Abstract
The abundance of Vibrio vulnificus in coastal environments has been linked to water temperature, while its relationship to salinity is less clear. We have developed a culture-independent, most-probable-number quantitative PCR approach to examine V. vulnificus population dynamics in Barnegat Bay, N.J. Based on the combined analysis of our results from Barnegat Bay and from the literature, the present data show that (i) V. vulnificus population dynamics are strongly correlated to water temperature and (ii) although the general trend is for V. vulnificus abundance to be inversely correlated with salinity, this relationship depends on salinity levels. Irrespective of temperature, high abundances of V. vulnificus are observed at 5 to 10 ppt, which thus appears to be the optimal salinity regime for their survival. At 20 to 25 ppt, V. vulnificus abundances show a positive correlation to salinity. Unsuccessful attempts to resuscitate V. vulnificus, combined with our inability to detect cells during the winter despite an assay adapted to detect viable but nonculturable (VBNC) cells, suggest that the decline and eventual disappearance of V. vulnificus from the water column during the winter months is due primarily to a significant reduction in population size and is not only the consequence of cells entering the VBNC state. These findings are in line with the hypothesis that the sediment serves as a refuge for a subpopulation of V. vulnificus over the winter and weather-driven mixing events during the spring initiate a summer bloom in the water column.
Vibrio vulnificus is a halophilic gamma proteobacterium endemic to temperate coastal waters. This bacterium is a major concern to public heath agencies and the shellfish industry because it is an opportunistic human pathogen found ubiquitously in the water column and sediment and as an intracellular symbiont of shellfish (5, 13, 49). Most environmental assessments of V. vulnificus have centered on coastal waters surrounding Gulf Coast states (23, 28, 30, 43, 46), and a few studies have reported the presence of this organism along the eastern (15, 41, 50) and western coasts (22) of the United States and the Mediterranean coast (3, 16). Together with laboratory experiments, these studies showed that V. vulnificus undergoes a striking seasonal fluctuation in estuarine waters, with water temperature being the major factor controlling the abundance of V. vulnificus. The highest abundances of this organism, both in the water column and in association with shellfish (43, 50), occur in warm waters (typically exceeding 20°C) of moderate salinity (ranging from 5 to 25 ppt) (8, 18, 20). The abundance as well as the incidence of V. vulnificus-related disease decreases with temperature to the point where it is usually undetectable during the winter months. Salinity has also been identified as playing a role in controlling V. vulnificus abundance, though its effect on V. vulnificus is less clear. Several studies have identified an inverse correlation between V. vulnificus abundance (50) and salinity, but others have suggested that the relationship is dependent on the range of salinity (30, 35).
Similar temperature-mediated dynamics have been shown in several other species of Vibrio. For example, the abundances of Vibrio cholerae and Vibrio parahaemolyticus, two human pathogens that occupy an ecological niche similar to that of V. vulnificus, also demonstrate a seasonal oscillation in estuaries (12, 19, 21, 25, 31). A previous paper describes a variation of two orders of magnitude in total Vibrio concentrations in Barnegat Bay, N.J., between the summer and the winter months (45). Identification of distinct warm-water and cold-water Vibrio populations in that study further established the importance of temperature in generating community patterns of abundance and distribution for the Vibrio genus. Despite overwhelming evidence linking temperature to V. vulnificus seasonal dynamics, it remains unresolved whether the decline in abundance during the winter is due directly to temperature-mediated mortality of V. vulnificus or to its host or vector or is the result of the population entering the viable but nonculturable (VBNC) state (10, 37, 42, 51). Because identification and enumeration of V. vulnificus in the past have relied primarily on culture-based techniques, cells in the VBNC state would have escaped detection.
Nucleic-acid-based detection and quantification methods allow enumeration of microorganisms independent of their culturability. PCR has been shown to be effective for detecting V. vulnificus in the VBNC state with the cytolysin gene as a target, albeit at reduced sensitivity compared to that for culturable cells (6). Quantitative PCR (QPCR) approaches, which offer increased sensitivity over fluorescence in situ hybridization for target quantitation, are increasingly being used to enumerate specific microbial populations (27, 32, 34, 44). Our laboratory's competitive QPCR approach using Vibrio-specific primers, followed by separation and quantification of the amplicons by constant denaturant capillary electrophoresis, enabled enumeration of co-occurring Vibrio species in Barnegat Bay (45). This approach provided insights into the seasonal succession of dominant Vibrio populations, but V. vulnificus was not detected in that study, likely because its population size was below the detection limit of the method, which, as for all coamplification techniques, was set at ∼100-fold less than the size of the most abundant population.
In the present paper, we expand on earlier findings and report on the population dynamics of V. vulnificus and its relationship to the physicochemical conditions of Barnegat Bay using a most-probable-number (MPN)-QPCR approach for cell enumeration. We also specifically address whether (i) the relationship between salinity and V. vulnificus abundance is dependent on salinity regime and (ii) the temperature-mediated population decline is due to entry of the population into the VBNC state, with the organisms thereby escaping detection by culture-based enumeration.
MATERIALS AND METHODS
Sample collection and processing.
Surface water samples from Barnegat Bay were collected at a town dock in Seaside Heights, N.J. (30°33′N, 74°1′W), between June 2001 and February 2003. Samples were collected bimonthly with the exception of the winter months (January, February, and March), during which samples were collected once a month. Water temperature and salinity were measured on-site with a field thermometer and a hand-held refractometer (Leica, Bannockburn, Ill.). The water samples were serially fractionated through 20-, 10-, and 5-μm-pore-size mesh nylon screens (Spectrum Laboratories, Inc., Rancho Dominguez, Calif.) to remove large particles and plankton. The seawater fraction filtered through 5-μm-pore-size mesh was used for all subsequent analyses. For DNA extraction and purification, 15 to 80 ml of fractionated seawater was filtered through 0.22-μm-pore-size polycarbonate filters (GE Osmonics, Minnetonka, Minn.) by gentle vacuum filtration and stored at −20°C immediately. Chlorophyll a concentration was determined from 10 or 15 ml of seawater filtered onto 0.45-μm-pore-size nitrocellulose filters (Millipore, Billerica, Mass.). The filters were frozen until extraction with acetone (24). The pigment concentration was measured with a TD700 fluorometer (Turner Designs, Sunnyvale, Calif.). The total bacterial abundance was enumerated from samples preserved in 3.7% formaldehyde by the acridine orange direct counting method (17).
Cultures and growth conditions.
The following bacterial strains were used in the present study: V. vulnificus (ATCC 27567), V. parahaemolyticus (ATCC 17802), Vibrio anguillarum (ATCC 19264 and ATCC 14181), Vibrio fischeri (ATCC MJ1), and Vibrio alginolyticus (ATCC 19208). All the cultures were grown in Luria-Bertani (LB) broth prepared with 25 ppt of artificial seawater (24) at either 37°C or room temperature, with gentle shaking.
V. vulnificus was induced into the VBNC state by using the methods of Linder and Oliver (29). Briefly, V. vulnificus was grown to mid-exponential phase, harvested via a series of centrifugation and wash steps to remove culture medium, and resuspended in artificial seawater. The cell suspension was kept at 4°C, and culturability was determined by plating aliquots of the culture on LB agar plates to check for colony formation. As the number of culturable cells decreased to <10 CFU ml−1, 5 to 10 ml of the cell suspension was filtered through a 0.22-μm-pore-size polycarbonate filter and transferred to LB agar plates to increase the detection limit of V. vulnificus. V. vulnificus was considered to have entered the VBNC state when <0.1 CFU ml−1 was detected by direct plating. In addition to culturability, the total number of bacteria in the cell suspension was monitored by the acridine orange direct counting method (17) with a Zeiss Axiovert epifluorescence microscope.
Subsamples of seawater (100 ml) from each sampling date were supplemented with 0.01% LB broth and incubated at 22°C in the dark for 48 h as an enrichment for V. vulnificus. Following incubation, 2 ml of the culture was filtered onto a 0.22-μm-pore-size polycarbonate filter for DNA extraction and purification followed by PCR (see below). This procedure was used to detect V. vulnificus and confirm the presence or absence of this bacterium in water samples as enumerated by MPN-QPCR (see below).
DNA extraction.
DNA was extracted and purified from filters using the PureGene cell and tissue DNA isolation kit (Gentra Systems, Minnesota, Minn.). The following modifications were made to the extraction protocol supplied by the manufacturer. Proteinase K was added to the cell lysis solution provided with the kit at a final concentration of 0.5 μg ml−1. Filters were incubated with this solution for 1 h at 55°C, followed by a 20-min incubation at 70°C to inactivate the proteinase K. During treatment of the samples with RNase A as suggested by the manufacturer, the samples were additionally treated with lysozyme at a final concentration of 1.0 mg ml−1 for 1 h at 37°C. Following the enzymatic treatments, cells were sonicated using a Microson Ultrasonic Cell Disruptor (Heat Systems Ultrasonic) at 3,500 cycles s−1 for three 10-s intervals to facilitate cell wall breakage. DNA isolated using the PureGene kit was further purified using the DNeasy extraction kit (QIAGEN Inc., Valencia, Calif.) to remove soluble substances carried over from the first round of extraction that occasionally inhibited DNA amplification by PCR.
Primers and PCR conditions.
Two sets of primers that target the eubacterial 23S rRNA gene were used in a nested format for the V. vulnificus MPN-QPCR assay. The primers for the first round of PCR were V23S108F (5′-CGATTTCCGAATGGGGAAACCC-3′), which was modified from a primer used by Anthony et al. (2), and V23S478R (5′-TTGCCTTTCCCTCAGGTACT-3′) (2). These primers target the 23S rRNA gene (rDNA) of a wide range of eubacteria, including Vibrio spp., and produce a 360-bp fragment after PCR. The product from the first round of PCR is then used as a template for the second round of PCR with the primers Vvul23S138F (5′-TAAGCCAGTATCATTGAGT-3′) and Vvul23S296 (5′-ACCGTTCGTCTAACACAT-3′) (4), which specifically target V. vulnificus 23S rDNA and yield a 160-bp fragment. Primer specificity was assessed by (i) nucleotide BLAST analysis (1), (ii) PCR amplification of DNA extracted from cultures of six Vibrio species (see “Cultures and growth conditions”), and (iii) comparing the melting curve and melting temperature of PCR products amplified from pure V. vulnificus DNA with those from environmental samples using the melt-curve analysis function within the iCycler thermal cycler (Bio-Rad Laboratories, Hercules, Calif.). The PCR conditions for the primers were optimized by varying the annealing temperatures from 58 to 68°C and from 48 to 54°C for the first and second rounds of PCR, respectively, and the MgCl2 concentrations from 2.0 to 2.8 mM.
MPN-QPCR assay.
Detection limits for the MPN-QPCR assay were established for culturable and VBNC V. vulnificus cells by using DNA extracted from pure cultures as well as from seawater samples spiked with known numbers of V. vulnificus. To test the PCR detection limit in the presence of background environmental DNA and potential PCR inhibitors, 80 ml of Barnegat Bay seawater was filtered through a 3.0-μm-pore-size polycarbonate filter. The use of 3.0-μm-pore-size polycarbonate filters excluded most of the prokaryotic cells and, thus, endogenous V. vulnificus. The filters were then spiked with a series of 10-fold dilutions of V. vulnificus cells (from 1 to 107 cells ml−1) harvested from mid-exponential-phase cultures, and DNA was extracted from the filters for PCR amplification. A filter that was not spiked with V. vulnificus served as the control. The same procedure was repeated with 1-, 2-, and 3-month-old cultures of V. vulnificus following induction of the VBNC state. The spiked samples (with culturable V. vulnificus only) were also used to examine the accuracy of the MPN-QPCR assay for enumerating V. vulnificus. For this purpose, extracted DNA was serially diluted, amplified, and then scored (see below).
DNA extracted from filters was unamended and/or diluted 1:10, 1:20, 1:40, 1:60, 1:80, or 1:100 in a 50-μg ml−1 glycogen solution, and 2 μl of each dilution was used as the template in PCR (0.025 U of Taq polymerase, 200 μM [each] deoxynucleoside triphosphates, 2.0 mM MgCl2, 1× buffer A [all PCR constituents were from Promega], and a 100 nM concentration of each primer in a total reaction volume of 20 μl). The series of dilutions from each sampling date were amplified in triplicate. Two microliters of the product from the first round of PCR was then used as the template for the second round of PCR with fresh reaction mixtures. The cycling conditions for PCR were as follows: for the first round, 94°C (4 min), 40 cycles of 94°C (1 min) and 65°C (1 min), and 65°C (10 min) (2); for the second round, 94°C (4 min), 35 cycles of 94°C (30 s), 49°C (1 min), and 72°C (1 min), and 72°C (10 min). All PCR were performed on an iCycler thermal cycler (Bio-Rad Laboratories). Amplification products were run in a 2.0% agarose gel containing ethidium bromide at 0.5 μg ml−1. The numbers of positive reactions (those that produced 160-bp amplification products) and negative reactions were scored for each sample.
Data analysis.
MPN calculations were performed using the program developed by Briones and Reichardt (7). Cell abundances were natural log transformed to achieve normal distribution prior to statistical analysis. Correlations between V. vulnificus abundance and the environmental parameters measured were determined using Spearman's coefficient of rank correlation (rs). Linear regressions were then conducted for each of the environmental factors, followed by a standardized multiple regression analysis. All the statistical analyses were performed using the SPSS software package (SPSS Inc., Chicago, Ill.).
A data set containing V. vulnificus abundances in a wide range of temperatures and salinities from this study and six other published studies that surveyed the abundance of V. vulnificus in coastal waters (Table 1) was compiled for comprehensively analyzing temperature and salinity effects on V. vulnificus across different environments and temperature and salinity regimes. V. vulnificus abundance was plotted as a function of temperature and salinity, and its abundance was correlated to temperature according to distinct salinity ranges as well as to the entire salinity range in the data set, and vice versa, using Spearman's rank correlation. For the analyses, salinity was divided into 5 to 10, 10 to 15, 15 to 20, 20 to 25, and >25 ppt, and temperature was divided into 0 to 10, 10 to 20, and 20 to 30°C.
TABLE 1.
Studies from which V. vulnificus abundances and corresponding temperature and salinity measurements were obtained for comparative analysis with the seasonal data from this study
| Sampling site(s) | Year of study | Method of identification and enumerationa | V. vulnificus abundance (cells ml−1) | Reference |
|---|---|---|---|---|
| Chesapeake Bay, Del. | 2002 | Fluorescent oligonucleotide probe direct counting procedure | 2-10,000 | Heidelberg et al. (15) |
| Charlotte Harbor, Fla. | 2001 | Membrane filtration followed by identification and enumeration on CPC agar | 50-3,000 | Lipp et al. (30) |
| Mobile, Ala. | 1997 | Most-probable-number assay with immunoassay identification | 0-251 | DePaola et al. (13) |
| Chesapeake Bay, Del. | 1996 | Plating with oligonucleotide probe identification and enumeration | 0-200 | Wright et al. (50) |
| Mobile Bay and Mississippi Sound, Ala. | 1994 | Most-probable-number assay with CPC agar identification | 32-2,511 | DePaola et al. (11) |
| Apalachicola Bay, Fla. | 1982 | Most-probable-number assay with TCBS agar plates and API identification kit | 0-7,000 | Tamplin et al. (43) |
CPC, colistin-polymyxin B-cellobiose; TCBS, thiosulfate-citrate-bile salt-sucrose.
RESULTS
DNA purification from environmental samples.
DNA isolation using the PureGene DNA extraction kit enabled detection of V. vulnificus in water samples spiked with as few as one or two target cells as enumerated by MPN-QPCR. Consequently, further examination of extraction efficiency with control DNA was not performed. Modifications to the DNA extraction protocol, however, were made to ensure that the same degree of DNA extraction efficiency was achieved for VBNC cells. Kondo et al. (26) demonstrated that, while in the VBNC state, the gram-negative bacterium V. cholerae possessed a thicker peptidoglycan layer than normal cells. Their observation led us to include a step in our DNA isolation protocol whereby samples were first treated with proteinase K to permeabilize the outer membrane and then treated with lysozyme, which specifically cleaves peptidoglycan cross-links, to disrupt the unusually thick cell walls of VBNC cells. Several samples collected during the summer of 2002 were not amplifiable even with the DNA purification protocols described above. For those samples, environmental DNA isolated by the PureGene method was subjected to an additional DNA purification step using the QIAGEN DNeasy kit. This step removed PCR inhibitors carried over from the first round of DNA purification without compromising the detection limit of the MPN-QPCR assay (data not shown).
MPN-QPCR assay specificity and detection limits.
The MPN-QPCR protocol employed in this study involves two rounds of PCR amplification, first with a pair of 23S rDNA-targeted universal eubacterial primers and then with a pair of V. vulnificus-specific primers in a nested format. The V23S108F-V23S478R primer pair targeted a variety of eubacteria, based on Ribosomal Database Project and GenBank database searches, and produced a strong 360-bp amplicon with V. vulnificus. The V. vulnificus-specific primer set amplified a 160-bp product in the second round of PCR from V. vulnificus only and not from the four other Vibrio species tested. The melting-curve profiles and melting temperatures of the 160-bp DNA fragment amplified from a pure culture of V. vulnificus and from Barnegat Bay water samples were identical, thus confirming the specificity of the V. vulnificus primer set (data not shown). This result also indicates that the cultured strain is representative of V. vulnificus in nature.
The detection limit of the nested PCR protocol was tested with Barnegat Bay water samples spiked with V. vulnificus in order to simulate the background in which the MPN-QPCR assay was to be used to enumerate V. vulnificus. The detection limit was normalized to the cell number, as opposed to the rDNA copy number, per milliliter so that V. vulnificus abundance is comparable to those measurements obtained from other environments as reported in the literature. V. vulnificus was detected by nested PCR at the entire range of cell numbers, from 1 to 107 cells ml−1, added to the environmental samples. In contrast, V. vulnificus could only be detected in samples spiked with more than 2.0 × 104 cells ml−1 (data not shown) when a single round of PCR with the specific primers was performed. The nested PCR approach thus achieved an approximately 10,000-fold increase in sensitivity, with a lower detection limit of 1 cell ml−1. A detection limit of 1 cell ml−1 was also achieved with 1-month-old cultures of V. vulnificus in the VBNC state, but the limit of detection increased with the age of the culture. After 2 months in the VBNC state, the detection limit increased by 10-fold, and by 3 months, the detection limit increased by 1,000-fold (data not shown). This decrease in detection sensitivity mirrors our inability to resuscitate them from the VBNC state using standard resuscitation techniques (36, 38, 40) after 2 months of incubation at 4°C (data not shown). As such, cells in laboratory cultures or field samples that could not be resuscitated were considered nonviable and no longer relevant to be detected in our study.
V. vulnificus population dynamics.
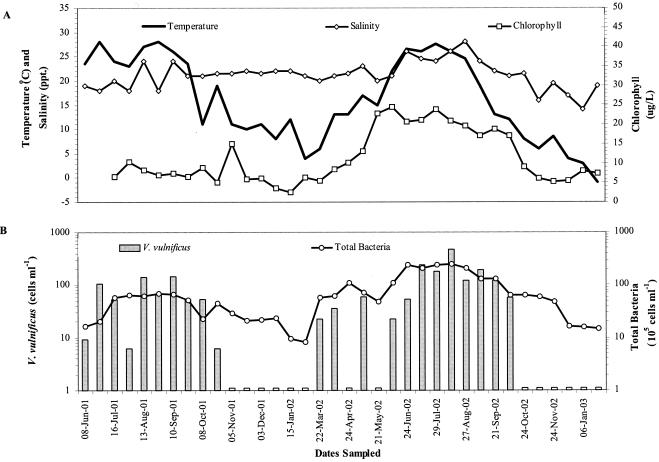
The abundance of V. vulnificus in Barnegat Bay fluctuated seasonally and ranged from undetectable to 480 cells ml−1 (Fig. 1). The water temperature during the time when V. vulnificus was detected averaged 21.2 ± 6.5°C. The highest seasonal abundance, from 6 to 480 cells ml−1, was observed during the summer months (June 21 through September 21). The average abundance of V. vulnificus in the summer of 2002 was 2.5-fold higher than in 2001 (Table 2). The average summer water temperature for each year was approximately 25°C, but salinity, chlorophyll a, and total bacteria in the summer of 2002 were significantly higher than in 2001 (by Student's t test, P was <0.005 for salinity; P was <0.001 for chlorophyll a and total bacteria). V. vulnificus was not detected during the late autumn (November) to winter months (December 21 through March 21), when the average water temperature was 8.0 ± 4.4°C for both sampling years. During the autumn (September 21 through December 21) and spring (March 21 through June 21) months, V. vulnificus abundance was intermediate between the summer and winter abundances or undetectable. The water temperature at this time of the year averaged 12.6 ± 4.2°C.
FIG. 1.
Seasonal fluctuations of temperature, salinity, and chlorophyll a (A) and total bacteria and V. vulnificus abundance (B) in Barnegat Bay, N.J. Open bars indicate sampling dates on which V. vulnificus was not detected.
TABLE 2.
Comparison of annual summer V. vulnificus abundance, total bacteria, chlorophyll a concentration, salinity, and temperaturea
| Study period | V. vulnificus (cells ml−1) | Total bacteria (cells ml−1) | Chlorophyll (μg liter−1) | Salinity (ppt) | Temperature (°C) |
|---|---|---|---|---|---|
| Summer 2001 | 85.7 ± 53.6 | 5.7 ± 1.8 × 106 | 7.9 ± 1.4 | 20.3 ± 2.9 | 26.0 ± 2.1 |
| Summer 2002 | 210.9 ± 146.6 | 2.1 ± 0.4 × 107 | 20.5 ± 2.2 | 25.4 ± 1.6 | 24.9 ± 3.1 |
Values are averages ± 1 standard deviation of measurements collected from June 21 to September 21. P values indicate levels of significance as determined by Student's t test and are <0.005 for salinity and <0.001 for chlorophyll a and total bacteria except for V. vulnificus and temperature measurements, which are not significant.
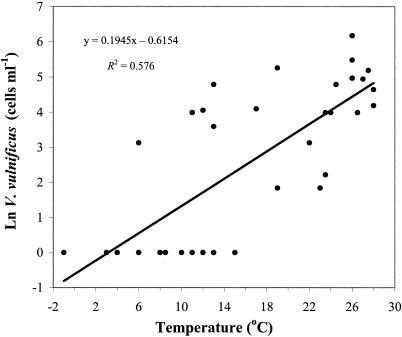
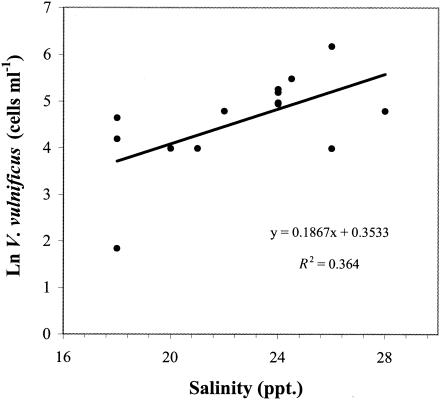
The abundance of V. vulnificus in Barnegat Bay was positively correlated to the water temperature (rs = 0.775; P < 0.001), total bacterial abundance (rs = 0.684; P < 0.001), chlorophyll a concentration (rs = 0.537; P < 0.005), and salinity (rs = 0.499; P < 0.005) over the entire sampling period (Table 3). Seasonal changes in water temperature explained nearly 60% of the variation in V. vulnificus abundance (Fig. 2). Multiple regression analysis including salinity, chlorophyll a, and total bacteria as variables revealed that these factors did not significantly contribute to the seasonal variation in V. vulnificus abundance. Temperature and chlorophyll a, but not salinity, together explained 70% of the variation observed in total bacterial abundance (P < 0.001). Thus, although chlorophyll a and total bacteria are strongly correlated with V. vulnificus abundances and are thus important indices for prediction, they are not directly responsible for the variability observed in V. vulnificus abundance because of the parallel effect of temperature on chlorophyll a, total bacteria, and V. vulnificus. During the summer, when temperature did not vary by more than 3°C, salinity was positively correlated to V. vulnificus and accounted for 36% of the fluctuation in abundance (P < 0.05) (Fig. 3).
TABLE 3.
Correlation between V. vulnificus abundance, total bacterial abundance, chlorophyll, salinity, and temperature in Barnegat Bay, N.J., over a 21-month period as determined by using Spearman's rank correlation coefficient (rs)a
| Parameter | Spearman's rank correlation coefficient for:
|
|||
|---|---|---|---|---|
| V. vulnificus | Total bacteria | Chlorophyll | Salinity | |
| Total bacteria (cells ml−1) | 0.684 | |||
| Chlorophyll (μg liter−1) | 0.537* | 0.719 | ||
| Salinity (ppt) | 0.499* | 0.569 | 0.392* | |
| Temperature (°C) | 0.775 | 0.587 | 0.511 | 0.398* |
*, P <0.005. For all unmarked values, P is <0.001.
FIG. 2.
Relationship between V. vulnificus abundance and temperature in Barnegat Bay from June 2001 to February 2003. For linear regression analysis, the limit of detection (1 cell ml−1) was used for samples in which V. vulnificus was not detected.
FIG. 3.
Relationship between the summer abundances of V. vulnificus and salinity in Barnegat Bay (June 21 to September 21 of 2001 and 2002).
Enrichment of water samples under conditions that promote the resuscitation of V. vulnificus from the VBNC state confirmed the presence or absence of V. vulnificus in the field samples and, thus, the robustness of our MPN-QPCR assay. V. vulnificus was detected in all the enrichment cultures started with field samples in which V. vulnificus could be enumerated by QPCR. In contrast, attempts to recover V. vulnificus from water samples during cold-weather months by either enrichment of water samples with LB broth or incubation at room temperature for 24 or 48 h (36, 48, 50) were unsuccessful, with the exception of two field samples collected in the fall and spring.
Combined analysis of temperature and salinity effects on V. vulnificus abundance.
The potential for temperature and salinity effects on V. vulnificus to vary according to different temperature and salinity regimes was examined using data from this study and an extended data set compiled from the literature (Table 1). V. vulnificus abundance is on the whole positively correlated to temperature (rs = 0.310; P < 0.001) and negatively correlated to salinity (rs = −0.451; P < 0.050) over the entire range of temperatures and salinities analyzed in the data set. However, at different salinity levels, the relationship does not follow the overall trend (Fig. 4). In the range of 5 to 10 ppt, V. vulnificus abundance is not correlated to temperature. High abundances are observed in this salinity range irrespective of temperature (range of 10 to 32°C). At 20 to 25 ppt, V. vulnificus abundance is positively correlated to salinity (rs = 0.354; P < 0.050).
FIG. 4.
V. vulnificus abundance as a function of temperature and salinity. The data was compiled from this study and the studies summarized in Table 1. Only abundances that are >1 cell ml−1 are plotted on this graph. The sizes of the bubbles are proportional to the relative abundance of V. vulnificus, which is grouped into nine categories: 1 to 50, 51 to 100, 101 to 250, 251 to 500, 501 to 750, 751 to 1,000, 1,001 to 2,500, 2,501 to 5,000, and >5,000 cells ml−1.
DISCUSSION
Most studies of V. vulnificus population dynamics to date have adopted a purely culture-based approach or have combined culturing with DNA probes to confirm positive cultures (28, 30, 33, 39, 41, 46, 50). Here we used a culture-independent QPCR approach for enumerating V. vulnificus to expand our understanding of the seasonal and annual population dynamics of V. vulnificus in Barnegat Bay, N.J., an estuary in which biotic factors (e.g., recurring episodes of brown tide and other algal blooms) and abiotic fluctuations (in particular, salinity and temperature) have a great impact on microbial community dynamics.
The MPN-QPCR strategy employed in this study provided increased detection sensitivity combined with quantitative precision that is as sensitive as previously published microbiological enumeration methods, such as monitoring of CFU and MPN analysis, but avoids the biases associated with selective culturing. The assay detected V. vulnificus in amplification reactions that contained as few as three target cells in a complex background of environmental DNA and particulate material from Barnegat Bay. This template concentration corresponds to 1 V. vulnificus cell ml−1 of spiked water samples. It has been argued that MPN-PCR estimates are only semiquantitative because all MPN procedures provide estimates and not absolute quantification. However, several aspects of our MPN-QPCR approach compensate for the lower precision. First, targeting multicopy genes, such as the rRNA gene, instead of single-copy genes increases detection sensitivity. The V. vulnificus genome was recently sequenced and annotated, showing the presence of nine copies of the 23S rRNA gene per genome (9). Second, primers flanking relatively short regions of the 23S rRNA gene for the first and second rounds of PCR (resulting in 360- and 160-bp fragments, respectively) were used, thereby allowing the PCR to be more efficient, because short amplicons are typically amplified at a much greater efficiency than long regions. Third, the incorporation of two rounds of PCR into a nested PCR format increased the detection sensitivity by 10,000-fold. Finally, environmental contaminants or PCR inhibitors present in a sample are minimized as a result of dilution in an MPN-based assay. The abundances measured in the Barnegat Bay samples were comparable to those in samples from similar estuarine environments, which suggests that the QPCR method used here provides reliable estimates of V. vulnificus abundance (28, 30, 41, 46, 50).
An important aspect of the development of the V. vulnificus assay was the ability to detect cells in the VBNC state, given the potential of these cells for resuscitation (5, 14, 38). Our MPN-QPCR method detected VBNC cells as effectively as culturable cells in the mid-exponential phase of growth. However, following a prolonged VBNC state (2 months or more of incubation at 4°C in the laboratory), the assay's ability to detect V. vulnificus decreased dramatically. This phenomenon coincided with our inability to resuscitate the cells from the VBNC state. Assuming that the failure to resuscitate the cells is an indication of loss of viability, our inability to detect such cells is due to cell death and gradual DNA degradation following loss of viability, which results in a lack of amplification. This sequence of events, in which the VBNC response of V. vulnificus is characterized by a loss of cultivability while cellular integrity and intact RNA and DNA (and thus possible viability) are maintained, followed by a gradual degradation of nucleic acids and reduction of viability, has been hypothesized by Weichart et al. (47) and Rice et al. (42). Based on these considerations, we assumed V. vulnificus to be absent or below the assay detection limit of 1 cell ml−1 in the water samples if they were not detected by our assay.
Previous studies that analyzed the relationship between salinity and V. vulnificus abundance have been contradictory. Kaspar and Tamplin (20) found that the survival of V. vulnificus was adversely affected by elevated salinities (>25 ppt). In field surveys, Wright et al. (50) showed that the abundance of V. vulnificus in the Chesapeake Bay was inversely correlated to salinity, while others did not reveal a significant relationship between V. vulnificus abundance and salinity (18, 41). Motes et al. (35) and Lipp et al. (30), on the other hand, found that the abundance of V. vulnificus in the Gulf of Mexico was positively correlated to salinity when the salinity was less than 15 ppt and inversely correlated to salinity when the salinity was greater than 15 ppt. The lack of consensus among the various studies may be due to the fact that the studies were conducted at sites with different salinity regimes and that temperature and the effects of salinity on V. vulnificus may be interdependent, as has been proposed by Kaspar and Tamplin (20). Based on the combined analysis of field data from this study and from the literature (Fig. 4), we found that the general trend is for V. vulnificus abundance to be inversely correlated with salinity. However, this relationship depends on the salinity level. Most notably, at 5 to 10 ppt, the survival of V. vulnificus appears to be optimal—high abundances of V. vulnificus are observed irrespective of temperature (range of 10 to 32°C). At 20 to 25 ppt, V. vulnificus abundance shows a positive correlation to salinity (Fig. 4). Interestingly, we demonstrated in this study that V. vulnificus abundance is positively correlated to salinity in Barnegat Bay, whereby 75% of the samples in which V. vulnificus was detected measured between 20 and 28 ppt (Table 2 and Fig. 1). The finding that the highest abundance of V. vulnificus documented in the field thus far falls in the range of 5 to 10 ppt is somewhat surprising, because this salinity range is at the lower limit of growth for V. vulnificus (20). While we have no mechanistic explanation for these observations, the results illustrate the complexity of physicochemical interactions on V. vulnificus population dynamics in the environment.
The seasonal pattern of abundance observed for V. vulnificus in Barnegat Bay, characterized by maximum population abundances during the summer months and their decline to undetectable levels during the cold winter months, applies across broad geographic and ecological areas (Fig. 4). Our results corroborate the finding of a recent study of V. vulnificus in a North Carolina estuary by Pfeffer et al. (41) that showed that water temperature accounts for almost 50% of the variability in V. vulnificus abundances (Fig. 3), thus establishing temperature as the major factor determining the seasonal population dynamics of the species. The water temperature threshold at which V. vulnificus could be detected was approximately 12°C, below which this organism was mostly undetectable in our study site and in various coastal environments in which the organism has been monitored by others (Fig. 4). This temperature threshold is in agreement with the temperature cutoff of 13°C that was empirically determined in previous laboratory experiments for the growth of V. vulnificus (20).
Two general hypotheses have been proposed to explain the temperature-mediated decline of V. vulnificus. The first hypothesis asserts that cold temperatures induce the population to enter a VBNC state until late spring to early summer, when warm temperatures resuscitate the cells and activate their growth (37, 40). The observed decline in population abundance and eventual disappearance from the water column may be indirectly due to the VBNC cells escaping detection by culture-based enumeration methods, which have been the traditional methods for quantifying V. vulnificus. The second hypothesis proposes that the population overwinters in the floc zone at the sediment-water interface, and in early spring, the cells are resuspended in the water column by weather-related perturbation of the water column (46). Our data from Barnegat Bay suggest that the VBNC-state hypothesis does not completely explain the disappearance of V. vulnificus from the water column during the winter. Repeated, unsuccessful attempts to resuscitate V. vulnificus combined with our inability to detect V. vulnificus during the winter months, despite using an assay adapted to detect VBNC cells, indicate that the population was absent from the water column in the winter or, if they are present in the VBNC state, that they are not a significant part of the community. Further data from our study suggest that the decline and eventual disappearance of V. vulnificus during the winter may be more consistent with the second hypothesis, which proposes that the sediment may serve as a refuge for a subpopulation of V. vulnificus over the winter. Levels of V. vulnificus in the sediment have been shown to be up to two orders of magnitude higher than those in the water column (30, 46). During the period of the present study, V. vulnificus was detected in one winter water sample with a temperature of 6°C. This sampling date coincided with a winter storm, which caused bottom mixing of the water column and caused the water sample to be visibly murky with particulate material. Although our sampling was limited during this storm event and sediment samples were not analyzed, it is possible that numbers of viable cells of V. vulnificus remained elevated in Barnegat Bay sediment during the winter and that mixing events during the spring resuspended these cells from the sediment, resulting in elevated numbers of viable cells in the water column. This process may indeed be the mechanism by which the water column is seeded with V. vulnificus during the spring, thus initiating the summer bloom in the water column (46).
Using culture-independent QPCR, our study has refined previous findings on the effects of temperature and salinity on V. vulnificus. We demonstrate that temperature and salinity are strong determinants of V. vulnificus abundance and population dynamics in coastal waters. In addition, we have shown that the population decline and eventual disappearance of V. vulnificus during the winter months are due primarily to a significant reduction of V. vulnificus abundance in the water column and are not solely the consequence of cells entering the VBNC state. The application of QPCR approaches for detecting small-scale changes in population abundance allows precise monitoring of oscillations in microbial populations and enables links between their population dynamics and prevailing physicochemical condition.
Acknowledgments
This work was supported by research grants to E.L. and M.F.P. from Seagrant and NOAA and a graduate research assistant fellowship to M.A.R. from the Biology Department, Temple University.
We thank Jonathan Vandergrift for general technical assistance.
REFERENCES
- 1.Altschul, S. F., M. L. Madden, J. Z. Schaffer, Z. Zheng, M. Web, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, R. M., T. J. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of the 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias, C. R., R. Aznar, J. Pujalte, and E. Garay. 1998. A comparison of strategies for the detection and recovery of Vibrio vulnificus from marine samples of the western Mediterranean coast. Syst. Appl. Microbiol. 21:128-134. [DOI] [PubMed] [Google Scholar]
- 4.Aznar, R., W. Ludwig, R. I. Amann, and K. H. Schleifer. 1994. Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of Vibrio vulnificus with rRNA-target oligonucleotide probes. Int. J. Syst. Bacteriol. 44:330-337. [DOI] [PubMed] [Google Scholar]
- 5.Barer, M. R., R. J. Smith, R. P. Cooney, and P. T. Kinnitt. 2000. Relationships between culturability, activity and virulence in pathogenic bacteria. J. Infect. Chemother. 6:108-111. [DOI] [PubMed] [Google Scholar]
- 6.Brauns, L. A., M. Hudson, and J. D. Oliver. 1991. Use of polymerase chain reaction in detection of culturable and nonculturable Vibrio vulnificus cells. Appl. Environ. Microbiol. 57:2651-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briones, A. M., and W. Reichardt. 1999. Estimating microbial population counts by ′most probable number' using Microsoft Excel. J. Microbiol. Methods 35:157-161. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, P. J., R. J. Steffan, A. DePaola, J. W. Foster, and A. K. Bej. 1999. Adaptive responses to cold temperature in Vibrio vulnificus. Curr. Microbiol. 38:168-175. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., K. Wu, C. Chang, H. Tsai, T. Liao, Y. Liu, H. Chen, A. Shen, J. Li, T. Su, C. Shao, C. Lee, L. Hor, and S. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colwell, R. R. 2000. Viable but nonculturable bacteria: a survival strategy. J. Infect. Chemother. 6:121-125. [DOI] [PubMed] [Google Scholar]
- 11.DePaola, A., G. M. Capers, and D. Alexander. 1994. Densities of Vibrio vulnificus in the intestines of fish from the U.S. Gulf Coast. Appl. Environ. Microbiol. 60:984-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePaola, A., J. L. Nordstrom, J. C. Bowers, J. G. Wells, and D. W. Cook. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69:1521-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePaola, A., S. McLeroy, and G. McManus. 1997. Distribution of Vibrio vulnificus phage in oyster tissues and other estuarine habitats. Appl. Environ. Microbiol. 63:2464-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer-Le Saux, M., D. Hervio-Heath, S. Loaec, R. R. Colwell, and M. Pommepuy. 2002. Detection of cytotoxin-hemolysin mRNA in nonculturable populations of environmental and clinical Vibrio vulnificus strains in artificial seawater. Appl. Environ. Microbiol. 68:5641-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heidelberg, J. F., K. B. Heidelberg, and R. R. Colwell. 2002. Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol. 68:5488-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hervio-Heath, D., R. R. Colwell, A. Derrien, A. Robert-Pillot, J. M. Fournier, and M. Pommepuy. 2002. Occurrence of pathogenic vibrios in coastal areas of France. J. Appl. Microbiol. 92:1123-1135. [DOI] [PubMed] [Google Scholar]
- 17.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Høi, L., J. L. Larsen, I. Dalsgaard, and A. Dalsgaard. 1998. Occurrence of Vibrio vulnificus in Danish marine environments. Appl. Environ. Microbiol. 64:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, S. C., and W. Fu. 2001. Seasonal abundance and distribution of Vibrio cholerae in coastal waters quantified by a 16S-23S intergenic spacer probe. Microb. Ecol. 42:540-548. [DOI] [PubMed] [Google Scholar]
- 20.Kaspar, C. W., and M. L. Tamplin. 1993. Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl. Environ. Microbiol. 59:2425-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaysner, C. A., C. Abeyta, Jr., M. M. Wekell, A. DePaola, Jr., R. F. Stott, and J. M. Leitch. 1987. Incidence of Vibrio cholerae from estuaries of the United States West Coast. Appl. Environ. Microbiol. 53:1344-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaysner, C. A., C. Abeyta, Jr., M. M. Wekell, A. DePaola, Jr., R. F. Stott, and J. M. Leitch. 1987. Virulent strains of Vibrio vulnificus isolated from estuaries of the United States West Coast. Appl. Environ. Microbiol. 53:1349-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly, M. T. 1982. Effects of temperature and salinity on Vibrio (Beneckea) vulnificus occurrence in a Gulf Coast environment. Appl. Environ. Microbiol. 44:820-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemp, P. F., B. F. Sherr, E. B. Sherr, and J. J. Cole. 1993. Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 25.Kenyon, J. E., D. R. Piexoto, B. Austin, and D. C. Gillies. 1984. Seasonal variation in numbers of Vibrio cholerae (non-O1) isolated from California coastal waters. Appl. Environ. Microbiol. 47:1243-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondo, K., A. Takade, and K. Amako. 1994. Morphology of the viable but nonculturable Vibrio cholerae as determined by freeze fixation technique. FEMS Microbiol. Lett. 123:179-184. [DOI] [PubMed] [Google Scholar]
- 27.Leser, T. D., M. Boye, and N. B. Hendriksen. 1995. Survival and activity of Pseudomonas sp. strain B13(FR1) in a marine microcosm determined by quantitative PCR and an rRNA-targeting probe and its effect on indigenous bacterioplankton. Appl. Environ. Microbiol. 61:1201-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, M., D. A. Payne, and J. R. Schwarz. 2003. Intraspecific diversity of Vibrio vulnificus in Galveston Bay water and oysters as determined by randomly amplified polymorphic DNA PCR. Appl. Environ. Microbiol. 69:3170-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linder, K., and J. D. Oliver. 1989. Membrane fatty acid and virulence changes in the viable but nonculturable state of Vibrio vulnificus. Appl. Environ. Microbiol. 55:2837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipp, E. K., C. Rodriquez-Palacios, and J. B. Rose. 2001. Occurrence and distribution of the human pathogen Vibrio vulnificus in a subtropical Gulf of Mexico estuary. Hydrobiologia 460:165-173. [Google Scholar]
- 31.Louis, V. R., E. Russek-Cohen, N. Choopun, I. N. G. Rivera, B. Gangle, S. C. Jiang, A. Rubin, J. A. Patz, A. Huq, and R. R. Colwell. 2003. Predictability of Vibrio cholerae in Chesapeake Bay. Appl. Environ. Microbiol. 69:2773-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon, W. J. 2001. TaqMan PCR for detection of Vibrio cholerae O1, O139, non-O1, and non-O139 in pure cultures, raw oysters, and synthetic seawater. Appl. Environ. Microbiol. 67:4685-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miceli, G. A., W. D. Watkins, and S. R. Rippey. 1993. Direct plating procedures for enumerating Vibrio vulnificus in oysters (Crassostrea virginica). Appl. Environ. Microbiol. 59:3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michotey, V., V. Méjean, and P. Bonin. 2000. Comparison of methods for quantification of cytochrome cd1-denitrifying bacteria in environmental marine samples. Appl. Environ. Microbiol. 66:1564-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motes, M. L., A. DePaola, D. W. Cook, J. E. Veazey, J. C. Hunsucker, W. E. Garthright, R. J. Blodgett, and S. J. Chirtel. 1998. Influence of water temperature and salinity on Vibrio vulnificus in North Gulf and Atlantic Coast oysters (Crassotrea virginica). Appl. Environ. Microbiol. 64:1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilsson, L., J. D. Oliver, and S. Kjelleberg. 1991. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. J. Bacteriol. 173:5054-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver, J. D. 1995. The viable but non-culturable state in the human pathogen Vibrio vulnificus. FEMS Microbiol. Lett. 133:203-208. [DOI] [PubMed] [Google Scholar]
- 38.Oliver, J. D., and R. Bockian. 1995. In vivo resuscitation, and virulence towards mice, of viable but nonculturable cells of Vibrio vulnificus. Appl. Environ. Microbiol. 61:2620-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver, J. D., K. Guthrie, J. Preyer, A. Wright, L. M. Simpson, R. Siebeling, and J. G. Morris, Jr. 1992. Use of colistin-polymyxin B-cellobiose agar for isolation of Vibrio vulnificus from the environment. Appl. Environ. Microbiol. 58:737-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliver, J. D., F. Hite, D. McDougald, N. L. Andon, and L. M. Simpson. 1995. Entry into, and resuscitation from, the viable but nonculturable state by Vibrio vulnificus in an estuarine environment. Appl. Environ. Microbiol. 61:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeffer, C. S., M. F. Hite, and J. D. Oliver. 2003. Ecology of Vibrio vulnificus in estuarine waters of eastern North Carolina. Appl. Environ. Microbiol. 69:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, S. A., D. McDougald, and S. Kjelleberg. 2000. Vibrio vulnificus: a physiological and genetic approach to the viable but nonculturable response. J. Infect. Chemother. 6:115-120. [DOI] [PubMed] [Google Scholar]
- 43.Tamplin, M., E. Rodrick, N. J. Blake, and T. Cuba. 1982. Isolation and characterization of Vibrio vulnificus from two Florida estuaries. Appl. Environ. Microbiol. 44:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tay, T.-L., H. Hemond, L. Krumholz, C. M. Cavanaugh, and M. F. Polz. 2001. Population dynamics of two toluene degrading species in a contaminated stream assessed by quantitative PCR. Microb. Ecol. 41:124-131. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. R., M. A. Randa, L. A. Marcelino, A. Tomita-Mitchell, E. Lim, and M. F. Polz. 2004. Diversity and dynamics of a North Atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70:4103-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanoy, R. W., M. L. Tamplin, and J. R. Schwarz. 1992. Ecology of Vibrio vulnificus in Galveston Bay oysters, suspended particulate matter, sediment and seawater: detection by monoclonal antibody-immunoassay-most probable number procedures. J. Ind. Microbiol. 9:219-223. [Google Scholar]
- 47.Weichart, D., D. McDougald, D. Jacobs, and S. Kjelleberg. 1997. In situ analysis of nucleic acids in cold-induced nonculturable Vibrio vulnificus. Appl. Environ. Microbiol. 63:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitesides, M. D., and J. D. Oliver. 1997. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. Appl. Environ. Microbiol. 63:1002-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong, H. C., W. R. Sheih, and Y. S. Lee. 1993. Toxigenic characterization of Vibrios isolated in foods available in Taiwan. J. Food Prot. 56:980-982. [DOI] [PubMed] [Google Scholar]
- 50.Wright, A. C., R. T. Hill, J. A. Johnson, M. C. Roghman, R. R. Colwell, and J. G. Morris. 1996. Distribution of Vibrio vulnificus in the Chesapeake Bay. Appl. Environ. Microbiol. 62:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto, H. 2000. Viable but nonculturable state as a general phenomenon of non-spore-forming bacteria, and its modeling. J. Infect. Chemother. 6:112-114. [DOI] [PubMed] [Google Scholar]