in absolute ethanol, and let dry. |
| Sediment | Wash in 0.2 M HCl for 10 min at RT. Incubate in 0.1% active DEPC in PBS for 12 min at RT and then wash in PBS and MilliQ water. Incubate in lysozyme (5 mg ml−1 in TE) for 30 min at RT and wash in MilliQ water. Incubate in proteinase K (1 μg ml−1 in TE) for 15 min at RT. Wash three times in MilliQ water, wash in absolute ethanol, and let dry. |
| Tissue sections | Deparaffinize. Rehydrate in absolute ethanol and 70% ethanol. Incubate in 0.1% active DEPC in PBS for 12 min at RT and then wash in PBS and MilliQ water. Incubate in proteinase K (7 μg ml−1 in TE) for 15 min at RT. Wash in MilliQ water. Dehydrate in 70% ethanol and absolute ethanol and let dry. |
| Hybridization | Prehybridize in hybridization buffer without probe for 1 h at hybridization temperature. Denature probe in hybridization buffer at 80°C for 5 min. Add to cells or tissue sections (final concn, 250 ng μl−1) and hybridize overnight at 58°C. |
| Posthybridization washes | Wash in 1× SSC-50% formamide for 1 h at hybridization temperature. Wash in 0.2× SSC-0.01% SDS for 30 min at hybridization temperature. |
| Immunocytochemical analysis | Block in PBS-0.5% blocking reagent at RT for 30 min. Incubate with antibody (0.75 U ml−1) in PBS-1% blocking reagent at 37°C for 1 h. Wash three times in PBS. |
| CARD | Incubate in amplification buffer containing fluorescence-labeled tyramide (0.25-0.5 μg ml−1) for 5 min at RT. Wash in PBS, MilliQ water, and absolute ethanol. |
Parameters to be optimized for each type of sample are shown in bold type.
FISH of 16S rRNA.
For subsequent 16S rRNA hybridization with HRP-labeled oligonucleotide probes, the anti-DIG-HRP antibody was inactivated by incubation with 0.01 M HCl for 10 min at RT. Preparations were washed with PBS and MilliQ water. FISH with HRP- or fluorochrome-labeled oligonucleotide probes was performed as described previously (24, 25). For 16S rRNA FISH of sediment, we used probe EUB338 I-III (7) for the detection of Bacteria, probe ALF968 (10) for the detection of α-Proteobacteria, probe Gam42a (20) for the detection of γ-Proteobacteria, and probe MTMC701 (4) for detection of the γ-proteobacterial methylotrophic genera Methylomonas, Methylobacter, Methylococcus, and Methylomicrobium. All probes were HRP labeled and detected with Alexa546-labeled tyramide. For 16S rRNA FISH of B. azoricus symbionts, we used Cy5-labeled probe Baz_meth_845I for methanotrophic symbionts and Cy3-labeled probe Baz_thio_193 for thiotrophic symbionts (C. Bergin and N. Dubilier, unpublished data).
Microscopic evaluation.
Spotted cells and tissue sections were covered with 4′,6′-diamidino-2-phenylindole (DAPI)-amended mounting solution and evaluated on a Axioplan II microscope (Carl Zeiss, Jena, Germany) equipped with an HBO 100-W Hg vapor lamp, with appropriate filter sets for Cy3 and Alexa546 (Chroma, Brattleborough, Conn.), fluorescein and Alexa488 (Zeiss09; Zeiss), and DAPI (Zeiss01; Zeiss) fluorescence, and with ×40, ×64, and ×100 Plan Apochromat objectives. Images of triple hybridizations of B. azoricus gills and double hybridizations of sediment were taken with a confocal laser scanning microscope (LSM510; Zeiss, Göttingen, Germany) equipped with helium and neon lasers (633 and 543 nm) and an argon laser (488 nm).
RESULTS AND DISCUSSION
The success of ISH for mRNAs depends strongly on the integrity of the target mRNAs in the cell. Another requirement is a powerful reporter system capable of revealing small numbers of probe-mRNA hybrids while keeping background staining as low as possible. Variables that influence the sensitivity and reproducibility of the ISH technique include (i) the effects of cell fixation on target mRNA preservation and accessibility to probes, (ii) the type and the quality of probes, (iii) the efficiency of hybrid formation, (iv) the stability of hybrids formed in situ during posthybridization treatments, (v) the method of detection of hybrids, and (vi) background noise masking the hybridization signal. A step-by-step-protocol optimized for the detection of pmoA mRNA is shown in Table 1. Here we describe and discuss variables that had to be optimized.
Sample pretreatment.
Major problems with in situ detection of mRNA are a low abundance of target molecules (usually there would be fewer than 100 copies of a particular mRNA) and their degradation by intracellular RNases. Acetylation has been recommended for the majority of mRNA ISH protocols to inactivate RNases and to decrease the background signal (12). More recently, Braissant and Wahli (5) introduced carbethoxylation of tissue sections by DEPC treatment. Carbethoxylation irreversibly inactivates RNases, which otherwise might be reactivated under renaturation conditions, e.g., during digestion with proteinase K. Since carbethoxylation is also easier to perform than acetylation, we chose DEPC treatment for our protocol. As in the study by Braissant and Wahli (5), DEPC treatment led to stronger and more consistent hybridization signals. A time series of DEPC treatment was performed, and 12 min of incubation at RT was found to be optimal (data not shown).
Hybridization temperature.
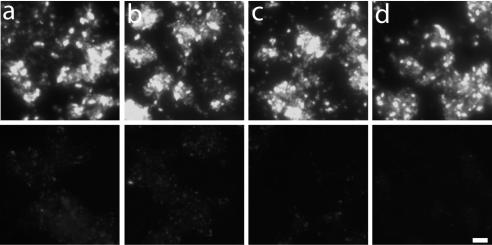
The specific hybridization temperature used depends on the length and the complexity of probes. A clone expressing pmoA of B. azoricus symbionts was hybridized at 50, 54, 58, and 62°C with the respective sense and antisense pmoA probes as well as the sense and antisense pmoA probes for sediment methanotrophs. Very low hybridization signals were observed with both control probes at 50 and 54°C. At 58°C, the signal of the control probes disappeared completely, whereas the signal of both antisense probes remained high even at a hybridization temperature of 62°C (shown for symbiont pmoA in Fig. 2). All hybridizations were therefore performed at 58°C.
FIG. 2.
Hybridization of an E. coli clone expressing pmoA of B. azoricus symbionts with a pmoA antisense probe (upper panels) and a pmoA control probe (lower panels) at different hybridization temperatures: 50°C (a), 54°C (b), 58°C (c), and 62°C (d). Bar, 10 μm.
Controls.
For the detection of gene expression in environmental microorganisms, at least two controls are needed. (i) The specificity of a probe for induced and noninduced target organisms must be shown. This option might often be unavailable for microorganisms in environmental samples. Instead, the probe must be tested with induced and noninduced expression clones. A similar strategy was recently described for the optimization of hybridization temperatures for rRNA-targeted oligonucleotide probes for uncultured Bacteria (26). (ii) A control probe which is the reverse complement to the antisense probe must be used for all hybridizations of environmental samples. This control is needed to ensure that the signal obtained by the antisense probe is due to hybridization and not due to “sticking” of the probe to any kind of material in the sample.
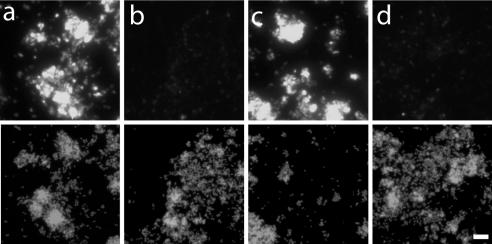
The α-proteobacterial methylotroph M. echinoides was grown with or without copper in the medium to favor or repress pmoA expression. It is known that copper is a cofactor of pMMO (14). In the presence of copper depletion, many methylotrophic Bacteria down-regulate pmoA mRNA expression but up-regulate sMMO. As expected, M. echinoides cells grown with copper showed strong fluorescence with the respective pmoA mRNA antisense probe, whereas cells grown without copper showed only a very low hybridization signal (Fig. 3).
FIG. 3.
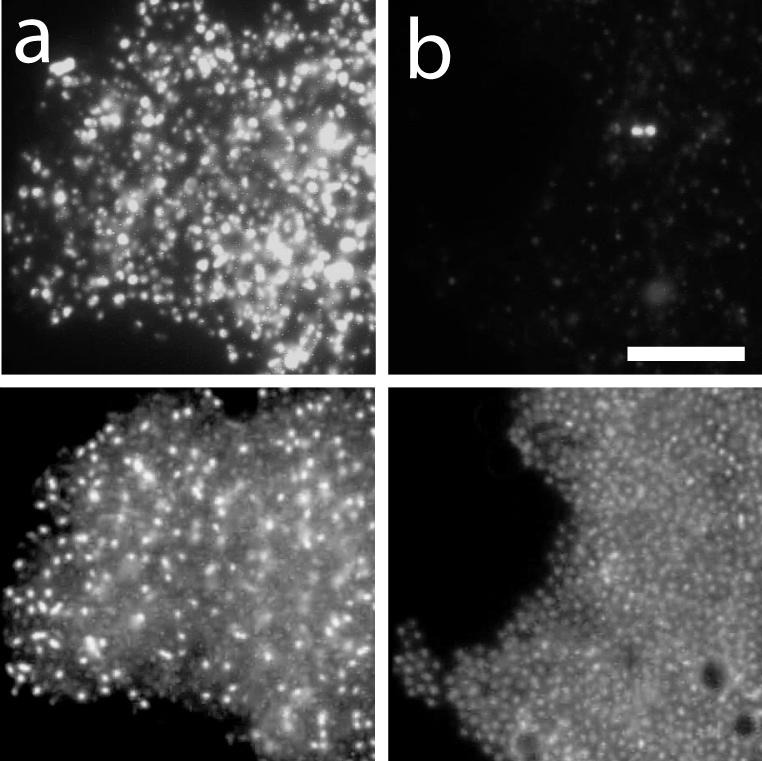
Hybridization of M. echinoides with an antisense probe targeting pmoA mRNA. (Upper panels) Probe signal. (Lower panels) DAPI staining. (a) Grown with Cu2+ for pmoA expression. (b) Grown without Cu2+ to inhibit pmoA expression. Bar, 10 μm.
The specificity of the pmoA mRNA antisense probe for B. azoricus symbionts was tested with an expression clone. Antisense probes showed strong hybridization signals (Fig. 4a), whereas no signals were observed in uninduced cells (Fig. 4b). This expression clone was also used to test the pmoA mRNA probes for sediment methanotrophs, which were also positive in induced cells (Fig. 4c) and negative in uninduced cells (Fig. 4d). These results imply that pmoA of B. azoricus symbionts is closely related to pmoA sequences in the sediment samples. The antisense probe for pmoA mRNA of M. echinoides did not hybridize with this expression clone.
FIG. 4.
Analysis of E. coli clone expressing pmoA of B. azoricus symbionts. (Upper panels) Hybridized cells. (Lower panels) DAPI-stained cells. Induced cells (a and c) and noninduced cells (b and d) were hybridized with an antisense probe targeting pmoA of B. azoricus symbionts (a and b) and an antisense probe mixture targeting pmoA in Eckernförde sediment (c and d). Bar, 10 μm.
Cell fixation and permeabilization.
The extent of fixation and the extent of permeabilization are very important considerations (40). Formaldehyde is a cross-linking fixative most appropriate for ISH of tissues and cell suspensions. However, overfixation can inhibit the penetration of probes and antibodies (5). We used different fixatives for gill tissues (4% paraformaldehyde) and cell suspensions (2% formaldehyde).
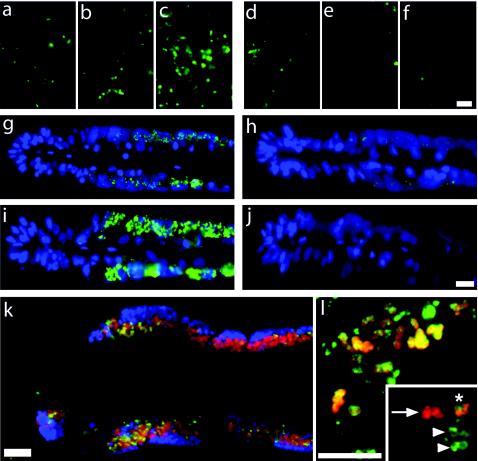
Upon fixation with formaldehyde, cell permeabilization is required to enhance probe penetration and hence the strength of the hybridization signal. Cell permeability that is too low can lead to false-negative or even false-positive results (Fig. 5a to j). However, overdigestion must also be avoided, since it results in the loss of target molecules, poor cell morphology, or even the loss of cells. In order to ensure the diffusion of transcript probes and antibodies into target cells and to prevent the diffusion of mRNA out of the cells, it is necessary to test different fixation and permeabilization procedures in order to find the optimum for the respective type of sample.
FIG. 5.
(a to f) Effect of permeabilization on hybridization signals in sediment pretreated with 0.1% DEPC. Each triple panel depicts antisense probe staining (a to c) and control probe staining (d to f). (a and d) Nonpermeabilized. (b and e) Permeabilized with lysozyme. (c and f) Permeabilized with lysozyme followed by proteinase K digestion. Bar, 10 μm. (g to j) Effect of permeabilization on hybridization signals in cross sections of B. azoricus gills. pmoA mRNA hybridization after treatment with different proteinase K concentrations is shown. Each double panel depicts hybridization with an antisense probe (g and i) and a control probe (h and j). (g and h) Proteinase K at 0.3 μg ml−1. (i and j) Proteinase K at 7 μg ml−1. Bar, 20 μm. (k) Triple hybridization of B. azoricus gill sections in a composite of three images: red, methanotrophic symbionts hybridized with 16S rRNA-targeted, Cy5-labeled probe Baz_meth_845I; blue, thiotrophic symbionts hybridized with 16S rRNA-targeted, Cy3-labeled probe Baz_thio_193; green, pmoA mRNA antisense probe. Bar, 10 μm. (l) Double hybridization of sediment microbes in a composite of two images: red, 16S rRNA hybridized with probe MTMC701-HRP for the detection of Methylomonas, Methylobacter, Methylococcus, and Methylomicrobium; green, pmoA mRNA. Images were taken with a confocal laser scanning microscope and represent a reconstruction of 21 optical z slices. Bar, 10 μm. (Inset) Reconstruction of four optical z slices. The arrow indicates cells with only a 16S rRNA FISH signal; the arrowheads show cells with only a pmoA mRNA FISH signal; the asterisk indicates cells with both FISH signals.
For optimization of permeabilization, sediment and pure cultures were treated with lysozyme (5 mg ml−1) for 15 and 30 min. Also, mild digestion with proteinase K (0.01 to 2 μg ml−1) was tested, either after lysozyme treatment or alone. Pure cultures showed the best hybridization signals after 30 min of lysozyme treatment (data not shown); proteinase K digestion did not improve mRNA FISH signal intensities and was therefore omitted. Hybridization of pmoA mRNA in sediment showed the brightest signals with the antisense probe after 30 min of lysozyme treatment followed by digestion with proteinase K at 1 μg ml−1 (Fig. 5c). In suboptimally permeabilized sediment, false-positive signals were observed (Fig. 5d). Background fluorescence compromised the detection of pmoA-expressing cells in sediment and was even present when probes were omitted during hybridization and no antibody was used. It is possible that endogenous peroxidases were reactivated during posthybridization washes. Therefore, the sediment was washed with different HCl concentrations (0.01 to 0.5 M) for 10 min at RT. A concentration of 0.2 M HCl was found to be optimal (data not shown). However, hydrolysis of mRNA due to acid treatment must be taken into consideration.
For sections of B. azoricus gills, a proteinase K concentration series ranging from 0 to 10 μg ml−1 was performed (Fig. 5g to j), and 7 μg ml−1 was found to be optimal. Lower proteinase K concentrations resulted in a very low signal with the antisense probe and a background signal with the control probe. Higher proteinase K concentrations caused a loss of hybridization signals and background fluorescence in the host tissue.
ISH.
Hybridization buffers contain reagents to maximize nucleic acid duplex formation and inhibit nonspecific binding of probes. Formamide was used to lower the optimal hybridization temperature to minimize cell damage and encourage specific probe binding. Formamide is also a nuclease inhibitor and allows the salt concentration to be adjusted to physiological osmolarity. Detergents such as SDS and heterologous nucleic acids inhibit background signals due to charge or nonspecific interactions with nucleic acids. Prehybridization without a probe in the hybridization buffer proved to be crucial to avoid high background fluorescence (5, 30, 40).
Several approaches can increase the hybridization rate. The probe concentration should be in excess of the target mRNA concentration, resulting in a “probe-driver” condition. However, a probe concentration that is too high can result in an unacceptably high background signal. Different probe concentrations (0.01 to 1,000 ng ml−1) were tested. A concentration of 250 ng ml−1 was found to be optimal (data not shown). Dextran sulfate was included in the buffer as a high-molecular-weight “volume-excluding” polymer, in effect concentrating the probe into a smaller physical space and therefore increasing the hybridization rate (16, 37).
Probes and probe labeling.
The labeling of transcript probes also influences the quality of hybridization. Significantly more DIG can be incorporated into the probe (1 in 15 nucleotides) (13) by using chemical labeling with platinum-linked DIG than by using enzymatic labeling (1 in 25 nucleotides) (15). We therefore used chemically labeled probes in this study.
Oligonucleotide probes combined with CARD are sometimes used to detect mRNAs in cell cultures and tissues (30). We tested this approach and found that the sensitivity and the signal-to-noise ratio were much lower than those of mRNA FISH with long transcript probes (data not shown). Another argument against the use of oligonucleotides for the in situ detection of mRNA in environmental samples is that short probes might hybridize to other mRNAs. Since environmental genomics is still in its infancy, we cannot predict the specificity of a highly sensitive mRNA FISH approach with oligonucleotide probes. Therefore, the use of transcript probes is recommended. It is very unlikely that a transcript probe will hybridize to mRNA that is not closely related to the target mRNA.
Immunocytochemical analysis.
To test for nonspecific antibody binding, sediment samples were hybridized without a probe. Different blocking reagent concentrations (0.5, 1, and 3%) and different blocking times (30 min to 15 h) at RT and 4°C were tested. We found that 0.5% blocking reagent in PBS and 30 min of blocking time at RT were sufficient (data not shown).
Different antibody concentrations from 1.5 to 0.15 U ml−1 were tested. A decrease in the FISH signal intensity was observed at concentrations below 0.75 U ml−1. At higher concentrations, prolonged washing was required to remove excess antibody (data not shown). Therefore, a concentration of 0.75 U ml−1 was used for subsequent immunochemical detection.
For CARD, different amplification buffers and tyramides were tested. A commercial fluorescein-labeled tyramide signal amplification kit (TSA plus; Perkin-Elmer) gave lower FISH signal intensities than custom fluorescein-labeled tyramide in PBS (pH 7.6)-0.0015% H2O2 (data not shown). IPBA and high concentrations of NaCl enhance the deposition of some fluorescence-labeled tyramides by HRP (3). Each enhancer (2.2 M NaCl and 20 mg of IPBA per 1 mg of tyramide) as well as the combination of both was tested for all tyramides used. Substantial increases in FISH signal intensities for fluorescein-, Alexa488-, and Alexa546-labeled tyramides were observed. Background fluorescence could be decreased by the addition of 0.1% blocking reagent and 10% dextran sulfate to the amplification buffer. The combination of enhancers, a low concentration of custom-labeled tyramide, dextran sulfate, and blocking reagent during the amplification reaction increased the signal-to-noise ratio (data not shown).
Simultaneous mRNA FISH and rRNA FISH.
Using a combination of mRNA FISH and 16S rRNA FISH, we could show that the pmoA antisense probe specifically hybridized only with the methanotrophic symbionts in B. azoricus gills (Fig. 5k).
In sediment samples, methanotrophic γ-proteobacteria were enriched during incubation with methane, as revealed by 16S rRNA FISH with probe MTMC701, targeting Methylomonas, Methylobacter, Methylococcus, and Methylomicrobium. A BLAST (1) search for pmoA sequences derived from sediment DNA showed that most sequences (43 out of 49) were closely related to Bathymodiolus symbiont pmoA (about 90% sequence similarity), explaining why the heterologous pmoA mRNA antisense probe for sediment methanotrophs also targeted B. azoricus symbiont pmoA. The remaining sequences were more closely related to pmoA of Methylobacter and Methylococcus species (data not shown).
In sediment samples, all cells hybridizing with the pmoA antisense probe could be simultaneously hybridized with a probe targeting the 16S rRNA of Bacteria (data not shown). Many cells hybridizing with the pmoA antisense probe also hybridized with probe MTMC701 (Fig. 5l), showing that the majority of pmoA-expressing cells belonged to methylotrophic γ-proteobacteria.
Based on 16S rRNA analysis, the closest relatives of the methanotrophic symbionts of B. azoricus are Methylobacter and Methylomonas species (Bergin and Dubilier, unpublished), which were enriched during incubation of sediment samples with methane. pmoA sequences of Methylobacter, Methylomonas, and Methylococcus species are more closely related (nucleic acid similarity, 80%) to each other than to species of Methylocystis (nucleic acid similarity, 64%), an α-proteobacterial methylotroph (6). Due to low sequence similarity, the probe targeting pmoA mRNA of B. azoricus symbionts did not hybridize with M. echinoides cells (data not shown). In sediment samples, no α-proteobacteria could be detected by rRNA FISH, and no cells hybridized with the antisense probe for M. echinoides pmoA mRNA.
Ludwig et al. (19) used transcript probes (250 nucleotides) to target the 23S rRNA in bacterial pure cultures. In this study, the threshold of mismatch discrimination was found to be between 85 and 78% sequence similarity. However, the specificity of transcript probes strongly depends on the hybridization conditions, and fine-tuning of probe specificity should be done when discrimination between closely related target sequences is needed.
The presence of a pmoA mRNA FISH signal directly proves transcription of the respective gene and strongly supports the presence of active methanotrophic bacteria. However, the intensity of the FISH signal will at best provide semiquantitative information on the number of target molecules per cell. Several factors other than the amount of mRNA influence the FISH signal intensity. Immunodetection of hapten-labeled probes and tyramide signal amplification depends not only on the concentration of target molecules but also on the quality of the antibody as well as on the concentration and labeling of the tyramide. Furthermore, the amount of mRNA is not directly correlated with the amount or activity of the enzyme.
Few protocols have been introduced for FISH of mRNA in pure cultures (2, 11, 36). Wagner et al. (36) could detect high-copy-number mRNA in Listeria and combined that with 16S rRNA hybridization. Hurek et al. (17) detected nifH expression in an Azoarcus sp. growing in kallar grass by using transcript probes for hybridization. However, a true negative control was not used, since the control probe had a higher complexity and therefore a higher melting temperature than the antisense probe. Bakermans and Madsen (2) performed mRNA FISH for bacteria from contaminated groundwater. In that study, oligonucleotide probes were used and hybridization signals were close to background levels.
We present a sensitive and reliable FISH protocol that allows for the simultaneous detection of mRNA and rRNA in bacteria from different environmental samples. The only parameters that need to be adjusted for each sample type are fixation and permeabilization of target cells.
Acknowledgments
We are grateful to M. Krüger for sediment samples and M. echinoides cultures, C. Cavanaugh and Z. P. McKiness for B. azoricus specimens, C. Bergin for 16S rRNA probes and tissue sectioning, N. Dubilier for pmoA clones, and J. Pernthaler for encouraging us not to give up on method development. M. Tummers is acknowledged for the introduction to radioactive mRNA ISH. We thank the crews of the RV Littorina and the RV L'Atalante for assistance.
This work was supported by the Max Planck Society.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bakermans, C., and E. L. Madsen. 2002. Detection in coal tar waste-contaminated groundwater of mRNA transcripts related to naphthalene dioxygenase by fluorescent in situ hybridization with tyramide signal amplification. J. Microbiol. Methods 50:75-84. [DOI] [PubMed] [Google Scholar]
- 3.Bobrow, M. N., K. E. Adler, and A. Roth. April 2002. Enhanced catalyzed reporter deposition. U.S. patent 6,372,931 B1.
- 4.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 5.Braissant, O., and W. Wahli. 1998. A simplified in situ hybridization protocol using non-radioactively labeled probes to detect abundant and rare mRNAs on tissue sections. Biochemica 1:10-16. [Google Scholar]
- 6.Costello, A. M., and M. E. Lidstrom. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 8.Farquharson, M., R. Harvie, and A. M. McNcol. 1990. Detection of messenger RNA using a digoxigenin end-labeled oligonucleotide probe. J. Clin. Pathol. 43:423-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerfen, C. R. 1989. Quantification of in situ hybridization histochemistry for analysis of brain function, p. 79-97. In P. M. Conn (ed.), Methods in neuroscience. Academic Press, San Diego, Calif.
- 10.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn, D., R. I. Amann, and J. Zeyer. 1993. Detection of mRNA in Streptomyces cells by whole-cell hybridization with digoxigenin-labeled probes. Appl. Environ. Microbiol. 59:2753-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi, S., I. C. Gillam, A. D. Delaney, and G. M. Tener. 1978. Acetylation of chromosome squashes of Drosophila melanogaster decreases the background in autoradiographs from hybridization with [125I]-labeled RNA. J. Histochem. Cytochem. 26:677-679. [DOI] [PubMed] [Google Scholar]
- 13.Heetebrij, R. J., E. G. Talman, M. A. van Velzen, R. P. M. van Gijswijk, S. S. Snoeijters, M. Schalk, J. Wiegant, F. van de Rijke, R. M. Kerkhoven, A. K. Raap, H. J. Tanke, and H.-J. Houthoff. 2003. Platinum(II)-based compounds as nucleic acid labeling reagents: synthesis, reactivity, and applications in hybridization assays. Chem. Biochem. 4:573-583. [DOI] [PubMed] [Google Scholar]
- 14.Heyer, J., V. F. Galcenko, and P. F. Dunfield. 2002. Molecular phylogeny of type II methane-oxidizing bacteria isolated from various environments. Microbiology 148:2831-2846. [DOI] [PubMed] [Google Scholar]
- 15.Höltke, H.-J., and C. Kessler. 1990. Non-radioactive labeling of RNA transcripts in vitro with the hapten digoxigenin (DIG): hybridization and ELISA-based detection. Nucleic Acids Res. 18:5843-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hrabovszky, E., and S. L. Petersen. 2002. Increased concentrations of radioisotopically-labeled complementary ribonucleic acid probe, dextran sulfate, and dithiothreitol in the hybridization buffer can improve results of in situ hybridization histochemistry. J. Histochem. Cytochem. 50:1389-1400. [DOI] [PubMed] [Google Scholar]
- 17.Hurek, T., T. Egener, and B. R. Hurek. 1997. Divergence in nitrogenases of Azoarcus spp., Proteobacteria of the beta subclass. J. Bacteriol. 179:4172-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.John, H. A., M. L. Birnstiel, and K. W. Jones. 1969. RNA-DNA hybrids at the cytological level. Nature 223:582-587. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig, W., S. Dorn, N. Springer, G. Kirchhof, and K. H. Schleifer. 1994. PCR-based preparation of 23S rRNA-targeted group-specific polynucleotide probes. Appl. Environ. Microbiol. 60:3234-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 21.Morris, R. G., M. J. Arends, P. E. Bishop, K. Sizer, E. Duvall, and C. C. Bird. 1990. Sensitivity of digoxigenin- and biotin-labeled probes for detection of human papillomavirus by in situ hybridization. J. Clin. Pathol. 43:800-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murrel, C. J., B. Gilbert, and I. R. McDonald. 2000. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 173:325-332. [DOI] [PubMed] [Google Scholar]
- 23.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition (CARD) for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pernthaler, A., J. Pernthaler, and R. Amann. Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms. In G. E. A. Kowalchuk (ed.), Molecular microbial ecology manual, in press. Kluwer Academic Press, Dordrecht, The Netherlands.
- 25.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 26.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. A. Stahl. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 27.Singer, R. H., and D. C. Ward. 1982. Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinylated nucleotide analog. Proc. Natl. Acad. Sci. USA 79:7331-7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speel, E. J. M. 1999. Detection and amplification systems for sensitive, multiple-target DNA and RNA in situ hybridization: looking inside cells with a spectrum of colors. Histochem. Cell Biol. 112:89-113. [DOI] [PubMed] [Google Scholar]
- 29.Speel, E. J. M., P. Saremaslani, P. Komminoth, and A. H. N. Hopman. 1997. Card signal amplification: an efficient method to increase the sensitivity of DNA and mRNA in situ hydridization. Am. J. Pathol. 151:1499. [Google Scholar]
- 30.Speel, E. J. M., P. Saremaslani, J. Roth, A. H. N. Hopman, and P. Komminoth. 1998. Improved mRNA in situ hybridization on formaldehyde-fixed and paraffin-embedded tissue using signal amplification with different haptenized tyramides. Histochem. Cell Biol. 110:571-577. [DOI] [PubMed] [Google Scholar]
- 31.Stolyar, S., M. Franke, and M. E. Lidstrom. 2001. Expression of individual copies of Methylococcus capsulatus Bath particulate methane monooxygenase genes. J. Bacteriol. 183:1810-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tchawa, Y. M., P. F. Dunfield, P. Ricke, J. Heyer, and W. Liesack. 2003. Wide distribution of a novel pmoA-like gene copy among type II methanotrophs and its expression in Methylocystis strain SC2. Appl. Environ. Microbiol. 69:5595-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Corput, M. P. C., R. W. Dirks, R. P. M. van Gijlswijk, F. M. van de Rijke, and A. K. Raap. 1998. Fluorescence in situ hybridization using horseradish peroxidase-labeled oligodeoxynucleotides and tyramide signal amplification for sensitive DNA and mRNA detection. Histochem. Cell Biol. 110:431-437. [DOI] [PubMed] [Google Scholar]
- 34.Van Gijlswijk, R. P. M., H. J. M. A. A. Zijlmans, J. Wiegant, M. N. Bobrow, T. J. Erickson, K. E. Adler, H. J. Tanke, and A. K. Raap. 1997. Fluorochrome-labeled tyramides: use in immuno-cytochemistry and fluorescence in situ hybridization. J. Histochem. Cytochem. 45:375-382. [DOI] [PubMed] [Google Scholar]
- 35.Van Heusden, J., P. de Jong, F. Ramaekers, H. Bruwiere, M. Borgers, and G. Smets. 1997. Fluorescein-labeled tyramide strongly enhances the detection of low bromodeoxyuridine incorporation levels. J. Histochem. Cytochem. 45:315-320. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, M., M. Schmid, S. Juretschko, K. H. Trebesius, A. Bubert, W. Goebel, and K. H. Schleifer. 1998. In situ detection of a virulence factor mRNA and 16s rRNA in Listeria monocytogenes. FEMS Microbiol. Lett. 160:159-168. [DOI] [PubMed] [Google Scholar]
- 37.Wahl, G. M., M. Stern, and G. R. Starck. 1979. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc. Natl. Acad. Sci. USA 76:3683-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittenbury, R., K. C. Phillips, and J. F. Wilkinson. 1970. Enrichments, isolation and some properties of methane utilizing bacteria. J. Gen. Microbiol. 61:205-218. [DOI] [PubMed] [Google Scholar]
- 39.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In K.-H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 40.Yang, H., I. B. Wanner, S. D. Roper, and N. Chaudhari. 1999. An optimized method for in situ hybridization with signal amplification that allows the detection of rare mRNAs. J. Histochem. Cytochem. 47:431-445. [DOI] [PubMed] [Google Scholar]