Abstract
We have characterized the expression pattern of a gene, ddcA, involved in initial colonization of corn seeds by Pseudomonas putida KT2440. The ddcA gene codes for a putative membrane polypeptide belonging to a family of conserved proteins of unknown function. Members of this family are widespread among prokaryotes and include the products of a Salmonella enterica serovar Typhimurium gene expressed during invasion of macrophages and psiE, an Escherichia coli phosphate starvation-inducible gene. Although its specific role is undetermined, the presence of ddcA in multicopy restored the seed adhesion capacity of a KT2440 ddcA mutant. Expression of ddcA is growth phase regulated, being maximal at the beginning of stationary phase. It is independent of RpoS, nutrient depletion, or phosphate starvation, and it is not the result of changes in the medium pH during growth. Expression of ddcA is directly dependent on cell density, being also stimulated by the addition of conditioned medium and of seed exudates. This is the first evidence suggesting the existence of a quorum-sensing system in P. putida KT2440. The potential implication of such a signaling process in seed adhesion and colonization by the bacterium is discussed.
Fluorescent pseudomonads are among the bacteria most frequently found in association with plants, either as pathogens, epiphytes (i.e., living on leaves and other aerial parts of the plant), or rhizosphere-colonizing organisms (i.e., surviving on roots and the soil area under their influence). Certain rhizosphere pseudomonads have a beneficial role on plant growth, either by mobilizing nutrients or by exerting biocontrol activities that lead to the displacement of plant pathogenic bacteria or fungi. These characteristics have prompted an increasing interest in the mechanisms involved in bacterial colonization of the rhizosphere. Efficient rhizosphere colonization is one of the keys to the success of biocontrol processes (6, 7). Elements known to participate in bacterial root colonization include NADH dehydrogenase I, flagellar motility, chemotaxis, lipopolysaccharide, and type IV pili (12, 16, 47, 48, 50, 51). Mutants of Pseudomonas fluorescens unable to synthesize certain vitamins and amino acids also show reduced colonization capacity, indicating that the amounts of these compounds found in the rhizosphere are not sufficient to overcome some auxotrophies (28). The ability to utilize different nutrients, such as lysine and organic acids, is also important for competitive root colonization by Pseudomonas putida and P. fluorescens, respectively (18, 28).
Regulatory elements involved in root colonization by P. fluorescens have also been identified. They include the two-component system colR/colS and a site-specific recombinase (13, 14). Site-specific recombinases cause DNA rearrangements, which might correlate with the phenotypic variability observed in P. fluorescens populations isolated from roots (40). Other regulatory processes may be mediated by cell-cell communication via quorum-sensing signaling. Quorum sensing is an intercellular communication system mediated by small molecules referred to as autoinducers, which regulate bacterial gene expression when a certain concentration of the signal molecule is reached. Quorum sensing plays a role in plant pathogenesis, and interferences between plant signals and quorum sensing have been observed (3, 33). Quorum sensing mediated by N-acylhomoserine lactones (AHLs) has been shown to take place in microcolonies associated with plant roots (43), but its actual role in these root-associated populations remains to be defined.
Compared to the available information regarding rhizosphere colonization, much less is known with respect to the elements and mechanisms that determine bacterial colonization of the spermosphere (the surface of plant seeds and the surrounding soil areas when sown). However, seeds are one of the principal ways of propagation of plant-associated bacteria, constitute a reservoir of plant pathogens, and are the most effective vehicle to introduce plant-beneficial inoculants in the field for agricultural purposes or for rhizoremediation (24, 31). Determinants of seed colonization have been studied in Pseudomonas spp. and in Enterobacter cloacae. In E. cloacae, the importance of amino acids and peptides as carbon sources during spermosphere colonization has been established (36, 37). Carbohydrate utilization is also a fundamental trait in the interaction of E. cloacae with seeds; a pfkA mutant, defective in phosphofructokinase, a key glycolytic enzyme, is impaired in seed colonization (38). The importance of nutrients released by germinating seeds for bacterial colonization is also reflected by the fact that seedling-associated Pseudomonas chlororaphis cells are mainly found in the area around the cotyledon and emerging embryo, where nutrient release is presumably maximal (46).
Mutants defective in adhesion to seeds have been isolated in P. fluorescens (9, 10) and in Pseudomonas putida KT2440 (17), a derivative of P. putida mt-2 (20) which efficiently colonizes the spermosphere and the rhizosphere of a number of plants (17, 19, 31). In the first case, two of the three mutants isolated were deficient in the synthesis of flagellin, the structural component of the flagellum (9, 10). In P. putida KT2440, a number of genes involved in attachment to corn seeds have been identified (17). Among these lapA, a gene coding for one of the largest bacterial proteins described to date, also present in P. fluorescens WCS365 and essential for adhesion to seeds and for biofilm formation, has been recently characterized (23).
We have now studied a gene involved in seed colonization by P. putida KT2440, whose expression responds to cell density and to seed exudates. This constitutes the first evidence that P. putida KT2440 may possess an as-yet-uncharacterized quorum-sensing system and suggests that cell-cell and cell-host communications are important for spermosphere colonization.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Strains and plasmids used in this work are listed in Table 1. P. putida KT2440R is a spontaneous rifampin-resistant derivative of KT2440 obtained in our laboratory. This strain presents a point mutation (A to G) in nucleotide 1562 of the rpoB gene, within the so-called Rif region, which results in amino acid 521 changing from aspartic acid to glycine.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Features | Source or reference |
|---|---|---|
| P. putida strains | ||
| KT2440 | Plasmid-free derivative of natural isolate mt-2 | 20 |
| KT2440R | Rifr derivative of KT2440 | This work |
| mus-5 | KT2440 ddcA::mini-Tn5[Km1] insertional mutant | 17 |
| EU5 | KT2440 ddcA::Km null mutant | This work |
| C1R1 | KT2440 rpoS::luxAB null mutant | 34 |
| KT2440 relA | KT2440 relA::Km null mutant | 44 |
| KT2440 relA spoT | KT2440 relA::Km spoT::Gm null mutant | 44 |
| E. coli strain | ||
| DH5α | Host strain for cloning | PRCCa |
| Plasmids | ||
| pGB1 | Cloning vector, replicates in E. coli and Pseudomonas; Tcr Apr | 4 |
| pME5 | 1.8-kb EcoRV/SalI fragment containing ddcA cloned into pGB1 | This work |
| pMP220 | Promoter-probe vector, broad host range; Tcr | 41 |
| pME510 | 880-bp EcoRI/PstI fragment from pME51 cloned in pMP220 (ddcA::lacZ) | This work |
| pUC18Not | Cloning vector, pUC18 derivative with polylinker flanked by NotI sites | 22 |
| p34S-Km3 | Plasmid carrying a Kmr cassette flanked by duplicated restriction sites | 15 |
| pME51 | 871-bp EcoRV/BamHI fragment (upstream region and first 11 codons of ddcA) cloned in pUC18Not | This work |
| pME52 | 597-bp BamHI/SalI fragment (last 20 codons and downstream region of ddcA) cloned into pUC18Not | This work |
| pME512 | BamHI/SalI fragment from pME52 cloned into pME51 | This work |
| pME512K | BamHI fragment containing Kmr cassette from p34S-Km3 cloned into pME512 | This work |
| pKNG101 | Suicide vector for gene replacement; Smr; sacB | 25 |
Pseudomonas Reference Culture Collection (http://www.eez.csic.es/prcc/).
Escherichia coli cultures were grown at 37°C in Luria-Bertani (LB) medium (39). P. putida cultures were grown at 30°C in LB or in M9 minimal medium (39) supplemented with 1 mM MgSO4, 50 μM FeCl3 or iron citrate, and trace metals, with glucose (20 mM) or citrate (10 or 20 mM) as a carbon source. Where indicated, low-phosphate minimal medium (10 mM NaCl, 30 mM KCl, 20 mM NH4Cl, 0.5 mM Na2HPO4, buffered with Tris-HCl at pH 7.5) was used. When appropriate, antibiotics were added at the following concentrations (in micrograms per milliliter): rifampin (Rif), 20; kanamycin (Km), 25; tetracycline (Tc), 15; streptomycin (Sm), 25.
Sequence analysis.
Sequence data of the KT2440 genome were obtained from The Institute for Genomic Research (www.tigr.org) and analyzed with the Omiga 2.0 software package (Oxford Molecular). Comparisons with the databases were done with the BLAST programs (1) at www.ncbi.nlm.nih.org. Protein localization and topology predictions were obtained with the Omiga 2.0, PredictProtein (http://cubic.bioc.columbia.edu/predictprotein/), and PSORT (http://psort.ims.u-tokyo.ac.jp/) programs. Alignments were done with CLUSTALW (45) and refined manually.
Molecular biology techniques.
Plasmid and chromosomal DNA preparations and agarose gel electrophoresis were done following standard procedures (2, 39). Restriction enzymes, DNA ligase, and shrimp alkaline phosphatase (Roche and New England BioLabs) were used following the manufacturers' instructions. The DIG-DNA labeling and detection kit (Roche) was used for Southern hybridization. PCRs were done with Taq DNA polymerase (Pharmacia) on a Perkin-Elmer GeneAmp PE2400. Sequencing was done on an ABI Prism 3100 automated sequencer with the BigDye kit (Applied Biosystems), as recommended by the manufacturer.
Cloning of ddcA and obtainment of a ddcA::Km mutant and a ddcA::lacZ fusion.
A DNA fragment containing ddcA was isolated after PCR amplification with oligonucleotides MUS5-1 (5′-TTGATGATGGCGTGGT-3′) and MUS5-2 (5′-ATTTCCGTGACCCACA-3′) and digested with EcoRV and SalI. This restriction renders a 1.8-kb fragment containing ddcA, which was cloned in the SmaI/SalI sites of the shuttle vector pGB1 (4) to create plasmid pME5.
A ddcA mutant was constructed by gene replacement via homologous recombination. Two fragments corresponding to upstream and downstream regions of ddcA were PCR amplified with the oligonucleotide pairs MUS5-1 and MUS5-4 (5′-CCGTGCAGGATCCTTGCGCA-3′) and MUS5-2 and MUS5-3 (5′-CTGGCGTTCTGGATCCTGGT-3′). The first pair amplifies a 930-bp fragment comprising the upstream region and first 11 codons of ddcA and introduces a BamHI site (underlined); the second pair amplifies a 660-bp fragment corresponding to the 3′ end and downstream region of ddcA, also introducing a BamHI site. The resulting fragments were digested with EcoRV/BamHI and BamHI/SalI, respectively, and cloned into pUC18Not (22), giving rise to plasmids pME51 and pME52. The BamHI/SalI fragment of pME52 was introduced in pME51 to give pME512. A BamHI fragment carrying a kanamycin resistance cassette from plasmid p34S-Km3 (15) was then cloned into pME512, yielding plasmid pME512K. After digestion of pME512K with NotI, the ddcA::Km construct was ligated to plasmid pKNG101, a suicide vector unable to replicate in Pseudomonas sp. that allows the generation of double recombination events (25). These can be selected after loss of both the streptomycin resistance marker and the sacB gene (which causes growth inhibition in the presence of sucrose) contained in the plasmid (25). The ligation mixture was introduced in P. putida KT2440 by electroporation. Clones in which a double recombination event had taken place were selected on LB plates containing kanamycin and 7% (wt/vol) sucrose and further tested for plasmid loss on plates with streptomycin. Kanamycin- and sucrose-resistant, streptomycin-sensitive clones were checked by Southern hybridization, PCR, and sequencing to confirm replacement of ddcA by ddcA::Km. One of these clones was selected and named EU5.
A ddcA::lacZ transcriptional fusion was constructed by cloning an EcoRI/PstI fragment from pME51, containing the upstream region and first 11 codons of ddcA, in the promoter-probe vector pMP220 (41), giving rise to plasmid pME510.
Seed adhesion assays.
Bacterial adhesion to corn seeds was tested essentially as previously described (17), with the difference that, instead of overnight cultures, cells grown in LB for 6 to 8 h (to an optical density at 600 nm [OD660] of ≈3) were used. Cultures were diluted in M9 medium to an OD660 of ≈1 (which corresponds to 109 cells/ml), and 5 μl was inoculated in 1 ml of M9 medium with one surface-sterilized, hydrated seed, in triplicate. Serial dilutions were plated to estimate the initial number of bacteria inoculated per seed. After 1 h, seeds were washed and attached bacteria were recovered by vortexing in the presence of glass beads (17). Dilutions were plated in selective medium to quantify the number of cells that had attached to the seeds. Results shown are the average of four independent experiments. Student's t test was used to determine the significance of the differences observed between strains.
Obtainment of seed exudates and conditioned medium.
Seed exudates were obtained from surface-sterilized seeds and incubated overnight at room temperature in M9 (20 seeds in 20 ml), in a closed flask. Exudates were filter sterilized and used immediately or stored at −20°C for subsequent use. Conditioned medium was obtained from cultures of KT2440 grown in LB for 6 to 8 h, to an OD660 of 3.5. Cells were removed by centrifugation, and the medium was filter sterilized and used immediately or stored overnight at 4°C.
Measurement of β-galactosidase activity.
β-Galactosidase activity was assayed during growth in LB or minimal medium as described elsewhere (30). Data are given in Miller units. Unless otherwise specified, overnight cultures were inoculated (1:100 dilution) in fresh medium and grown for 1 h, diluting 1:2 every half hour, and then allowed to proceed for another hour prior to initiating the collection of samples. This was done to ensure proper dilution of accumulated β-galactosidase after overnight growth. Experiments were done at least three times and, unless otherwise stated, a representative result is shown.
RESULTS
Role of ddcA in seed colonization.
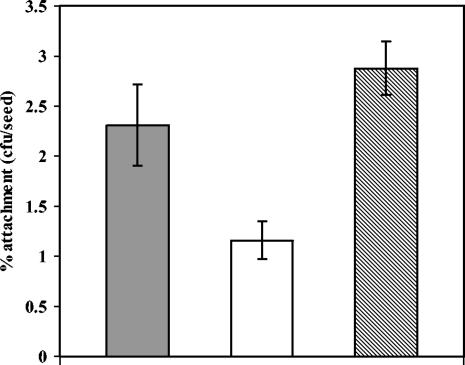
Strain mus-5 is a derivative of P. putida KT2440 obtained by random transposon mutagenesis with mini-Tn5[Km1], which was selected for screening mutants defective in adhesion to corn seeds (17). The recent completion of the genome sequence of KT2440 (32) has allowed unequivocal identification of the gene interrupted by the transposon insertion as a 468-bp open reading frame starting with the alternative initiation codon GTG (GenBank accession number AF182512; TIGR accession number PP4615), which we have named ddcA for density-dependent colonization (see below). However, as described previously (17), the seed adhesion defect of mus-5 is relatively modest, since the percentage of inoculated cells that attached to a seed in 1 h was approximately half that of the parental strain. Such a moderate defect could be due to the transposon insertion rendering a partially functional protein, given its position close to the 3′ end of the gene. To test this possibility, a ddcA null mutant was constructed by replacement of the gene with a kanamycin resistance cassette that has no transcriptional terminators (15), as described in Materials and Methods. Attachment of this mutant, named EU5, to corn seeds was reduced with respect to the parental strain (Fig. 1) to a similar extent as in the case of mus-5.
FIG. 1.
Seed colonization by KT2440 (gray bar), ddcA derivative EU5 (open bar), and EU5 harboring the complementing plasmid pME5 (hatched bar). Assays were done with early stationary phase cultures, as described in the text. Data correspond to the percentage of attached cells recovered from seeds after 1 h of incubation versus the total number of inoculated cells and are the averages and standard deviations of four independent assays (three seeds per assay). The difference between the attachment data obtained with EU5 and those obtained with the two other strains was statistically significant (P < 0.01).
Complementation studies were done to confirm the implication of ddcA in seed colonization. Plasmid pME5, containing ddcA (see Materials and Methods), was introduced into strain EU5. Experiments were performed to test adhesion of the complemented strain to corn seeds (Fig. 1). Plasmid pME5 restored the seed adhesion capacity of the ddcA mutant to levels slightly above those of the parental strain. Similar results were obtained when pME5 was introduced in strain mus-5 (data not shown).
DdcA is a putative membrane polypeptide belonging to a family of conserved proteins.
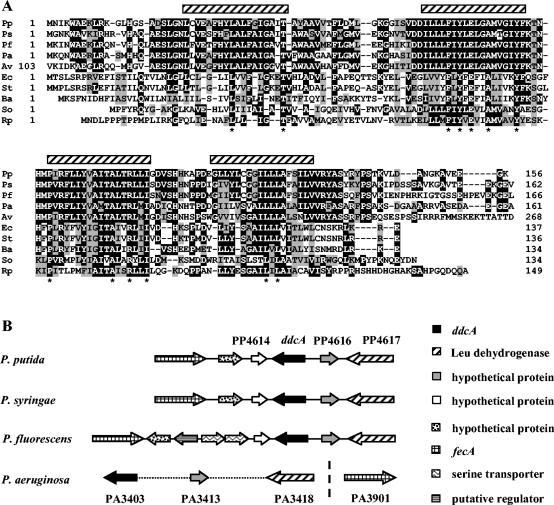
The predicted DdcA polypeptide is 156 amino acids long. Its hydrophobicity profile and secondary structure predictions suggest it is a membrane protein with four transmembrane domains (Fig. 2A). Comparison of the protein sequence with the databases revealed similarities with a family of putative membrane proteins of unknown function. This family includes, among others, the product of psiE, a phosphate starvation-inducible gene of E. coli (26, 53), and Salmonella enterica serovar Typhimurium Mig-7, encoded by an open reading frame that was identified as being expressed during invasion of macrophages by this bacterium (49). Genes coding for related proteins are widespread among bacteria, being highly conserved in Pseudomonas spp. (Fig. 2A). The sizes of these related proteins are very similar in all cases except for in Azotobacter vinelandii, where it is much larger. The organization of the chromosomal region where ddcA is located is also rather well conserved in soil and plant-associated Pseudomonas spp. (Fig. 2B).
FIG. 2.
(A) Alignment of DdcA of P. putida (Pp, 156 amino acids [aa]) with other proteins of the family, from P. aeruginosa (Pa; 161 aa), P. fluorescens (Pf; 166 aa), P. syringae (Ps; 162 aa), E. coli (Ec; 137 aa), S. enterica serovar Typhimurium (St; 136 aa), Bacillus anthracis (Ba; 134 aa), Shewanella oneidensis (So; 134 aa), Rhodopseudomonas palustris (Rp; 149 aa), and a fragment of a 312-aa protein of A. vinelandii (Av). Identical residues present in a majority of proteins are shaded in black, and conserved residues are shaded in gray. Residues conserved in all 10 proteins are marked with an asterisk. Hatched rectangles indicate the predicted transmembrane regions in DdcA. (B) Comparison of the genomic regions containing ddcA homologs in different Pseudomonas species.
Expression of ddcA is growth phase regulated and independent of RpoS, nutrient depletion, or pH changes during growth.
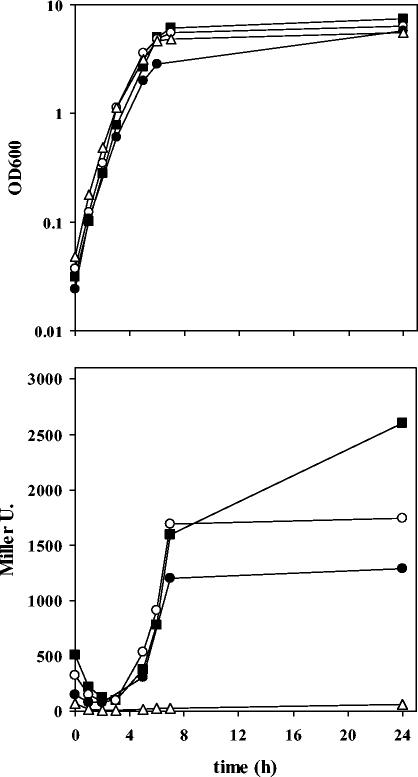
A transcriptional fusion of an ∼850-bp DNA fragment containing the upstream region and first 11 codons of ddcA with the reporter gene lacZ was constructed using the promoter-probe vector pMP220 (41), as described in Materials and Methods. The resulting plasmid, pME510, was introduced in KT2440, and β-galactosidase activity was analyzed during growth in LB. As shown in Fig. 3, expression of the ddcA::lacZ fusion was induced at the end of the logarithmic phase, when growth was decelerating.
FIG. 3.
Expression of a ddcA::lacZ fusion in multicopy (bottom panel) in cultures of KT2440 (wild type), mus-5 (ddcA mutant), and C1R1 (rpoS mutant) grown in LB (growth curves are shown in the top panel). Open circles, KT2440(pME510); closed circles, mus-5(pME510); closed squares, C1R1(pME510); open triangles, KT2440 harboring pMP220 (promoterless lacZ), used as a negative control.
Such an induction pattern made us consider the possibility of ddcA being under the control of the sigma factor RpoS. This alternative sigma factor regulates the expression of a wide number of genes upon entry into stationary phase (34). However, this does not seem to be the case for ddcA. A P. putida KT2440 rpoS mutant carrying pME510 showed a pattern of expression similar to that of the wild type and even higher levels of induction, since the increase in β-galactosidase activity did not stop after cells entered stationary phase (Fig. 3), as is the case in the wild-type strain. This behavior is similar to what has been observed in growth phase-regulated, rpoS-independent promoters in E. coli (5). Intriguingly, when pME510 was introduced in mutant mus-5, a slight but consistent reduction in the maximal activity levels of the ddcA::lacZ fusion was observed (Fig. 3).
The possibility that ddcA expression was triggered by nutrient depletion was also tested. KT2440 (pME510) cultures were grown in LB or LB diluted 1:3 and in minimal medium with citrate. Results are shown in Fig. 4. Growth in dilute LB resulted in reduced β-galactosidase activity with respect to cultures grown in LB, which indicated that nutrient depletion was not responsible for ddcA induction. Results obtained in minimal medium further supported this idea. When KT2440(pME510) was grown in M9 with 10 mM citrate as carbon and energy source, β-galactosidase activity was significantly reduced with respect to the activity observed in LB. This could indicate that growth on citrate had a negative effect on expression of ddcA, or it could be the result of the fact that cultures grown in citrate reach a much lower cell density than those grown in LB. To define which of these possibilities was true, expression of the ddcA::lacZ fusion was analyzed in cultures grown in the presence of 20 mM citrate, instead of 10 mM. Under these conditions, cultures reached higher cell densities and a concomitant increase in β-galactosidase activity was observed (Fig. 4). When glucose was used as a carbon source, the maximal expression of ddcA::lacZ was higher than in citrate but lower than in LB (Table 2).
FIG. 4.
Growth (top) and β-galactosidase activities (bottom) of KT2440(pME510) cultures in LB (open squares), LB diluted 1:3 (closed squares), or in minimal medium with 10 mM citrate (open circles) or 20 mM citrate (closed circles).
TABLE 2.
Growth and β-galactosidase activity of KT2440 harboring pME510 grown in different mediaa
| Medium | Max. OD660 | Max. activity (Miller units) | Fold induction |
|---|---|---|---|
| LB | 6 | 1,734 ± 166 | 17.5 |
| M9 + glucose | 3 | 965 ± 181 | 10 |
| M9 + citrate 10 mM | 1.5 | 150 ± 50 | 1.5 |
| M9 + citrate 20 mM | 2.4 | 450 ± 45 | 4.5 |
| LB + HEPES pH 7 | 4.8 | 1,503 ± 121 | 15.5 |
| LB + Na2HPO4 | 5.8 | 1,560 ± 50 | 16 |
| LPb | 0.38 | 55 ± 23 | 0.5 |
Data are the averages of at least three independent experiments.
Low phosphate Tris medium, with glucose as carbon source.
Given that expression of psiE is induced by phosphate starvation in E. coli, we decided to test expression of ddcA under conditions of phosphate limitation. When incubated in low-phosphate medium, P. putida KT2440 harboring pME510 grew very poorly, up to an OD660 of ≈0.4, and expression of ddcA::lacZ was practically abolished (Table 2). In contrast, the induction pattern was identical in cultures grown in LB or in LB supplemented with phosphate (10 mM). These data suggest that phosphate limitation does not have a significant influence on ddcA expression.
Induction of ddcA::lacZ expression is also independent of pH changes that take place during growth in LB, presumably as the result of the excretion of excess ammonia and amines, which cause a pH increase in the medium. When KT2440 harboring pME510 was grown in LB buffered with 0.1 M HEPES at pH 7.0, the induction pattern was similar to that of cultures grown in unbuffered medium, although the maximal β-galactosidase activity levels were slightly reduced and cultures consistently reached lower turbidity (Table 2).
Expression of ddcA responds to cell density, conditioned medium, and seed exudates.
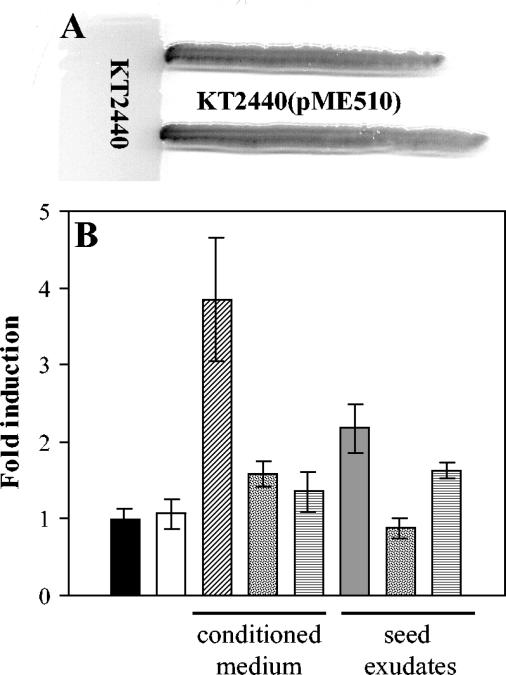
The previous data suggested that ddcA expression was dependent on cell density and that a certain threshold had to be reached for full induction of ddcA. Such correlation between culture turbidity and ddcA::lacZ expression (Fig. 3 and Table 2) led us to hypothesize that ddcA might be under the control of a quorum-sensing mechanism. Initial evidence in this respect was obtained by streaking P. putida cells harboring pME510 from a culture growing exponentially in liquid LB medium, next to P. putida KT2440 cells from a late-log culture, on a plate containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). After overnight incubation, the blue was more intense in the area where both cultures were in contact (Fig. 5).
FIG. 5.
(A) Quorum-sensing-dependent expression of ddcA::lacZ. Cells of KT2440 harboring pME510 were grown in liquid medium to mid-exponential phase (3 h) and streaked on an LB plate containing X-Gal, in contact with KT2440 cells that had been grown to early stationary phase (7 h). (B) Induction of ddcA::lacZ expression in the presence of seed exudates and conditioned medium. Cells from a culture of KT2440(pME510) grown in LB to an OD660 of 0.3 were collected by centrifugation and inoculated in fresh LB (black bar), LB diluted 1:10 (open bar), LB mixed 1:1 with seed exudates (gray bar), or conditioned medium (hatched bar) obtained from the filtered supernatant of a late-log culture of KT2440. The effects of incubating conditioned medium and seed exudates at low (horizontally striped bars) or high (dotted bars) pH for 2 h before mixing with the culture were also tested. Data are shown as fold induction after 1 h of incubation with respect to the β-galactosidase activity of the culture immediately before being collected.
Activity of the ddcA::lacZ fusion was also studied in liquid medium in the presence of conditioned medium. A mid-exponential culture (OD660 = 0.3, when only basal levels of ddcA::lacZ expression could be observed) of KT2440 harboring pME510 was centrifuged, resuspended in M9, and mixed (1:1) with either fresh LB, LB diluted 1:10, or cell-free medium obtained from a KT2440 culture grown in LB to early stationary phase. A fourfold increase in β-galactosidase activity was observed after 1 h of incubation with conditioned medium, but not with LB or dilute LB (Fig. 5).
All these data suggest the existence of a quorum-sensing system in P. putida KT2440 with a role in spermosphere colonization via DdcA. To date, there is no evidence of this phenomenon in KT2440. As a preliminary characterization of the nature of the signaling molecule(s) involved, culture supernatant of KT2440 was subjected to different treatments prior to its addition to KT2440(pME510) cells. An increase in β-galactosidase activity similar to that observed with the untreated supernatant was observed after 15 min at 100°C or treatment with 10 μg of proteinase K/ml for 1 h. Although bioassays with reporter strains that respond to different AHLs produced negative results (data not shown), it seemed possible that KT2440 produced an AHL molecule that was not recognized by these reporters, a phenomenon that is not uncommon (27). Since AHL-type molecules suffer hydrolization of the lactone ring at high pH and are stable at low pH, we decided to test the effects of raising or lowering the pH of conditioned medium (to 9.5 or 4.5, respectively). In both cases, after incubating for 2 h under these conditions and then readjusting the pH to 7.5, the stimulatory effect upon expression of ddcA::lacZ was practically abolished (Fig. 5). Such sensitivity to low pH suggests that AHL molecules may not be involved in the induction of expression of ddcA.
Different reports have suggested the possibility of cross-communication between host and bacterial signaling systems (33, 42), and the synthesis of molecules that mimic quorum-sensing signals has been observed in different plants (3). We wanted to determine if ddcA responded not only to cell-cell signaling but also to signals released by corn seeds. Exponentially growing cultures of KT2440(pME510) were centrifuged, and cells were resuspended in LB mixed 1:1 with seed exudates. As shown in Fig. 5, addition of seed exudates had a moderately stimulatory effect on expression of ddcA::lacZ, which was increased twofold after 1 h of incubation at 30°C. As in the case with conditioned medium, incubation of seed exudates at acidic or basic pH for 2 h resulted in a decrease of their stimulatory activity (Fig. 5).
Role of the stringent response in ddcA expression.
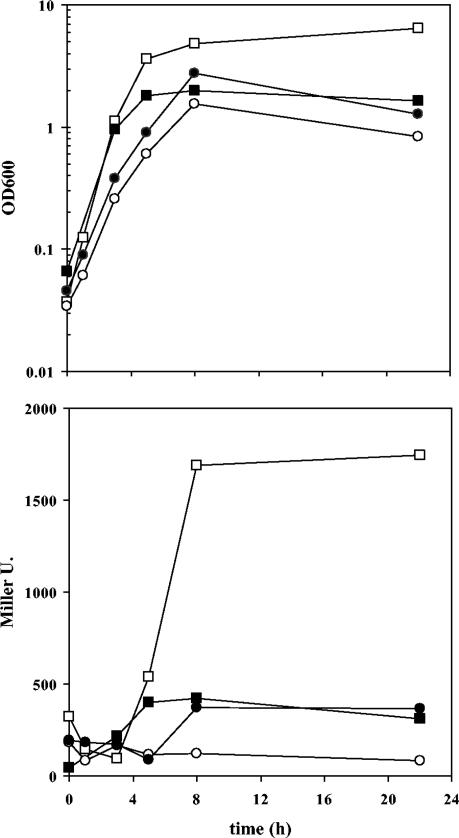
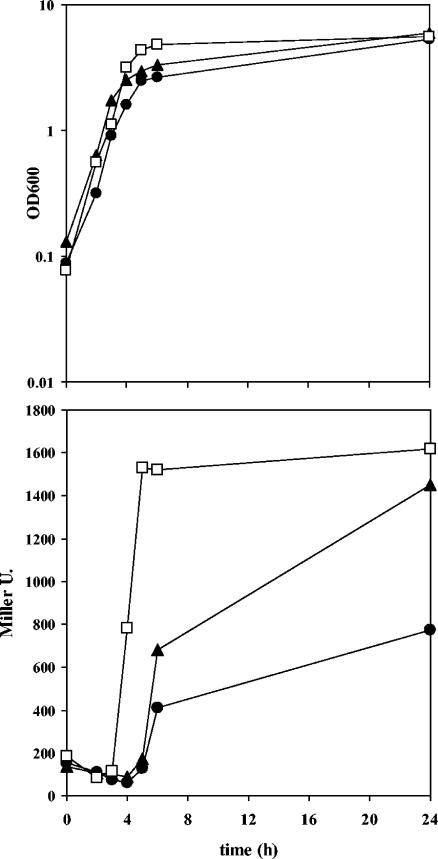
It has been reported that in Pseudomonas aeruginosa, quorum sensing is influenced by the stringent response, which is mediated by the relA and spoT genes (52). We therefore tested the effect of mutations in these genes on the expression of ddcA. Plasmid pME510 was introduced into KT2440 relA::Km and in KT2440 relA::Km spoT::Gm (44), and β-galactosidase activity was monitored during growth in LB. In both cases, expression of ddcA::lacZ was induced when cultures reached an OD600 of >2.5, but to a lesser extent than in the wild-type strain. The maximal levels of activity were lower in the relA mutant and were reduced by 50% in the double mutant compared to the wild-type activity (Fig. 6). Thus, a functional stringent response appears to be necessary for full expression of ddcA. Yet, it is important to note that these two mutants showed differences in growth with respect to KT2440 (Fig. 6), which might also explain the altered pattern of expression of ddcA in these backgrounds.
FIG. 6.
Growth (top) and β-galactosidase activities (bottom) of KT2440 (open squares), KT2440 relA (closed triangles), and KT2440 relA spoT (closed circles), each harboring pME510, in LB.
DISCUSSION
A number of physiological processes are regulated in bacteria by small diffusible signaling molecules, or autoinducers, produced in response to population density. This regulatory mechanism is known as quorum sensing, and it can be mediated by different signal molecules. In most gram-negative bacteria where it has been described, it is mediated by AHLs, although other molecules such as quinolones or cyclic dipeptides also act as quorum-sensing signals (11, 29). AHLs are found in different bacteria and regulate processes such as bioluminescence or production of virulence factors (33). A role for quorum sensing in biofilm formation has also been postulated (8, 35). Cell-cell communication has been recently shown to take place during bacterial colonization of the tomato rhizosphere, and the production of AHLs in rhizosphere populations has been reported (43). In the soil and rhizosphere-colonizing bacterium P. putida KT2440, no production of AHLs has been detected to date, and there was no genetic evidence that this microorganism possesses a quorum-sensing system (32). The results presented here indicate that KT2440 does respond to population density via a still-unidentified signaling mechanism. Furthermore, this potential quorum-sensing system plays a role in the interaction of KT2440 with plants, since a mutant defective in ddcA, a gene whose expression is cell density dependent, shows reduced ability to colonize corn seeds. We have done a search in the genome sequence of KT2440 that has revealed the absence of genetic loci homologous to those involved in the synthesis of quorum-sensing signals in other organisms. The only exception is a gene similar to hdtS, which in P. fluorescens F113 has been proposed to participate in the synthesis of certain AHLs (27). The role of this gene in P. putida KT2440, and its potential involvement in signal synthesis and/or ddcA expression, remains to be elucidated. The data obtained when conditioned medium was incubated at acidic pH suggest ddcA expression might not respond to a typical AHL-type signal, since these are generally stable at low pH. On the other hand, the sensitivity of conditioned medium to prolonged incubation at high pH might seem paradoxical, since pH increases during growth of KT2440 in LB. However, pH only reaches values above 8.5 when the culture is well into stationary phase, and at that time there is no further increase in ddcA expression.
Expression of ddcA might also be partly modulated by the stringent response, since in a relA spoT double mutant the induction of a ddcA::lacZ fusion was reduced by 50% with respect to that observed in the wild-type strain. Such a correlation between cell density-dependent regulation and the stringent response has been reported for P. aeruginosa, where overexpression of relA results in premature, cell density-independent synthesis of autoinducer and expression of transcriptional regulators involved in quorum sensing (52). However, growth of the relA spoT mutant of KT2440 is significantly affected, which makes it difficult to interpret the actual role of the stringent response in expression of ddcA.
Genes similar to ddcA are widespread in prokaryotes, but their specific physiological roles remain unknown. In E. coli, psiE is expressed in response to phosphate starvation and negatively regulated by cyclic AMP and CRP (26, 53). It has been recently reported that a psiE mutant appears to be more sensitive to the lipophilic iron chelator 8-hydroxyquinoline (54). This does not seem to be the case for P. putida ddcA mutants (M. Espinosa-Urgel, unpublished observation) and, as deduced from the results presented here, ddcA is not induced by phosphate starvation. In S. enterica serovar Typhimurium, the homologous gene (mig-7) was identified as being preferentially expressed during infection of macrophages (49), suggesting that, like P. putida ddcA, it might also play a role in the interaction with eukaryotic cells. A link between bacterial cell-cell communication by autoinducers and host cell signaling by hormones has been recently proposed, suggesting that quorum sensing may participate in bacterium-host communication (42). It had been postulated that quorum sensing was a means for enterohemorrhagic E. coli to sense its location within the intestine and trigger virulence and colonization genes. However, in a mutant unable to synthesize autoinducer, expression of virulence genes can still occur in response to the host hormone epinephrine (42). The fact that ddcA responds to population density and is also induced by seed exudates suggests that a similar phenomenon takes place during seed colonization by P. putida and that, as in the case of symbiotic associations with Rhizobiaceae, bacterial as well as plant signals modulate the colonization process. It has been shown that certain red algae produce halogenated furanones (21), which interact with quorum-sensing signaling mediated by AHLs, and higher plants can also synthesize molecules that mimic bacterial quorum-sensing signals (3). Further research aimed at the identification of the specific molecules that cause the induction of ddcA expression will help clarify the potential role of cell-cell and plant-bacterium communication in the interaction of P. putida with plants.
Acknowledgments
We thank N. Muñoz for technical assistance, A. Hurtado for DNA sequencing, M. I. Ramos-González and A. Rojas for helpful discussions and sharing of unpublished results, and V. Shingler for the generous gift of the relA and relA spoT derivatives of KT2440.
This work was supported by grants BMC2001-0576 from the Ministerio de Ciencia y Tecnología and QLK3-CT-2000-0170 from the European Commission. M.E.-U. is the recipient of a grant from the Ramón y Cajal Program.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Bauer, W. D., and J. B. Robinson. 2002. Disruption of bacterial quorum sensing by other organisms. Curr. Opin. Biotechnol. 13:234-237. [DOI] [PubMed] [Google Scholar]
- 4.Bloemberg, G. V., G. A. O'Toole, B. J. J. Lugtenberg, and R. Kolter. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 63:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohannon, D. E., N. Connell, J. Keener, A. Tormo, M. Espinosa-Urgel, M. M. Zambrano, and R. Kolter. 1991. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of σ70. J. Bacteriol. 173:4482-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull, C. T., D. M. Weller, and L. S. Thomashow. 1991. Relationship between root colonisation and suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens strain 2-79. Phytopathology 81:954-959. [Google Scholar]
- 7.Chin-A-Woeng, T. F. C., G. V. Bloemberg, I. H. M. Mulders, L. C. Dekkers, and B. J. J. Lugtenberg. 2000. Root colonisation is essential for biocontrol of tomato foot and root rot by the phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391. Mol. Plant-Microbe Interact. 13:1340-1345. [DOI] [PubMed] [Google Scholar]
- 8.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeFlaun, M. F., A. S. Tanzer, A. L. McAteer, B. Marshall, and S. B. Levy. 1990. Development of an adhesion assay and characterization of an adhesion-deficient mutant of Pseudomonas fluorescens. Appl. Environ. Microbiol. 56:112-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeFlaun, M. F., B. Marshall, E.-P. Kulle, and S. B. Levy. 1994. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl. Environ. Microbiol. 60:2637-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degrassi, G., C. Aguilar, M. Bosco, S. Zahariev, S. Pongor, and V. Venturi. 2002. Plant growth-promoting Pseudomonas putida WCS358 produces and secretes four cyclic dipeptides: cross-talk with quorum sensing bacterial sensors. Curr. Microbiol. 45:250-254. [DOI] [PubMed] [Google Scholar]
- 12.Dekkers, L. C., A. J. van der Bij, I. H. Mulders, C. C. Phoelich, R. A. Wentwoord, D. C. Glandorf, C. A. Wijffelman, and B. J. J. Lugtenberg. 1998. Role of the O-antigen of lipopolysaccharide, and possible roles of growth rate and of NADH:ubiquinone oxidoreductase (nuo) in competitive tomato root-tip colonization by Pseudomonas fluorescens WCS365. Mol. Plant-Microbe Interact. 11:763-771. [DOI] [PubMed] [Google Scholar]
- 13.Dekkers, L. C., C. C. Phoelich, L. van der Fits, and B. J. J. Lugtenberg. 1998. A site-specific recombinase is required for competitive root colonization by Pseudomonas fluorescens WCS365. Proc. Natl. Acad. Sci. USA 95:7051-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekkers, L. C., C. J. Bloemendaal, L. A. de Weger, C. A. Wijffelman, H. P. Spaink, and B. J. J. Lugtenberg. 1998. A two-component system plays an important role in the root-colonizing ability of Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 11:45-56. [DOI] [PubMed] [Google Scholar]
- 15.Dennis, J. J., and G. J. Zylstra. 1998. Improved antibiotic-resistance cassettes through restriction site elimination using Pfu DNA polymerase PCR. BioTechniques 25:772-776. [DOI] [PubMed] [Google Scholar]
- 16.Dörr, J., T. Hurek, and B. Reinhold-Hurek. 1998. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol. Microbiol. 30:7-17. [DOI] [PubMed] [Google Scholar]
- 17.Espinosa-Urgel, M., A. Salido, and J. L. Ramos. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182:2363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinosa-Urgel, M., and J. L. Ramos. 2001. A Pseudomonas putida aminotransferase involved in lysine catabolism is induced in the rhizosphere. Appl. Environ. Microbiol. 67:5219-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espinosa-Urgel, M., R. Kolter, and J. L. Ramos. 2002. Root colonization by Pseudomonas putida: love at first sight. Microbiology 148:341-343. [DOI] [PubMed] [Google Scholar]
- 20.Franklin, F. C. H., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta-cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Givskov, M., R. de Nys, M. Manefield, L. Gram, R. Maximilien, L. Eberl, S. Molin, P. Steinberg, and S. Kjelleberg. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signaling. J. Bacteriol. 178:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905-918. [DOI] [PubMed] [Google Scholar]
- 24.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S. K., S. Kimura, H. Shinagawa, A. Nakata, K. S. Lee, B. L. Wanner, and K. Makino. 2000. Dual transcriptional regulation of the Escherichia coli phosphate starvation-inducible psiE gene of the phosphate regulon by PhoB and the cyclic AMP (cAMP)-cAMP receptor protein complex. J. Bacteriol. 182:5596-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laue, B. E., Y. Jiang, S. R. Chhabra, S. Jacob, G. S. A. B. Stuart, A. Hardman, J. A. Downie, F. O'Gara, and P. Williams. 2000. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology 146:2469-2480. [DOI] [PubMed] [Google Scholar]
- 28.Lugtenberg, B. J. J., L. Dekkers, and G. V. Bloemberg. 2001. Molecular determinants of rhizosphere colonisation by Pseudomonas. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 29.McGrath, S., D. S. Wade, and E. C. Pesci. 2004. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 230:27-34. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Molina, L., C. Ramos, E. Duque, M. C. Ronchel, J. M. García, L. Wyke, and J. L. Ramos. 2000. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 32:315-321. [Google Scholar]
- 32.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. P. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Düsterhöft, B. Tümmler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 33.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos-González, M. I., and S. Molin. 1998. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J. Bacteriol. 180:3421-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riedel, K., M. Hentzer, O. Geisenberger, B. Huber, A. Steidle, H. Wu, N. Hoiby, M. Givskov, S. Molin, and L. Eberl. 2001. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249-3262. [DOI] [PubMed] [Google Scholar]
- 36.Roberts, D. P., A. M. Marty, P. D. Dery, I. Yucel, and J. S. Hartung. 1996. Amino acids as reduced carbon sources for Enterobacter cloacae during colonization of the spermospheres of crop plants. Soil Biol. Biochem. 28:1015-1020. [Google Scholar]
- 37.Roberts, D. P., P. D. Dery, and J. S. Hartung. 1996. Peptide utilization and colonization of corn, radish and wheat spermospheres by Enterobacter cloacae. Soil Biol. Biochem. 28:1109-1111. [Google Scholar]
- 38.Roberts, D. P., P. D. Dery, I. Yucel, J. Buyer, M. A. Holtman, and D. Y. Kobayashi. 1999. Role of pfkA and general carbohydrate catabolism in seed colonization by Enterobacter cloacae. Appl. Environ. Microbiol. 65:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Sánchez-Contreras, M., M. Martín, M. Villacieros, F. O'Gara, I. Bonilla, and R. Rivilla. 2002. Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J. Bacteriol. 184:1587-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 42.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sze, C. C., L. M. Bernardo, and V. Shingler. 2002. Integration of global regulation of two aromatic-responsive σ54-dependent systems: a common phenotype by different mechanisms. J. Bacteriol. 184:760-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tombolini, R., D. J. van der Gaag, B. Gerhardson, and J. K. Jansson. 1999. Colonization pattern of the biocontrol strain Pseudomonas chlororaphis MA342 on barley seeds visualized by using green fluorescent protein. Appl. Environ. Microbiol. 65:3674-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbull, G. A., J. A. W. Morgan, J. M. Whipps, and J. R. Saunders. 2001. The role of motility in the in vitro attachment of Pseudomonas putida PaW8 to wheat roots. FEMS Microbiol. Ecol. 35:57-65. [DOI] [PubMed] [Google Scholar]
- 48.Turnbull, G. A., J. A. W. Morgan, J. M. Whipps, and J. R. Saunders. 2001. The role of bacterial motility in the survival and spread of Pseudomonas fluorescens in soil and in the attachment and colonisation of wheat roots. FEMS Microbiol. Ecol. 36:21-31. [DOI] [PubMed] [Google Scholar]
- 49.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 50.Vande Broek, A., and J. Vanderleyden. 1995. The role of bacterial motility, chemotaxis, and attachment in bacteria-plant interactions. Mol. Plant-Microbe Interact. 8:800-810. [Google Scholar]
- 51.Vande Broek, A., M. Lambrecht, and J. Vanderleyden. 1998. Bacterial chemotactic motility is important for the initiation of wheat root colonisation by Azospirillum brasilense. Microbiology 144:2599-2606. [DOI] [PubMed] [Google Scholar]
- 52.van Delden, C., R. Comte, and A. M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wanner, B. L. 1986. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J. Mol. Biol. 191:39-58. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, L., X. H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]