Abstract
Type III protein secretion systems play a key role in the virulence of many pathogenic proteobacteria, but they also occur in nonpathogenic, plant-associated bacteria. Certain type III protein secretion genes (e.g., hrcC) have been found in Pseudomonas sp. strain SBW25 (and other biocontrol pseudomonads), but other type III protein secretion genes, such as the ATPase-encoding gene hrcN, have not been found. Using both colony hybridization and a PCR approach, we show here that hrcN is nevertheless present in many biocontrol fluorescent pseudomonads. The phylogeny of biocontrol Pseudomonas strains based on partial hrcN sequences was largely congruent with the phylogenies derived from analyses of rrs (encoding 16S rRNA) and, to a lesser extent, biocontrol genes, such as phlD (for 2,4-diacetylphloroglucinol production) and hcnBC (for HCN production). Most biocontrol pseudomonads clustered separately from phytopathogenic proteobacteria, including pathogenic pseudomonads, in the hrcN tree. The exception was strain KD, which clustered with phytopathogenic pseudomonads, such as Pseudomonas syringae, suggesting that hrcN was acquired from the latter species. Indeed, strain KD (unlike strain SBW25) displayed the same organization of the hrpJ operon, which contains hrcN, as P. syringae. These results indicate that the occurrence of hrcN in most biocontrol pseudomonads is not the result of recent horizontal gene transfer from phytopathogenic bacteria, although such transfer might have occurred for a minority of biocontrol strains.
Type III protein secretion systems (TTSS) are widely distributed among proteobacterial pathogens of plants (belonging to the genera Pseudomonas, Erwinia, Xanthomonas, and Ralstonia), animals, and humans (27). TTSS are specialized machineries for introduction of proteinaceous virulence factors directly into eukaryotic host cells, and mutations in the TTSS genes result in a loss of pathogenicity (22). The basic mechanism of protein secretion is the same even when bacterial species as divergent as the human plague-causing agents Yersinia spp. (18) and the plant pathogen Pseudomonas syringae (35) are compared. However, the proteins secreted can differ from one pathogen to the next (27). TTSS genes are often thought to have evolved from the genes encoding the flagellar export mechanism (17, 37, 42), but recent findings suggest instead that the two types of genes have a common ancestor (19).
In plant pathogens, functional TTSS are essential for induction of disease in susceptible host plants (3) and are encoded by hrc or hrp genes (17). In contrast, resistant plants develop a defense reaction called the hypersensitive response in the presence of a phytopathogen with a functional TTSS. The hypersensitive response is a local tissue necrosis, accompanied by the production of antimicrobial substances, which is aimed at preventing further spread of the infecting bacterium in the plant (32, 36).
In addition to phytopathogens, the existence of TTSS has also been demonstrated in plant endosymbionts, such as the nitrogen-fixing bacterium Rhizobium (15, 20, 39). TTSS mutants of Rhizobium are affected in nodulation ability and display altered host specificity (62). Therefore, it appears that TTSS can also be involved in beneficial prokaryote-eukaryote interactions. Recently, genes coding for a functional TTSS (e.g., hrcC) have been found in Pseudomonas strains capable of protecting a plant from disease (47), which raises the possibility that TTSS could also play a role in biocontrol interactions. The origin of TTSS genes in plant-beneficial bacteria is unknown, because so far the work has focused on pathogens (19). This issue is important, because (i) the molecular differences between biocontrol and phytopathogenic pseudomonads are poorly understood and (ii) until the work of Preston et al. (47) the presence of a TTSS was one criterion pointing to a pathogenic status. The organization and sequences of TTSS genes in biocontrol fluorescent Pseudomonas sp. strain SBW25 were similar to those in strain 61 of P. syringae (47), a taxon in which hrp genes are thought to be ancestral (55). This suggests that a horizontal gene transfer(s) took place from pathogenic to biocontrol pseudomonads. However, certain TTSS genes present in the pathogen P. syringae 61 were not present in the biocontrol strain SBW25; these genes include hrcN, which encodes a conserved peripheral membrane ATPase (46). Therefore, if the horizontal gene transfer hypothesis described above is valid, this finding could perhaps be explained by (i) incomplete gene transfer from a pathogenic pseudomonad or (ii) subsequent gene loss once the entire TTSS gene set was acquired by a biocontrol pseudomonad.
In this paper, we report the existence of TTSS sequences in a wide range of biocontrol pseudomonads. TTSS sequences were analyzed to determine the phylogenetic relationships between biocontrol fluorescent pseudomonads and plant pathogens, with the objective of assessing the likelihood of past transfer of a TTSS gene(s) between these plant-associated bacteria. Previous work on the occurrence of TTSS genes in nonpathogenic Pseudomonas has focused on strain SBW25 and hrcC (47), but as highlighted above, the range of TTSS genes in Pseudomonas sp. strain SBW25 is incomplete. Since our preliminary observations indicated that TTSS genes that were absent from strain SBW25 (e.g., hrcN) could be present in other biocontrol Pseudomonas strains along with hrcC, hrcN was preferred to the latter gene in the analysis. This choice was based on the assumption that biocontrol strains targeted in this way would display an organization of TTSS genes more comparable to that in their pathogenic counterparts. The phylogenetic analysis was done by using partial nucleotide sequences of the hrcN genes in a well-characterized collection of biocontrol fluorescent pseudomonads having worldwide origins (30, 51) and reference phytopathogenic proteobacteria (Table 1). In addition, the phylogenetic relationship between hrcN and structural genes for synthesis of the biocontrol compounds 2,4-diacetylphloroglucinol (Phl) and hydrogen cyanide (HCN) in biocontrol pseudomonads was assessed. Finally, the organization of key TTSS genes in one hrcN+ biocontrol strain was compared to that in the biocontrol strain SBW25 and the pathogen P. syringae.
TABLE 1.
Bacterial strains used in the study
| Strain(s) | Biocontrol or pathogenic propertiesa | Plant originb | Geographic origin |
hrcN analysis
|
Strain refer- ence | |
|---|---|---|---|---|---|---|
| PCR | Hybridi- zation | |||||
| Biocontrol Pseudomonas spp. strains | ||||||
| C*1A1, CM1′A2 | Cucumber (P. ultimum, P. sclerotioides), cotton (R. solani) | Cucumber | Switzerland | + | 16 | |
| K92.46, K94.08 | Cucumber (P. ultimum) | Cucumber | Switzerland | + | This study | |
| P97.38 | Cucumber (P. ultimum), tomato (F. oxysporum) | Cucumber | Switzerland | + | + | 64 |
| CHA0 | Tobacco (T. basicola), wheat, (G. graminis), cucumber (P. ultimum) | Tobacco | Switzerland | − | − | 59 |
| K93.2 | Cucumber (P. ultimum), tomato (F. oxysporum) | Tobacco | Switzerland | − | − | 64 |
| P12 | Tobacco (T. basicola), cucumber (P. ultimum) | Tobacco | Switzerland | + | 30 | |
| Pf1 | Cucumber (P. ultimum), tobacco (T. basicola) | Tobacco | Switzerland | − | + | 30 |
| K94.3 | Cucumber (P. ultimum), tomato (F. oxysporum) | Tomato | Switzerland | − | − | This study |
| TM1A3, TM1B2 | Cucumber (P. ultimum, P. sclerotioides), cotton (R. solani) | Tomato | Switzerland | + | 16 | |
| TM1′A4 | Cucumber (P. ultimum, P. sclerotioides), cotton (R. solani) | Tomato | Switzerland | + | + | 30 |
| TM1′A5 | Cucumber (P. ultimum, P. sclerotioides), cotton (R. solani) | Tomato | Switzerland | + | 30 | |
| K93.37 | Cucumber (P. ultimum) | Wheat | Switzerland | − | − | This study |
| K94.23 | Cucumber (P. ultimum), tomato (F. oxysporum) | Wheat | Switzerland | + | + | This study |
| K94.31 | Tomato (F. oxysporum) | Cucumber | Czech Republic | + | 64 | |
| K94.35 | Cucumber (P. ultimum), tomato (F. oxysporum) | Cucumber | Czech Republic | − | − | This study |
| K94.37 | Tomato (F. oxysporum) | Cucumber | Czech Republic | − | − | 64 |
| P97.30 | Cucumber (P. ultimum), tomato (F. oxysporum) | Wheat | Czech Republic | + | 64 | |
| DR54 | Sugar beet (P. ultimum) | Sugar beet | Denmark | − | + | 43 |
| F96.27 | Cucumber (P. ultimum), tomato (F. oxysporum) | Cucumber | Estonia | − | − | 64 |
| K94.2 | Cucumber (P. ultimum), tomato (F. oxysporum) | Cucumber | Italy | − | − | This study |
| K94.14 | Cucumber, (P. ultimum), tomato (F. oxysporum) | Cucumber | Italy | + | This study | |
| PINR3 | Cucumber (P. ultimum), tomato (F. oxysporum) | Tobacco | Italy | − | − | 30 |
| PILH1 | Cucumber (P. ultimum), tomato (F. oxysporum) | Tomato | Italy | + | + | 30 |
| PITR2 | Cucumber (P. ultimum), tomato (F. oxysporum) | Wheat | Italy | + | 30 | |
| F113 | Sugar beet (P. ultimum), potato (E. carotovora) | Sugar beet | Ireland | + | + | 14 |
| K94.56 | Cucumber (P. ultimum) | Cucumber | Romania | − | − | This study |
| SBW25 | Pea (P. ultimum) | Sugar beet | United Kingdom | − | − | 49 |
| P97.26 | Cucumber (P. ultimum), tomato (F. oxysporum) | Tomato | Bhutan | − | − | 64 |
| KD | Cucumber (P. ultimum), tomato (F. oxysporum) | Wheat | China | + | + | 56 |
| K94.26 | Cucumber (P. ultimum) | Cucumber | India | − | − | This study |
| K95.7 | Cucumber (P. ultimum), tomato (F. oxysporum) | Cucumber | Pakistan | − | − | This study |
| PGNR1, PGNR2, PGNR3, PGNR4, PGNL1 | Cucumber (P. ultimum), tomato (F. oxysporum) | Tobacco | Ghana | − | − | 30 |
| PF | Wheat (S. tritici) | Wheat | Texas | − | − | 34 |
| Pf-5 | Cotton (P. ultimum, R. solani), cucumber (P. ultimum) | Cotton | Texas | − | − | 25 |
| 2-79 | Wheat (G. graminis), Kentucky bluegrass (M. poae) | Wheat | Washington state | + | + | 65 |
| Q1-87, Q4-87, Q7-87, Q9-87, Q12-87, Q13-87, Q37-87, Q86-87, Q95-87, Q107-87, Q139-87 | Wheat (G. graminis) | Wheat | Washington state | + | 30 | |
| Q2-87 | Wheat (G. graminis) | Wheat | Washington state | + | + | 61 |
| Q65c-80, Q128-87 | Wheat (G. graminis) | Wheat | Washington state | + | 21 | |
| Q69c-80 | Wheat (G. graminis) | Wheat | Washington state | − | − | 21 |
| Pathogenic Pseudomonas spp. strains | ||||||
| P. caricapapayae LMG 2152 | Papaya (not documented) | Papaya | Brazil | + | 53 | |
| P. syringae ATCC 19310 | Unknown | Common lilac | United Kingdom | + | 57 | |
| P. syringae pv. tomato DC3000 | Tomato and Arabidopsis (bacterial speck) | Tomato | Unknown | + | 10 | |
| P. syringae pv. phaseolicola | Bean (halo blight) | Bean | Unknown | + | 9 | |
| Other Pseudomonas spp. strains | ||||||
| VS01 | Isolated from disease lesion of apple fruit | Apple | Switzerland | + | + | This study |
| VS02 | Isolated from disease lesion of apple fruit | Apple | Switzerland | + | This study | |
| P3 | Saprophytic, no biocontrol ability documented | Barley | Switzerland | + | 63 | |
| BE07, BE08 | Isolated from disease lesion of sugar beet leaf | Sugar beet | Switzerland | + | This study | |
| Pathogens other than pseudomonads | ||||||
| E. herbicola pv. gypsophilae | Gypsophila (crown gall) | Gypsophila | Unknown | + | 8 | |
| E. amylovora CNPB136 | Unknown | Unknown | Unknown | + | 6 | |
| E. amylovora 22716 | Apple (fire blight) | Apple | Switzerland | + | This study | |
| E. amylovora 22770 | Pear (fire blight) | Pear | Switzerland | + | + | This study |
| E. amylovora 23482 | Pyracantha (fire blight) | Firethorn | Switzerland | + | This study | |
| X. campestris ATCC 33913 | Rutabaga (not documented) | Rutabaga | United States | + | 44 | |
The corresponding pathogens are Erwinia carotovora subsp. carotovora (E. carotovora), Fusarium oxysporum f. sp. radicis-lycopersici (F. oxysporum), Gaeumannomyces graminis var. tritici (G. graminis), Magnaporthe poae (M. poae), Phomopsis sclerotioides (P. sclerotioides), Pythium ultimum (P. ultimum), Rhizoctonia solani (R. solani), Septoria tritici (S. tritici), and Thielaviopsis basicola (T. basicola).
All biocontrol pseudomonads were isolated from macerated roots or roots that were previously washed to remove the soil.
MATERIALS AND METHODS
PCR amplification and sequencing of hrcN.
The hrcN sequences of P. syringae pv. tomato DC3000 (accession number AF232004) and Erwinia amylovora strain CNPB136 (accession number L25828) were aligned by using LAlign software (26). The consensus sequence was used to design the degenerate 20-mer primers hrcN-5rR (forward) and hrcN-4r (reverse) (Table 2 and Fig. 1A). Primers were synthesized by MWG Biotech (Münchenstein, Switzerland).
TABLE 2.
Primers used in this work
| Primer | Sequence (5′-3′) | Use | Reference |
|---|---|---|---|
| hrcV and neighboring genes | |||
| HRCV-L | CCGGAATTCTGCG | Construction of hrcV probe | 58 |
| HRCV-R | ATTGTCATGTCGAT | Construction of hrcV probe | 58 |
| T3f2 | GGTTTAGCAGGTCGATAATC | Synthesis of KD-j | This study |
| prottss-1r | GTCGAGCTGACGAAGGAGAG | Synthesis of KD-j | This study |
| T3f5 | CTCGATCACTTATCCGGCTC | Synthesis of KD-l | This study |
| T3r8 | GGCCTTCATGATGACTTCCA | Synthesis of KD-l | This study |
| pKD2cV-1r | GCAATCGCCTAGTGGTGAAC | Synthesis of KD-vq and KD-vn | This study |
| hrpQ-1rR | CCGKTCAGTACGCGTAATTCA | Synthesis of KD-vq | This study |
| hrcN-1r | CTGGGCWGCGTGCTGGAYGG | Synthesis of KD-n | This study |
| hrcN-3rR | GGCAGCAGGGTGTAMACCGA | Synthesis of KD-vn | This study |
| hrcN-4r | CGAGCAGGAYTCGATGAACG | Partial hrcN sequencing | This study |
| hrcN-5rR | CCGGWYTGGTATTCACCCAG | Synthesis of KD-n, partial hrcN sequencing | This study |
| 16S rDNA gene | |||
| PH-16S | AAGGAGGTGATCCAGCCGCA | Sequencing of 16S rDNA gene | 12 |
| PA-16S | AGAGTTTGATCCTGGCTCAG | Sequencing of 16S rDNA gene | 12 |
| Intern 2a | GATGATCAGCCACAC | Sequencing of 16S rDNA gene | This study |
FIG.1.
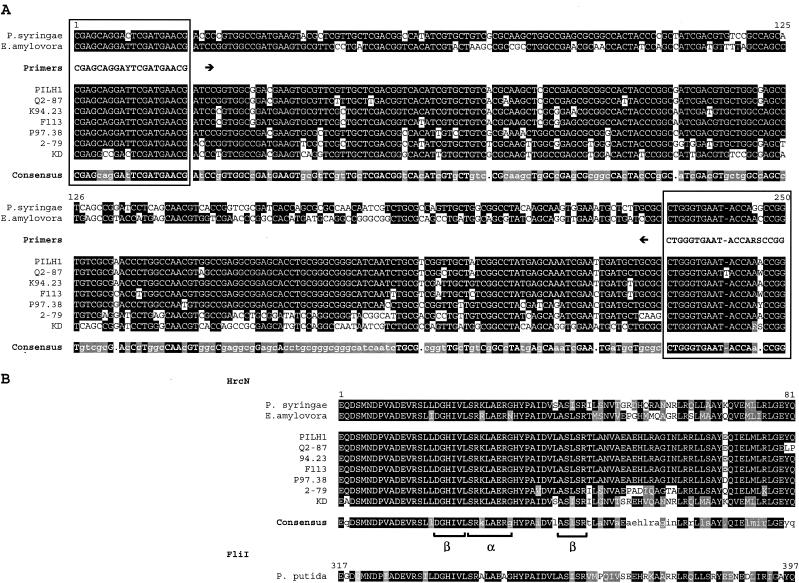
Alignment of partial hrcN nucleotide (A) and deduced HrcN amino acid (B) sequences of P. syringae pv. tomato DC3000, E. amylovora CNPB136, and selected biocontrol pseudomonads, including PILH1 (hrcN group 1), Q2-87 (hrcN group 2), K94.23 (hrcN group 3), F113 (hrcN group 4), P97.38 (hrcN group 6), 2-79 (hrcN group 8), and KD (hrcN group 10). The sites annealing to the PCR primers hrcN-4r (reverse) and hrcN-5rR (forward) are enclosed in boxes. The hrcN consensus sequence is indicated by uppercase letters (>90% identity), lowercase letters (between 50 and 90% identity), or a dot (<50% identity). The HrcN consensus sequence is indicated by a black background (100% identity) or a grey background (<100% identity but 100% similarity). Similarity was calculated by using a BLOSUM62 matrix (23) implemented in LALIGN (26). For each strain, amino acids are indicated by a black background if they are identical to the amino acids in the consensus sequence or by a grey background in case of homologous amino acids. The deduced FliI sequence of P. putida KT2440 is shown for reference.
The bacteria used in this study (Table 1) were grown at 27°C on King's B agar (31) or Luria-Bertani (LB) agar (54). PCR amplification was carried out in 20-μl reaction mixtures containing 5 μl of cell lysate (for pseudomonads), which was obtained by heating the cultures for 10 min at 99°C with 95 μl of lysis buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 0.1% Tween 20) in a PTC-100 thermal cycler (MJ Research, Waltham, Mass.), or 200 pg of genomic DNA of E. amylovora strain 22716, 22770, or 23482 (DNA kindly provided by E. Holliger, FAW Wädenswil, Wädenswil, Switzerland). Each PCR was performed in 1× PCR buffer containing 100 μM dATP, 100 μM dCTP, 100 μM dGTP, 100 μM dTTP, 0.07 U of Taq polymerase (Amersham Pharmacia Biotech, Piscataway, N.J.) per μl, and each primer at a concentration of 0.20 μM by using an initial denaturation step consisting of 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 60°C, and 1 min at 72°C and then a final elongation for 10 min at 72°C. The sizes of PCR products were checked by electrophoresis in 1.5% agarose.
PCR amplicons were purified from a PCR mixture by two washes with 100 μl of double-distilled water on a MultiScreen PCR plate (Millipore, Molsheim, France), resuspended in 30 μl of double-distilled water, and visually quantified by using an agarose gel. The cycle sequencing reaction was performed with 3 to 10 ng of purified PCR product by using an ABI PRISM BigDye Terminator v3.0 cycle sequencing kit (Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions and primers hrcN-4r and hrcN-5rR, each at a final concentration of 0.16 μM. Cycle sequencing products were cleaned by passage through water-swelled Sephadex G-50 columns (Amersham Biosciences, Uppsala, Sweden) on MultiScreen HV plates (Millipore) and were sequenced with an ABI PRISM 3100 genetic analyzer. The identities of the sequenced fragments were determined by BLASTN comparison with known sequences.
PCR amplification and sequencing of 16S rDNA.
PCR amplification of approximately 1.5 kb of the 16S ribosomal DNA (rDNA) gene (rrs) was performed by using the universal 20-mer primers PH-16S and PA-16S (12) (Table 2). PCR was done by using the conditions described above but with initial denaturation for 5 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 60°C, and 45 s at 72°C and a final elongation for 7 min at 72°C. Purification and sequencing of rrs amplicons were performed as described above for hrcN, except that an additional primer, Intern 2a (derived in this work from Intern 2 proposed by Johnsen et al. [28]) (Table 2), was used when it was needed to obtain complete double-stranded coverage of rrs. Sequences were obtained for 18 strains.
Phylogenetic inference.
DNA sequences were assembled by using the freeware Chromas, version 1.45 (http://www.technelysium.com.au/chromas.html; Technelysium Pty. Ltd., Helensvale, Australia), and were aligned with ClustalW (60). Sites with alignment gaps were excluded from the analysis. The Molecular Evolutionary Genetics Analysis (MEGA) program, version 2.1 (33), was used to calculate evolutionary distances and to infer trees based on the neighbor-joining (NJ) and maximum-parsimony (MP) methods. The maximum-likelihood (ML) tree for hrcN was calculated with the Phylogenetic Analysis Using Parsimony package (PAUP*, version 4.0; D. L. Swofford, Sinauer Associates, Sunderland, Mass.) by using a heuristic search. The pathogen Xanthomonas campestris was used as the outgroup in the rrs phylogenetic tree (NJ method), and the flagellar ATPase gene fliI of Pseudomonas putida KT2440 was used as the outgroup in the hrcN trees. The best evolutionary model was inferred by calculating log likelihood scores with the program ModelTest (45) implemented in PAUP*, version 4.0, and on this basis the number of nucleotide substitutions per site was estimated with the help of the Jukes-Cantor (JC) formula with the gamma parameter (29) (implemented in MEGA). The nodal robustness of the inferred trees was assessed by using 200 (ML method) or 1,000 (NJ and MP methods) bootstrap replicates.
DNA sequences were translated into predicted HrcN amino acid sequences, and an NJ tree was constructed with MEGA based on the number of differences or Poisson-corrected distances. MP analysis of protein sequences was also performed with MEGA by using a close-neighbor-interchange approach. FliI of P. putida KT2440 was used as the outgroup. Statistical support of the inferred trees was assessed by using 1,000 bootstrap replicates.
Detection of TTSS by hybridization.
Detection of the TTSS gene hrcV in Pseudomonas strains KD, K92.46, K94.14, K94.23, K93.37, CHA0, and P3 was carried out by hybridization to a digoxigenin-labeled probe which corresponded to positions 1376 to 2132 of hrcV in E. amylovora CNPB136 (accession number L25828) and was obtained by amplifying a 757-bp fragment from plasmid pCPP1103 (7) with primers HRCV-L and HRCV-R (Table 2), as described by Stuber et al. (58). Genomic DNA was purified by the Triton-Prep method, which consisted essentially of lysis of pelleted cells by 90 s of boiling in STET buffer (8% sucrose, 5% Triton X-100, 50 mM Tris-HCl, 50 mM EDTA; pH 8.0) containing 1 mg of lysozyme ml−1 and 0.1 mg of RNase ml−1, followed by phenol-chloroform extraction and precipitation of the DNA with 0.4 M lithium chloride. Purified DNA was digested for 2 h with 1.5 U of PstI and separated on 1.5% agarose. Southern blotting was done (54) by alkaline transfer of DNA onto a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech) by using an LCB 2016 Vacu Gene vacuum blotting unit (Pharmacia LKB Biotechnology AB, Bromma, Sweden). DNA was cross-linked on the membrane by UV irradiation. After overnight hybridization at 60°C, the blots were washed twice for 5 min at room temperature with 2× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate) containing 0.1% sodium dodecyl sulfate. Signals were detected by chemiluminescence by using a DIG luminescent detection kit for nucleic acids (Roche Diagnostic Corporation, Mannheim, Germany), in which disodium 3-(4-meth-oxyspiro{l,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenylphosphate was used as the substrate, and blots were exposed to Kodak Biomax MS film (Eastman Kodak Company, Rochester, N.Y.) and developed according to the manufacturer's instructions.
Analysis of hrcN by dot blot DNA hybridization was performed for all biocontrol strains for which PCR amplification of hrcN failed. The probe consisted of the 249-bp hrcN fragment that was amplified from strain KD by using primers hrcN-4r and hrcN-5rR and was labeled by using the ECL direct nucleic acid labeling and detection system (Amersham Biosciences). Detection was performed according to the manufacturer's instructions. The biocontrol pseudomonads PILH1, Q2-87, TM1′A4, K94.23, F113, P97.38, 2-79, and KD, the pathogenic Pseudomonas strain VS01, and E. amylovora 22770 were used as positive controls, and Escherichia coli strains XL10-Gold (Stratagene, Cedar Creek, Tex.) and DH5α (Invitrogen, Paisley, United Kingdom) were used as negative controls. All controls gave the expected results. All positive results were confirmed by a second hybridization with the hrcV probe described above.
Cloning of a 1.7-kb hrcV fragment from strain KD into plasmid pUK21.
A 1.7-kb PstI fragment from strain KD that hybridized with the hrcV probe was cloned in plasmid pUK21 by using the following procedure. Genomic DNA of strain KD was digested with PstI and electrophoresed on a 1.5% agarose, resulting in a smear. With the help of a 1-kb DNA ladder and a 0.16- to 1.77-kb RNA ladder (Invitrogen), the region containing fragments around 1.7 kb was identified and excised from the gel. The fragments were purified by using a QIAquick gel extraction kit (QIAGEN, Hilden, Germany) and ligated overnight at 4°C by using T4 DNA ligase (Stratagene) into the multiple cloning site of plasmid pUK19 (previously digested with PstI and dephosphorylated). The ligation reaction mixture was used to transform MAX Efficiency DH5α competent cells (Invitrogen) according to the manufacturer's instructions. White-blue screening was performed on LB agar containing kanamycin (50 μg ml−1), 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) (40 μg ml−1), and isopropyl-β-d-thiogalactopyranoside (IPTG) (0.5 mM) to identify colonies transformed with a plasmid containing an insert.
White colonies were subcultured overnight in LB agar containing kanamycin (50 μg ml−1) on 96-well microtiter plates. Ten microliters of each culture was lysed with 190 μl of 0.4 M NaOH-0.01 mM EDTA by heating at 95°C in a thermal cycler for 10 min. The samples were dot blotted onto a Hybond-N+ nylon membrane and hybridized with the hrcV probe, as described above. For positive colonies the plasmid was extracted by using the Wizard Plus SV Minipreps DNA purification system (Promega, Madison, Wis.). The insert was analyzed by restriction with PstI and by sequencing, which was performed by Microsynth GmbH (Balgach, Switzerland).
Partial sequencing of TTSS genes in the biocontrol organism Pseudomonas sp. strain KD.
Partial sequencing of the hrcJ operon of the biocontrol organism Pseudomonas sp. strain KD was carried out as follows. The nucleotide sequences of the hrpJ operons of P. syringae pv. tomato DC3000 (accession number AF232004) and E. amylovora CNPB136 (accession number L25828) were aligned by using LAlign software (26), and primers with low degeneracy were designed manually based on conserved nucleotide sequences. Primer pKD2cV-1r was designed based on sequencing results for the PstI fragment cloned into pUK18. PCRs were performed by using combinations of the different forward and reverse primers and a TGradient thermal gradient cycler (Biometra, Göttingen, Germany). Twelve different PCRs at different annealing temperatures were done to test each set of primers.
PCRs were performed directly with cell lysates of strain KD, which were prepared as described by Rezzonico et al. (52). Briefly, 5 μl of an overnight LB medium culture was mixed with 95 μl of lysis buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 0.1% Tween 20) and heated for 10 min at 99°C in a PTC-100 thermal cycler (MJ Research). PCR amplification was carried out in 20-μl reaction mixtures by using 5 μl of cell lysate and 1× PCR buffer (described above).
After an initial denaturation step consisting of 10 min at 95°C, there were 35 cycles consisting of 1 min at 95°C, 1 min of annealing (12 different temperatures ranging from 52 to 66°C were tested), and 3 min at 72°C, followed by final elongation for 10 min at 72°C. DNA fragments of the expected size were excised from 1.5% agarose electrophoresis gels, purified with a QIAquick gel extraction kit (QIAGEN), and sent to Microsynth GmbH for sequencing with the primers used for amplification of the corresponding PCR fragments. The sequences were analyzed by BLASTN and BLASTP (4).
Nucleotide sequence accession numbers.
Nucleotide sequence data reported here are available at the National Center for Biotechnology Information database under accession numbers AY456697 to AY456712, AY622219, and AY622220 for rrs, AY456994 to AY457036 for hrcN, and AY463491 for hrpL, hrpJ, hrcV, hrpQ, and part of hrcN in strain KD.
RESULTS
Distribution of hrcN sequences in biocontrol Pseudomonas spp.
Degenerate primers hrcN-4r and hrcN-5rR (Table 2) were designed from a comparison of the hrcN sequences of E. amylovora CNPB136 and P. syringae pv. tomato DC3000. They amplified a fragment that was about 250 bp long, as predicted, both in the biocontrol agent Pseudomonas sp. strain KD and the plant pathogens E. amylovora and P. syringae (Fig. 1A), which are known to have a TTSS. The following PCR conditions were the best compromise between reaction specificity and target detection for biocontrol pseudomonads: after an initial denaturation step of 10 min at 95°C, 30 cycles consisting of 30 s at 95°C, 30 s of annealing at 60°C, and 1 min at 72°C, followed by a final elongation for 10 min at 72°C.
In addition to strain KD, PCR amplification was also successful for 33 of 57 biocontrol fluorescent pseudomonads (Fig. 2 and Table 1). The identities of the amplified fragments were confirmed by sequencing followed by database comparison by using BLASTN. hrcN amplicons were also obtained with nonbiocontrol Pseudomonas strains P3, BE07, BE08, VS01, and VS02. All fragments of biocontrol pseudomonads analyzed showed the highest levels of similarity with previously sequenced hrcN genes, like the gene found in P. syringae pv. tomato DC3000 (the levels of nucleotide identity were between 72.5% for K94.08 and 85.7% for KD), and only moderate levels of similarity (less than 60% identity) with other ATPase genes, such as the flagellum-associated ATPase gene fliI of P. putida KT2440. BLASTP analysis of the deduced 81-amino-acid HrcN sequence in biocontrol pseudomonads revealed the presence (in the first 46 residues) of a conserved ATP synthase α/β family nucleotide-binding domain (CDD accession number pfam00006), which included one α helix and two β sheets (Fig. 1B). This family includes the α and β subunits of the flagellum-associated ATP synthase.
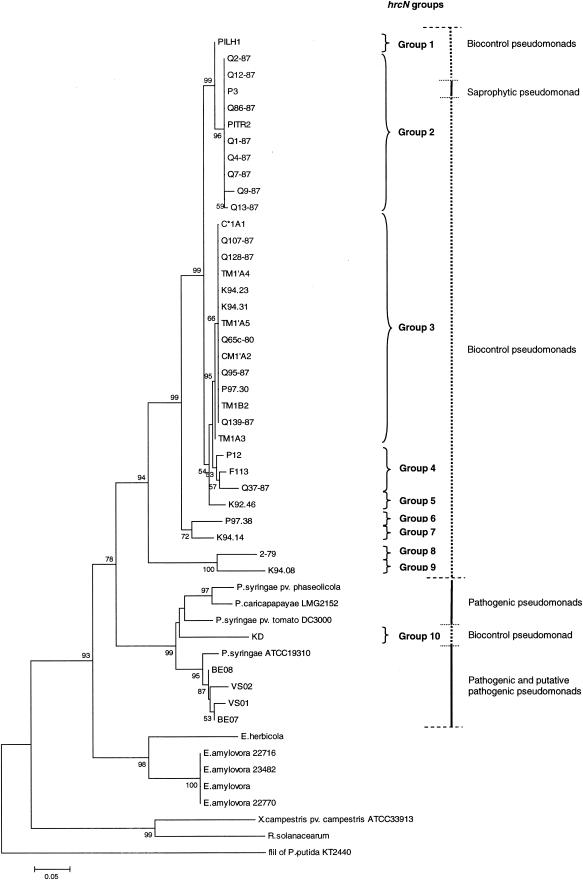
FIG.2.
Phylogenetic relationships based on partial hrcN sequences of biocontrol fluorescent pseudomonads and phytopathogenic bacteria belonging to the genera Pseudomonas, Erwinia, Xanthomonas, and Ralstonia. The distance tree was generated by the NJ method with the JC formula by using the flagellar ATPase gene fliI of P. putida KT2440 (accession number AE016790) as the outgroup. Nodal support was assessed by using 1,000 bootstrap replicates. Only bootstrap values greater than 50% are shown. Scale bar = 0.05 substitution per site. hrcN groups arbitrarily defined for biocontrol pseudomonads based on the topology of the hrcN tree are indicated on the right. The phytopathogenic bacteria included P. syringae pv. phaseolicola (accession number AJ430232), P. syringae pv. tomato DC3000 (AE016860), Erwinia herbicola (X99768), E. amylovora (L25828), X. campestris pv. campestris ATCC 33913 (AE012222), and Ralstonia solanacearum (AJ245811). Sequences were obtained in this study for Pseudomonas strains KD (AY456994), VS01 (AY456998), VS02 (AY456999), BE07 (AY457000), BE08 (AY457001), K94.23 (AY457002), C*1A1 (AY457003), TM1′A4 (AY457004), PITR2 (AY457005), K94.31 (AY457006), Q65c-80 (AY457007), CM1′A2 (AY457008), TM1′A5 (AY457009), Q1-87 (AY457010), TM1B2 (AY457011), Q7-87 (AY457012), Q86-87 (AY457013), Q9-87 (AY457014), Q12-87 (AY457015), Q128-87 (AY457016), P97.30 (AY457017), Q139-87 (AY457018), TM1A3 (AY457019), P97.38 (AY457020), F113 (AY457021), Q95-87 (AY457022), Q37-87 (AY457023), and 2-79 (AY457024), Pseudomonas caricapapayae LGM2152 (AY457025), Pseudomonas strains Q4-87 (AY457026), Q2-87 (AY457027), P12 (AY457028), P3 (AY457029), K92.46 (AY457030), K94.14 (AY457031), Q107-87 (AY457032), Q13-87 (AY457033), K94.08 (AY457034), and PILH1 (AY457035), P. syringae ATCC 19310 (AY457036), and E. amylovora 22716 (AY456995), 22770 (AY456996), and 23482 (AY456997). The G+C contents of the hrcN fragment studied were 58.3% ± 2.5% for the Erwinia cluster (n = 5), 60.4% ± 2.0% for the cluster comprised of established and putative pathogenic pseudomonads and strain KD (n = 9) (the G+C content of KD was 60.6%), and 61.7% ± 1.0% for the cluster containing all biocontrol pseudomonads except strain KD and the saprophytic pseudomonad P3 (n = 33). The values for the latter two clusters were significantly different (P < 0.05, as determined by the Mann-Whitney test).
The use of PCR enhancers, such as bovine serum albumin (25 μg ml−1) and dimethyl sulfoxide (5.5 mg ml−1), had no effect on PCR results in most cases. The exception was strain P97.26, which yielded an amplicon of the expected size only when both bovine serum albumin and dimethyl sulfoxide were added, but sequencing of this fragment failed. Several other primers were designed based on whole hrcN sequences, but when PCR products were obtained, it turned out that amplification was either nonspecific or specific only for some strains. Of the 24 strains that were hrcN negative by PCR, only 2 (DR54 and Pf1) hybridized to an hrcN probe. Both of these strains reacted to also an hrcV hybridization probe, but they did not yield any TTSS-related PCR amplicon.
There was no particular relationship between the occurrence of hrcN and the geographic origin of biocontrol pseudomonads, as the gene was found in strains from different continents. Similarly, the occurrence of hrcN was not linked with the type of plant host, as the gene was identified in biocontrol pseudomonads isolated from wheat, tobacco, cucumber, tomato, and sugar beet. hrcN was found in almost all groups of biocontrol pseudomonads described previously (30, 50, 64) based on the phylogeny of hcnBC, phlD, or rrs or the results of amplified 16S rDNA restriction analysis (ARDRA) or random amplified polymorphic DNA (RAPD) analysis. No amplification was obtained with strains belonging to the group containing strain CHA0, which can be distinguished from other biocontrol pseudomonads based on the analysis of RAPD markers (RAPD group 1) (30) or particular genes, including rrs (ARDRA group 1) (30), phlD (group PhlD1) (64), and hcnBC (group Hcn-4) (50), but as indicated above, hrcN hybridization was successful for one RAPD-1 ARDRA-1 strain (strain Pf1).
Phylogenetic analysis of hrcN in biocontrol Pseudomonas spp.
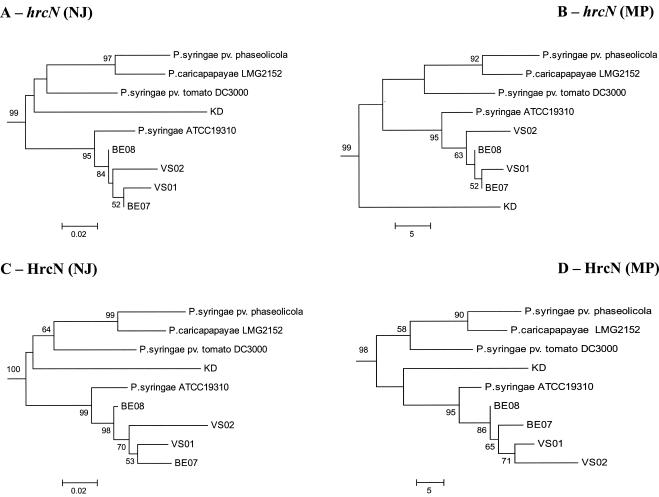
The hrcN alleles identified in biocontrol pseudomonads were similar to those in pathogenic pseudomonads (as determined by BLASTN analysis), yet most of them clustered separately from hrcN sequences of E. amylovora and pathogenic Pseudomonas strains in NJ phylogenetic comparisons (Fig. 2). Nevertheless, four strains (BE07, BE08, P3, and the biocontrol pseudomonad strain KD) had incongruent phylogenetic positions when hrcN and rrs trees were compared. Two strains belonging to the Pseudomonas fluorescens complex based on rrs properties (Fig. 3), i.e., strain KD (closely related to Pseudomonas corrugata and Pseudomonas brassicacearum) and strain BE08, as well as strain BE07 (belonging to the Pseudomonas aeruginosa complex), clustered with P. syringae when hrcN was considered (Fig. 2). P. putida P3 clustered with bacteria belonging to the P. fluorescens complex in the hrcN tree. The same findings were obtained when the MP or ML methods were used to construct the hrcN tree and when trees derived from deduced protein sequences were analyzed (data not shown). However, the internal position of KD within the P. syringae cluster in the hrcN and HrcN trees varied depending on the inference method (Fig. 4).
FIG.3.
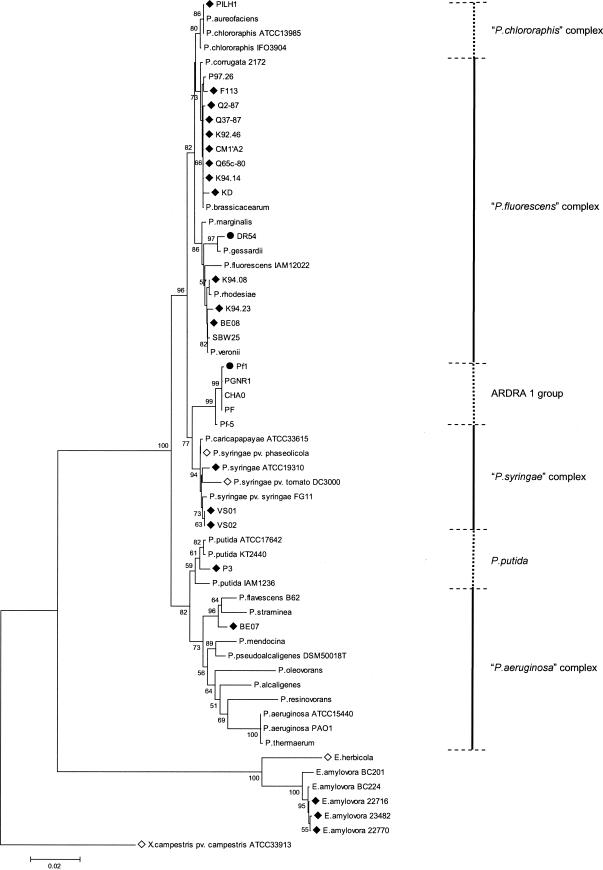
Phylogenetic relationships based on 16S rDNA sequences for biocontrol fluorescent pseudomonads and reference bacteria belonging to the genera Pseudomonas and Erwinia. The distance tree was generated by the NJ method with the JC formula by using X. campestris pv. campestris ATCC 33913 (accession number AE012540) as the outgroup. Nodal support was assessed by using 1,000 bootstrap replicates. Only bootstrap values greater than 50% are shown. Scale bar = 0.02 substitution per site. The P. chlororaphis, P. fluorescens, P. syringae, and P. aeruginosa complexes were those defined in Anzai et al. (5). Strains for which the hrcN sequence is available are indicated by a solid diamond for new sequences or an open diamond for hrcN sequences published previously (National Center for Biotechnology Information). The two strains positive by hrcN hybridization but hrcN negative as determined by PCR are indicated by solid circles. Sequences were obtained in this work for Pseudomonas strains KD (accession number AY456697), VS01 (AY456698), VS02 (AY456699), BE07 (AY456700), BE08 (AY456701), K92.46 (AY456702), K94.08 (AY456703), K94.14 (AY456704), K94.23 (AY456705), P3 (AY456706), P97.26 (AY456707), SBW25 (AY456712), and PILH1 (AY456708) and for E. amylovora 22716 (AY456709), 22770 (AY456710), and 23482 (AY456711). The other sequences used were the sequences for pseudomonad strains CHA0 (AJ278812), Q2-87 (AJ278813), F113 (AJ278814), CM1′A2 (AJ417068), Q37-87 (AJ417069), PGNR1 (AJ417071), Pf-5 (AJ417072), PF (AJ417073), Q65c-80 (AJ417074), DR54 (AY622219), and Pf1 (AY622220), P. chlororaphis ATCC 13985 (AF094722) and IFO3904 (D86004), P. aureofaciens (Z76656), P. corrugata 2172 (= ATCC 29736) (D84012), P. brassicacearum (AF100322), P. marginalis (AB021401), P. fluorescens IAM12022 (D84013), P. gessardii (AF074384), P. rhodesiae (AF064459), P. veronii (AB056120), P. caricapapayae ATCC 33615 (D84010), P. syringae pv. phaseolicola (AB001448) and ATCC 19310 (D84026), P. syringae pv. tomato DC3000 (AE016875), P. syringae pv. syringae FG11 (AY242068), P. putida KT2440 (AE016775), ATCC 17642 (AF094744), and IAM1236 (D84020), P. flavescens B62 (V01916), P. straminea (D084023), P. mendocina (D84016), P. pseudoalcaligenes (Z76675), P. oleovorans (D84018), P. alcaligenes (Z76653), P. resinovorans (Z76668), P. aeruginosa PAO1 (AE004844) and ATCC 15442 (AF094718), P. thermaerum (AB088116), Erwinia herbicola (U80202), and E. amylovora BC201 (AF141892) and BC224 (AF140339).
FIG. 4.
Relationship between the biocontrol organism Pseudomonas sp. strain KD and pathogenic (or putatively pathogenic) pseudomonads in the P. syringae cluster obtained by phylogenetic analysis of TTSS sequences. All trees were constructed by using the complete collection of strains shown in Fig. 2, but only the P. syringae cluster is shown. Trees were obtained for partial hrcN sequences by using the NJ (A) or MP (B) method and the flagellar ATPase gene fliI of P. putida KT2440 as the outgroup, as well as for deduced HrcN sequences by using the NJ (C) or MP (D) method and FliI of P. putida KT2440 as the outgroup. The JC formula was used for nucleotide sequences, and the Poisson correction was used for deduced amino acid sequences. Nodal support was assessed by using 1,000 bootstrap replicates, and only bootstrap values greater than 50% are shown. The scale bars indicate the number of substitutions per site (NJ trees) or the number of changes (MP trees).
hrcN groups were arbitrarily defined for biocontrol pseudomonads based on the topology of the NJ hrcN tree (Fig. 2). The hrcN pairwise distances were less than 0.018 base substitution per site within each group, with the exception of strains in group 4, which had pairwise distances ranging from 0.022 to 0.040 substitution per site. The same groups could be identified by using deduced protein sequences, and the numbers of amino acid substitutions per site were less than 0.049 within the groups. Two main hrcN groups (groups 2 and 3) were identified. Group 2 contained mostly biocontrol pseudomonads isolated from wheat in Washington state, along with a wheat isolate from Italy (strain PITR2) and another monocot strain (barley isolate P3, which has no biocontrol activity). Group 3 included biocontrol strains from wheat, cucumber, and tomato, which originated from Washington state, Switzerland, and the Czech Republic. Several groups were comprised of a single strain. These results were strongly supported by MP or ML analysis of hrcN and analysis of deduced HrcN sequences (by the NJ or MP method).
Certain strains used in this work can produce the biocontrol metabolite HCN and/or the biocontrol metabolite Phl and have been compared previously based on hcnBC or phlD properties (50, 51, 64). Here, a relationship was found between hrcN groups and previous Pseudomonas biocontrol groups derived from analysis of hcnBC, phlD, or RAPD markers (Table 3). For instance, hrcN group 2 contained strains belonging to groups PhlD3/Hcn-3 and PhlD7/Hcn-2, whereas hrcN group 3 included strains belonging to groups PhlD4 (or PhlD2) and Hcn-1. This relationship was not perfect; for example, certain strains belonging to hrcN groups 1, 2, and 4 belonged to a single hcnBC group (group Hcn-2). When hrcN groups and rrs properties were compared, it appeared that hrcN group 1 strains were found only in the rrs-defined Pseudomonas chlororaphis complex (Fig. 2 and 3). Within the P. fluorescens complex, hrcN groups 2, 4, 5, 7 and 10 were associated with the P. corrugata-P. brassicacearum cluster, and hrcN group 9 was associated with the Pseudomonas marginalis-P. fluorescens cluster, but hrcN group 3 was found in both clusters.
TABLE 3.
Relationship among hrcN, phlD, hcnBC, and RAPD groups for HCN+ Phl+ biocontrol pseudomonadsa
| hrcN group | Strain(s) | phlD group | hcnBC group | RAPD group |
|---|---|---|---|---|
| 1 | PILH1 | PhlD7 | Hcn-2 | 5 |
| 2 | P1TR2 | PhlD7 | Hcn-2 | 5 |
| Q1-87, Q4-87, Q7-87, Q9-87, Q12-87, Q13-87, Q86-87b | NDd | Hcn-3 | 4 | |
| Q2-87 | PhlD3 | Hcn-3 | 4 | |
| 3 | K94.31 | PhlD2 | Hcn-1 | ND |
| Q65c-80, Q95-87, Q107-87, Q128-87, Q139-87c | ND | Hcn-1 | 3 | |
| TM1A3, TM1′A4, TM1′A5, TM1B2, C*1A1, CM1′A2 | PhlD4 | Hcn-1 | 3 | |
| 4 | P12 | ND | Hcn-2 | 8 |
| Q37-87 | ND | Hcn-2 | 6 | |
| F113 | PhlD6 | Hcn-1 | 7 | |
| 6 | P97.38 | PhlD8 | Hcn-3 | ND |
phlD, hcnBC, and RAPD groups were determined by Wang et al. (64), Ramette et al. (51), and Keel et al. (30), respectively.
These strains produced the same phlD restriction pattern (HaeIII) as group PhlD3 strain Q2-87, but phlD was not sequenced (unlike Q2-87).
These strains produced the same phlD restriction pattern (HaeIII) as group PhlD4 strains TM1A3, TM1B2, and C*1A1, but phlD was not sequenced (unlike TM1A3, TM1B2, and C*1A1).
ND, not determined.
Organization of TTSS genes in the biocontrol organism Pseudomonas sp. strain KD.
Pseudomonas sp. strain KD was the only biocontrol strain for which incongruent data were observed when the hrcN and rrs trees were compared. If the horizontal gene transfer hypothesis is valid for this strain, it can be anticipated that in addition to having a similar hrcN allele, strain KD would display the same organization of TTSS genes as P. syringae. Therefore, the organization of TTSS genes was investigated in Pseudomonas sp. strain KD. This was done by using an hrcV probe that was derived from E. amylovora CNPB136 (as described by Stuber et al. [58]) and, as expected, hybridized with E. amylovora CNPB136 and Erwinia chrysanthemi (data not shown). A positive response was observed with strain KD (and with the hrcN+ biocontrol pseudomonads K92.46 and K94.14).
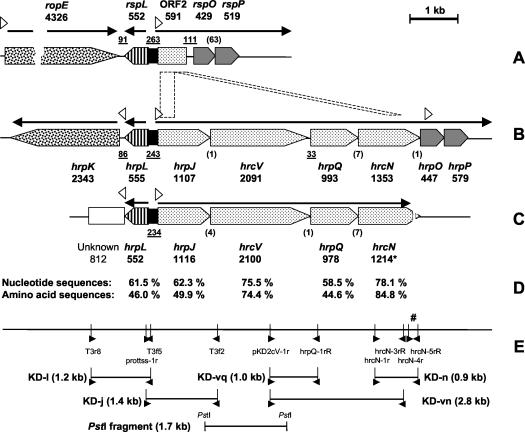
In strain KD, the hrcV probe hybridized with a PstI fragment that was approximately 1.7 kb long, which was subsequently cloned in plasmid pUK21 and sequenced. The insert was verified by digestion with PstI and dot blot hybridization to the hrcV probe, and its size was determined to be 1,718 bp based on sequencing results. The insert corresponded to a region that included the last 219 bp of hrpJ and the first 1,503 bp of hrcV, with the two genes overlapping by 4 bp (Fig. 5). The plasmid was designated pCBTypeIII.
FIG. 5.
(A to C) Comparison of the organization of TTSS genes in the biocontrol organism Pseudomonas sp. strain SBW25 (A), the phytopathogen P. syringae pv. tomato DC3000 (B), and the biocontrol organism Pseudomonas sp. strain KD (accession number AY463491) (C). (D) Levels of nucleotide and amino acid sequence identity between strains DC3000 and KD. (E) Positions of PCR primers used for sequencing. In panels A, B and C, the length of each gene (in base pairs) is indicated below its designation; one gene (hrcN) was sequenced only partially (indicated by an asterisk). The lengths of noncoding, intergenic gaps are underlined. The numbers of bases shared by overlapping genes are indicated in parentheses. The arrows above the genes represent the operons and the direction in which they are transcribed in DC3000. The open triangles indicate the positions and orientations of the hrp transcription boxes. In panel E, the positions and directions of primers used for sequencing are indicated by solid triangles. The number sign indicates the position of the hrcN-4r-hrcN-5rR fragment used for phylogenetic analysis.
Several degenerate PCR primer sets were designed (Table 2) based on sequence alignment of TTSS genes from P. syringae pv. tomato DC3000 and E. amylovora CNPB136, and they were tested in strain KD at 12 different annealing temperatures between 52 and 66°C. Fragment KD-vq (990 bp) (Fig. 5) was obtained with primers pKD2cV-1r and hrpQ-1rR by using an annealing temperature of 60°C. Fragment KD-n (902 bp) was obtained with primers hrcN-1r and hrcN-5rR at an annealing temperature of 62°C, and fragment KD-vn (2,816 bp) was obtained with primers pKD2cV-1r and hrcN-3rR at an annealing temperature of 58°C. Assembled together, the three PCR amplicons and the 1,718-bp PstI fragment from pCBTypeIII yielded a 4,499-bp sequence spanning from hrpJ to hrcN, which was homologous to the corresponding region in the hrpJ operon of P. syringae pv. tomato DC3000 (Fig. 5). This hrpJ operon is a 5,651-bp regulation unit, which includes hrpJ, hrcV, hrpQ, and hrcN.
Other primers (Table 2) were used to sequence upstream of the PstI fragment and recover the remaining 897 bp of hrpJ. Fragment KD-j (1,446 bp) was obtained with primers prottss-1r and T3f2 at an annealing temperature of 62°C, and fragment KD-l (1,242 bp) was obtained with primers T3r8 and T3f5 at an annealing temperature of 63°C (Fig. 5E). The presence of an open reading frame homologous (61.5% nucleotide identity) to the hrpL gene of DC3000 was demonstrated. hrpL encodes a TTSS-specific sigma factor necessary for recognition of hrp boxes and transcription of the corresponding TTSS genes.
Overall, it appears that the TTSS region studied in Pseudomonas sp. strain KD is organized in a similar manner in P. syringae pv. tomato DC3000 and in Pseudomonas sp. strains SBW25 and KD, except that in SBW25 part of the hrpJ operon is absent (Fig. 5A). For both DNA and deduced protein sequences, the levels of identity between strains KD and DC3000 varied according to the TTSS gene considered (Fig. 5D). With strain SBW25, comparisons could only be made for hrpL, and the levels of identity between KD and DC3000 (61.5% for nucleotides, 46.0% for amino acids) were higher than the levels of identity between KD and SBW25 (56.2% for nucleotides, 40.5% for amino acids). In contrast, the genes or sequences downstream of hrpL were different in the three strains. This position is often occupied by effector genes, which are usually poorly conserved (2, 11).
DISCUSSION
In this work, hrcN was found in about 60% of the biocontrol pseudomonads studied. This gene is not present in Pseudomonas sp. strain SBW25 (which was confirmed here), a strain that nevertheless contains other TTSS genes (47). One prominent group of biocontrol pseudomonads includes strains that produce HCN, Phl, and Plt (pyoluteorin) (previously designated ARDRA group 1 [30]), and 9 of the 10 strains from this group studied were hrcN negative. PCR with these 10 strains also failed when alternative PCR primers were used or when other TTSS genes were targeted. This indicates that for the most part a TTSS is absent from this phylogenetic group.
For a majority of hrcN+ biocontrol strains, the hrcN and rrs trees were largely congruent, which means that hrcN evolved in parallel with rrs and diverged sometime in the past in the corresponding taxa, rather than resulting from recent gene transfer. In this context, the hrcN alleles of most biocontrol pseudomonads differed clearly from those found in their phytopathogenic counterparts, in which, at least for P. syringae pathovars, the hrp gene cluster is considered to be ancestral (55). Two main groups of nonpathogenic pseudomonads were defined based on the hrcN tree (Fig. 2). Considering the geographic origins of the strains in each of these two groups, it appears that both groups have a cosmopolitan distribution worldwide. One of them was comprised only of monocot isolates, which might have ecological implications in terms of adaptation to the plant, whereas the other contained pseudomonads from monocots and dicots.
The ability to produce biocontrol metabolites (e.g., HCN and Phl) is widespread in biocontrol pseudomonads, and many strains included in this study have also been analyzed based on hcnBC and phlD sequences (50, 51). In the present work, there was some relationship between the hrcN phylogeny and the phylogenies based on phlD or hcnBC. The existence of this relationship can be explained by the fact that the phlD, hcnBC, and rrs phylogenies were highly congruent, which is also illustrated by the relationship among the hrcN, phlD, hcnBC, and RAPD groups (Table 3).
In Pseudomonas spp., the 16S rDNA-based phylogeny can be considered the species phylogeny (5, 40). Based on this assumption, incongruent results were obtained for four strains (BE07, BE08, P3, and KD) when the hrcV and rrs trees were compared, which raises the possibility of lateral gene transfer. This did not come as a surprise, as it has been established that TTSS genes are often present in pathogenicity islands, which are prone to lateral gene transfer (38). Strains BE07 and BE08 clustered with P. syringae based on hrcN phylogeny. Strain BE07 is probably a pathogen since it was isolated from a sugar beet disease lesion and displays 98.4% rrs identity with the walnut blight canker isolate Pseudomonas flavescens B62 (24) (accession number U01916), which is distinct from P. syringae. Therefore, this finding points to lateral transfer of TTSS genes between different pathogenic Pseudomonas species. The status of strain BE08 is less clear because the closest relatives of this sugar beet disease lesion isolate are bioremediation strains belonging to Pseudomonas veronii (1) (99.5% rrs identity; accession number AB056120) and biocontrol pseudomonads (including the hypersensitive response-inducing strain SBW25), both of which belong to the P. fluorescens complex. P. putida P3, which also contains hrcV (data not shown), clustered with biocontrol strains belonging to the P. fluorescens group based on hrcN, but this bacterium has no biocontrol capacity.
The case of Pseudomonas sp. strain KD is of particular interest since this strain was the only biocontrol strain that clustered among P. syringae pathogens in the hrcN tree. Strain KD belongs to the P. fluorescens complex. It produces HCN and a siderophore(s) but not Phl or Plt, and it efficiently suppresses Pythium ultimum on cucumber and Fusarium oxysporum f. sp. radicis-lycopersici on tomato (56). Although it can be hypothesized that this biocontrol bacterium acquired a TTSS gene(s) from P. syringae, no plasmid was found in strain KD when alkaline lysis was used (54), when the Wizard Plus SV Minipreps DNA purification system (Promega) was used, or when QIAGEN plasmid maxi and mega kits (QIAGEN) were used, indicating that hrcN is chromosomal. Despite the presence of a P. syringae-like TTSS gene(s), this strain did not elicit a hypersensitive response in tobacco or cucumber (data not shown) and is not known to cause any plant disease. When additional TTSS genes in strain KD were sought, it appeared that unlike Pseudomonas sp. strain SBW25, (i) other TTSS genes, such as hrpQ, were present and (ii) the organization of the hrpJ operon was the same as that in P. syringae. Thus, it is conceivable that KD acquired several TTSS genes, or even a whole pathogenicity island, from P. syringae. The position of KD within the P. syringae cluster differed somewhat in the MP hrcN tree compared with the two other hrcN trees and the HrcN trees, which might correspond to a long-branch attraction artifact (13).
This work was based on the assumption that gene transfer took place from phytopathogenic to biocontrol bacteria rather than the other way around, because individual pathogens were never found in biocontrol clusters in the TTSS trees, despite the fact that all available hrcN sequences from phytopathogens were considered. In contrast, one biocontrol pseudomonad (strain KD) clustered with its pathogenic counterparts. However, it must be kept in mind that very little has been done to date to analyze the organization and role of TTSS in plant-beneficial bacteria, and this area deserves further work. For instance, when the numbers of nonsynonymous substitutions per nonsynonymous site (dN) and the numbers of synonymous substitutions per synonymous site (dS) for hrcN were estimated by using the method of Nei and Kumar (41) and MEGA, it appeared that dS was significantly greater (as determined by Fisher exact tests) than dN, both for pathogenic pseudomonads (0.314 ± 0.070 versus 0.022 ± 0.008) and for biocontrol pseudomonads (0.218 ± 0.039 versus 0.031 ± 0.008 for hrcN groups 1 to 9 [i.e., without KD]; and 0.250 ± 0.043 versus 0.045 ± 0.009 for hrcN groups 1 to 10 [when KD was included]). For comparison, the dS and dN values for the five Erwinia strains studied were 0.246 ± 0.077 and 0.042 ± 0.014, respectively. These results indicate that hrcN is subjected to purifying selection in both types of pseudomonads, indicating that this gene has an important ecological role in biocontrol pseudomonads, as in pathogenic strains. Accordingly, the TTSS gene rscC/hrcC is expressed in Pseudomonas sp. strain SBW25 in the rhizosphere (48), and preliminary results obtained by using transcriptional fusions indicate that the TTSS genes hrpJ (from the same operon as hrcN) and hrpL are transcribed in Pseudomonas sp. strain KD under in vitro conditions (unpublished data). This suggests that TTSS genes are functional in biocontrol pseudomonads.
In conclusion, it appears that the presence of hrcN in biocontrol pseudomonads is ancient in most cases, but in the biocontrol strain Pseudomonas sp. strain KD a set of TTSS genes was probably acquired more recently from phytopathogenic P. syringae. Nothing is known about the possible role of TTSS in biocontrol, and it will be of interest to determine whether TTSS genes contribute to or interfere with the biocontrol activity of strain KD and other biocontrol pseudomonads.
Acknowledgments
We thank C. Binder (ETH Zürich) for technical help with cloning of hrcV and hybridization experiments, C. Prigent-Combaret (Université Lyon 1) for useful discussions, G. M. Preston (University of Oxford, Oxford, United Kingdom) for providing strain SBW25, T. H. Nielsen (Royal Veterinary and Agricultural University, Copenhagen, Denmark) for providing strain DR54, and E. Holliger (FAW Wädenswil) for the gift of purified DNA from Erwinia strains.
This work was supported by the Swiss National Foundation for Scientific Research (project 31-64048.00), the French Embassy in Switzerland (France-Switzerland research grant), and the PAI project Germaine de Staël.
REFERENCES
- 1.Ajithkumar, B., V. P. Ajithkumar, and R. Iriye. 2003. Degradation of 4-amylphenol and 4-hexylphenol by a new activated sludge isolate of Pseudomonas veronii and proposal for a new subspecies status. Res. Microbiol. 154:17-23. [DOI] [PubMed] [Google Scholar]
- 2.Alfano, J. R., A. O. Charkowski, W.-L. Deng, J. L. Badel, T. Petnicki-Ocwieja, K. van Dijk, and A. Collmer. 2000. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 97:4856-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfano, J., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Anzai, Y., H. Kim, J.-Y. Park, H. Wakabayashi, and H. Oyaizu. 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50:1563-1589. [DOI] [PubMed] [Google Scholar]
- 6.Beer, S. V., D. W. Bauer, X. H. Jiang, R. J. Laby, B. J. Sneath, Z. M. Wei, D. A. Wilcox, and C. H. Zumoff. 1991. The hrp gene cluster of Erwinia amylovora, p. 53-60. In H. Hennecke and D. P. S. Verma (ed.), Advances in molecular genetics of plant-microbe interactions. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Bogdanove, A. J., Z. M. Wei, L. Zhao, and S. V. Beer. 1996. Erwinia amylovora secretes harpin via a type III pathway and contains a homolog of yopN of Yersinia spp. J. Bacteriol. 178:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, N. A. 1934. A gall similar to crown gall, produced on Gypsophila by a new bacterium. J. Agric. Res. 48:1099-1112. [Google Scholar]
- 9.Burkholder, W. H. 1926. A new bacterial disease of the bean. Phytopathology 16:915-928. [Google Scholar]
- 10.Cuppels, D. A. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, W.-L., A. H. Rehm, A. O. Charkowski, C. M. Rojas, and A. Collmer. 2003. Pseudomonas syringae exchangeable effector loci: sequence diversity in representative pathovars and virulence function in P. syringae pv. syringae B728a. J. Bacteriol. 185:2592-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27:401-410. [Google Scholar]
- 14.Fenton, A. M., P. M. Stephens, J. Crowley, M. O'Callaghan, and F. O'Gara. 1992. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl. Environ. Microbiol. 58:3873-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-491. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, J., and G. Défago. 1991. Protection of cucumber plants against black root rot caused by Phomopsis sclerotioides with rhizobacteria, p. 57-62. In C. Keel, B. Koller, and G. Défago (ed.), Plant growth-promoting rhizobacteria—progress and prospects (IOBC/WPRS Bull XIV/8). IOBC/WPRS, Interlaken, Switzerland.
- 17.Galan, G., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 18.Goguen, J. D., J. Yother, and S. C. Straley. 1984. Genetic analysis of the low calcium response in Yersinia pestis Mu d1 (Ap lac) insertion mutants. J. Bacteriol. 160:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gophna, U., E. Z. Ron, and D. Graur. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151-163. [DOI] [PubMed] [Google Scholar]
- 20.Gottfert, M., S. Rothlisberger, C. Kundig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. Mol. Microbiol. 183:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison, L. A., L. Letendre, P. Kovacevich, E. Pierson, and D. Weller. 1993. Purification of an antibiotic effective against Gaeumannomyces graminis var. tritici produced by a biocontrol agent, Pseudomonas aureofaciens. Soil Biol. Biochem. 25:215-221. [Google Scholar]
- 22.He, S. Y. 1998. Type III protein secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 36:363-392. [DOI] [PubMed] [Google Scholar]
- 23.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrand, D. C., N. J. Palleroni, M. Hendson, J. Toth, and J. L. Johnson. 1994. Pseudomonas flavescens sp. nov., isolated from walnut blight cankers. Int. J. Syst. Bacteriol. 44:410-415. [DOI] [PubMed] [Google Scholar]
- 25.Howell, C. R., and R. D. Stipanovic. 1979. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69:480-482. [Google Scholar]
- 26.Huang, X., and W. Miller. 1991. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12:337-357. [Google Scholar]
- 27.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnsen, K., O. Enger, C. S. Jacobsen, L. Thirup, and V. Torsvik. 1999. Quantitative selective PCR of 16S ribosomal DNA correlates well with selective agar plating in describing population dynamics of indigenous Pseudomonas spp. in soil hot spots. Appl. Environ. Microbiol. 65:1786-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press Inc., New York, N.Y.
- 30.Keel, C., D. M. Weller, A. Natsch, G. Défago, R. J. Cook, and L. S. Thomashow. 1996. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62:552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 32.Klement, Z. 1982. Hypersensitivity, p. 149-177. In M. S. Mount and G. H. Lacy (ed.), Phytopathogenic prokaryotes. Academic Press Inc., New York, N.Y.
- 33.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 34.Levy, E., F. J. Gough, K. D. Berlin, P. W. Guiana, and J. T. Smith. 1992. Inhibition of Septoria tritici and other phytopathogenic fungi and bacteria by Pseudomonas fluorescens and its antibiotics. Plant Pathol. 41:335-341. [Google Scholar]
- 35.Lindgren, P. B., R. C. Peet, and N. J. H. Panopoulos. 1986. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity on bean plants and hypersensitive response on non-host plants. J. Bacteriol. 168:512-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindsay, W. P., C. J. Lamb, and R. A. Dixon. 1993. Microbial recognition and activation of plant defense systems. Trends Microbiol. 1:181-187. [DOI] [PubMed] [Google Scholar]
- 37.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propeller and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mecsas, J., and E. Strauss. 1996. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg. Infect. Dis. 2:271-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meinhardt, L. W., H. B. Krishnan, P. A. Balatti, and S. G. Pueppke. 1993. Molecular-cloning and characterization of a Sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 9:17-29. [DOI] [PubMed] [Google Scholar]
- 40.Moore, E. R. B., M. Mau, A. Arnscheidt, E. C. Böttger, R. A. Hutson, M. D. Collins, Y. van de Peer, R. de Wachter, and K. N. Timmis. 1996. The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst. Appl. Microbiol. 19:478-492. [Google Scholar]
- 41.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, England.
- 42.Nguyen, L., I. T. Paulsen, J. Tchieu, C. J. Hueck, and M. H. Saier, Jr. 2000. Phylogenetic analyses of the constituents of type III protein secretion systems. J. Mol. Microbiol. Biotechnol. 2:125-144. [PubMed] [Google Scholar]
- 43.Nielsen, T. H., C. Christophersen, U. Anthoni, and J. Søorensen. 1999. Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 87:80-90. [DOI] [PubMed] [Google Scholar]
- 44.Pammel, L. H. 1895. Bacteriosis of rutabaga (Bacillus campestris n. sp.). Bull. Iowa State College Agric. Exp. Stn. 27:130-134. [Google Scholar]
- 45.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 46.Pozidis, C., A. Chalkiadaki, A. Gomez-Serrano, H. Stahlberg, I. Brown, A. P. Tampakaki, A. Lustig, G. Sianidis, A. S. Politou, A. Engel, N. J. Panopoulos, J. Mansfield, A. P. Pugsley, S. Karamanou, and A. Economou. 2003. Type III protein translocase: HrcN is a peripheral ATPase that is activated by oligomerization. J. Biol. Chem. 278:25816-25824. [DOI] [PubMed] [Google Scholar]
- 47.Preston, G. M., N. Bertrand, and P. B. Rainey. 2001. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol. Microbiol. 41:999-1014. [DOI] [PubMed] [Google Scholar]
- 48.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 49.Rainey, P. B., and M. J. Bailey. 1996. Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol. Microbiol. 19:521-533. [DOI] [PubMed] [Google Scholar]
- 50.Ramette, A., M. Frapolli, G. Défago, and Y. Moënne-Loccoz. 2003. Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Mol. Plant-Microbe Interact. 16:525-535. [DOI] [PubMed] [Google Scholar]
- 51.Ramette, A., Y. Moënne-Loccoz, and G. Défago. 2001. Polymorphism of the polyketide synthase gene phlD in biocontrol fluorescent pseudomonads producing 2,4-diacetylphloroglucinol and comparison of PhlD with plant polyketide synthases. Mol. Plant-Microbe Interact. 14:639-652. [DOI] [PubMed] [Google Scholar]
- 52.Rezzonico, F., Y. Moënne-Loccoz, and G. Défago. 2003. Effect of stress on the ability of a phlA-based quantitative competitive PCR assay to monitor biocontrol strain Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 69:686-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robbs, C. F. 1956. Uma nova doença bacteriana do mamoeiro (Carica papaya L.). Rev. Soc. Bras. Agron. 12:73-76. [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Sawada, H., F. Suzuki, I. Matsuda, and N. Saitou. 1999. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J. Mol. Evol. 49:627-644. [DOI] [PubMed] [Google Scholar]
- 56.Sharifi-Tehrani, A., M. Zala, A. Natsch, Y. Moënne-Loccoz, and G. Défago. 1998. Biocontrol of soilborne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur. J. Plant Pathol. 104:631-643. [Google Scholar]
- 57.Skerman, V. B. D., V. McGowan, and P. H. A. Sneath. 1980. Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30:225-420. [Google Scholar]
- 58.Stuber, K., J. Frey, A. P. Burnens, and P. Kuhnert. 2003. Detection of type III secretion genes as a general indicator of bacterial virulence. Mol. Cell. Probes 17:25-32. [DOI] [PubMed] [Google Scholar]
- 59.Stutz, E. W., G. Défago, and H. Kern. 1986. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76:181-185. [Google Scholar]
- 60.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincent, M. N., M. N. Harrison, L. A. Brackin, J. M. Kovacevich, P. A. Mukerji, D. M. Weller, and E. A. Pierson. 1991. Genetic analysis of the antifungal activity of a soilborne Pseudomonas aureofaciens strain. Appl. Environ. Microbiol. 57:2928-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viprey, V., A. Del Greco, W. Golinowski, W. J. Broughton, and X. Perret. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381-1389. [DOI] [PubMed] [Google Scholar]
- 63.Voisard, C., C. Keel, D. Haas, and G. Défago. 1989. Cyanide production by Pseudomonas fluorescens helps suppress black root of tobacco under gnotobiotic conditions. EMBO J. 8:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, C., A. Ramette, P. Punjasamarnwong, M. Zala, A. Natsch, Y. Moënne-Loccoz, and G. Défago. 2001. Cosmopolitan distribution of phlD-containing dicotyledonous crop-associated biocontrol pseudomonads of worldwide origin. FEMS Microbiol. Ecol. 37:105-116. [Google Scholar]
- 65.Weller, D. M., and R. J. Cook. 1983. Suppression of take-all of wheat by seed treatment with fluorescent pseudomonads. Phytopathology 73:463-469. [Google Scholar]