Abstract
Atomic force microscopy (AFM) has emerged as a powerful technique for mapping the surface morphology of biological specimens, including bacterial cells. Besides creating topographic images, AFM enables us to probe both physicochemical and mechanical properties of bacterial cell surfaces on a nanometer scale. For AFM, bacterial cells need to be firmly anchored to a substratum surface in order to withstand the friction forces from the silicon nitride tip. Different strategies for the immobilization of bacteria have been described in the literature. This paper compares AFM interaction forces obtained between Klebsiella terrigena and silicon nitride for three commonly used immobilization methods, i.e., mechanical trapping of bacteria in membrane filters, physical adsorption of negatively charged bacteria to a positively charged surface, and glutaraldehyde fixation of bacteria to the tip of the microscope. We have shown that different sample preparation techniques give rise to dissimilar interaction forces. Indeed, the physical adsorption of bacterial cells on modified substrata may promote structural rearrangements in bacterial cell surface structures, while glutaraldehyde treatment was shown to induce physicochemical and mechanical changes on bacterial cell surface properties. In general, mechanical trapping of single bacterial cells in filters appears to be the most reliable method for immobilization.
During recent years, atomic force microscopy (AFM) has been increasingly used in the biosciences (5, 16). Theoretically, it combines the two most important aspects of studying structure-function relationships of biological specimens: it performs high-resolution imaging with a high signal-to-noise ratio on a molecular or submolecular scale and has the ability to operate in aqueous environments, allowing the observation of dynamic molecular events in real time and under physiological conditions. The AFM is surprisingly simple in its concept. A sharp tip located at the free end of a flexible cantilever scans over a surface. Interaction forces between the tip and the sample surface subsequently cause the cantilever to deflect. The deflection signal is acquired and digitized to provide a three-dimensional image of the surface.
Several biological specimens have been imaged, with lateral and vertical resolution on a nanometer and a subnanometer scale, respectively (9, 14, 23). However, when living microbial cell surfaces are imaged, the softness of the cell surface together with the high pressure over the contact area between the tip and the cell can prevent high-resolution imaging. Image contrast is indeed influenced by the probe's geometry, the imaging parameters, the surface topography, and the viscoelastic and physicochemical properties of the cell surface. Additional problems arise from friction and from lateral displacement of the organism under study, which makes immobilization strategies critical.
Beyond being an imaging device, the AFM has evolved as an instrument for measuring molecular interaction forces (21, 22). Biological interactions that have been investigated include antibody-antigen recognition, protein-ligand binding, and cDNA base pairing (2, 7, 12, 15, 18). It was further shown that AFM can be applied to measure interaction forces between bacteria and a substratum surface, including the contribution of bacterial polysaccharides to bacterium-surface interactions (10, 20). AFM has also been used to characterize, under aqueous conditions, the supramolecular organization of bacterial extracellular polymeric substances (EPS) adsorbed onto solid substrata (24). Moreover, AFM allowed the calculation of the turgor pressure of magnetotactic bacteria of the species Magnetospirillum gryphiswaldense, which was found to be between 85 and 150 kPa (1).
Immobilization of the organisms is critical not only when the AFM is used for imaging, but also when it is used as a force probe. For probing of the structure, function, and physicochemical and mechanical properties of bacterial cell surfaces under physiological conditions, it is required that immobilization does not affect the chemical and structural integrity of the cell surface. Yet the organisms under study must be firmly anchored in order to withstand the lateral forces of the scanning tip. Different approaches have been used for bacterial immobilization for AFM. For instance, poly-l-lysine or poly(ethyleneimide) (PEI) can be used to create positively charged glass surfaces, promoting an irreversible adhesion of bacteria (3, 26). Glass slides have also been treated with aminosilanes to immobilize bacteria through cross-linking carboxyl groups on their surfaces with amine groups coupled to the glass (11). Razatos et al. developed a procedure for coating the silicon nitride microscope tip with a confluent layer of bacteria in which a drop of glutaraldehyde-treated bacterial suspension was placed on a PEI-coated tip (20). Occasionally, organisms have been immobilized by mechanical trapping on membrane filters with a pore size that is slightly smaller than the dimensions of the bacterium (4, 13, 25). Rarely, minute glass beads bound to functional amino groups have been coated with bacteria and linked to the silicon nitride cantilever by use of a small amount of epoxy resin (19). Other methods for the immobilization of bacteria on surfaces for AFM may exist, but it is considered beyond the scope of this paper to give a comprehensive list of sample preparation techniques. Different immobilization strategies, however, are likely to yield different results by AFM, as not all methods preserve the integrity of the immobilized cells equally well.
Therefore, this paper compares the interaction forces obtained between Klebsiella terrigena and the silicon nitride tip of an AFM for three immobilization methods: (i) mechanical trapping, (ii) adsorption to positively charged glass, and (iii) fixation to the tip.
MATERIALS AND METHODS
Bacterial strain, growth conditions, and harvesting.
The gram-negative strain K. terrigena ATCC 33527, which occurs commonly in soil, water, grain, fruits, and vegetables, was used for this study. K. terrigena was grown aerobically in nutrient broth (Oxoid, Basingstoke, United Kingdom) at 37°C. For each experiment, the strain was inoculated from nutrient agar into a batch culture. This culture was used to inoculate a second culture that was grown for 16 h prior to harvesting. The bacteria were harvested by centrifugation (5 min at 10,000 × g), washed twice with demineralized water, and resuspended in water or in 0.25 mM potassium phosphate buffer at pH 6.8.
Sample preparation.
The immobilization of K. terrigena was performed by three different methods, as follows. (i) Bacterial cells were suspended in water to a concentration of 105 per ml, after which 10 ml of this suspension was filtered through an Isopore polycarbonate membrane (Millipore) with a pore size of 0.8 μm, i.e., slightly smaller than the bacterial dimensions, to immobilize the bacteria through mechanical trapping (17). Filtration was carried out by placing a filter on a vacuum filtration flask, after which bacteria were added to the top of the filter and a vacuum was applied for approximately 10 s. After filtration, the filter was carefully fixed with double-stick tape onto a glass slide and transferred to the AFM. (ii) Bacteria were also attached through electrostatic interactions (physical adsorption) to a glass slide that had been positively charged by adsorption of poly-l-lysine hydrobromide. For coating of the glass surface with poly-l-lysine hydrobromide, the glass was cleaned by sonication for 2 min in 2% RBS35 surfactant solution in water (Omnilabo International BV, Breda, The Netherlands), rinsed thoroughly with tap water, dipped in methanol, and rinsed again with demineralized water, after which a drop of 0.01% (wt/vol) poly-l-lysine hydrobromide solution was added. After air-drying, the slide was rinsed with demineralized water and dipped into the bacterial suspension in water. After 15 min, the bacterium-coated slide was rinsed with demineralized water to remove loosely attached bacteria and was then transferred to the AFM (11). (iii) Finally, bacteria were immobilized by glutaraldehyde fixation onto the silicon nitride tip of the AFM. This method required a pretreatment of both the K. terrigena cells and the AFM cantilevers (Park Scientific Instruments, Mountain View, Calif.). The bacteria were first treated with 2.5% (vol/vol) glutaraldehyde solution (pH adjusted to 6.8) for 2.5 h at 4°C. After glutaraldehyde fixation, the bacteria were washed in 0.25 mM potassium phosphate solution and pelleted by centrifugation at 10,000 × g for 5 min. For preparation of the AFM cantilevers, a drop of 1% (vol/vol) PEI solution was adsorbed onto the cantilevers for 2.5 h. The cantilevers were subsequently rinsed in demineralized water and stored at 4°C. A bacterial pellet was manually transferred onto the PEI-coated silicon nitride tip by use of a micromanipulator while the procedure was viewed under an optical microscope. The bacterium-covered tip was further treated with a drop of glutaraldehyde (2.5% [vol/vol]) at 4°C to strengthen and anchor the pellet onto the tip. After incubation for 1 to 2 h, the cantilevers were rinsed in demineralized water and transferred to the AFM (20).
AFM.
AFM measurements were made at room temperature in a 0.25 mM potassium phosphate solution at pH 6.8 under an optical microscope (Nanoscope III digital instrument). V-shaped silicon nitride cantilevers from Park Scientific Instruments, with a spring constant of 0.06 N/m and a probe curvature of ∼50 nm, were used. Individual force curves were collected over the tops of trapped and physically adsorbed bacteria at randomly selected locations, with z-displacements of 100 to 200 nm at z-scan rates of ≤1 Hz. Similarly, force curves were collected from between the bacterium-coated AFM tip and silicon nitride sheets (Onstream, Eindhoven, The Netherlands). The slopes of the retraction force curves in the region where the probe and sample were in contact were used to convert the voltage into a cantilever deflection. The conversion of deflection into force was carried out as previously described (13).
Approach curves were fitted to an exponential function by which the interaction force F is described as follows:
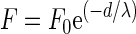 |
where F0 is the force at zero separation distance between the interacting surfaces, d is the separation distance, and λ is the decay length of the interaction force F. Retraction curves only showed a single adhesion peak in several cases. The percentage of occurrence of an adhesion peak, its magnitude, and the distance at which the adhesion peak appeared were recorded and averaged.
The results represent averages of at least 150 force-distance curves taken over 5 to 10 different organisms, with measurements at 10 different locations per organism.
RESULTS
Figure 1 shows AFM deflection images of K. terrigena immobilized by mechanical trapping in an Isopore polycarbonate membrane (Fig. 1a) or attached through electrostatic interactions to a positively charged glass slide (Fig. 1b) as well as a scanning electron micrograph of bacteria immobilized onto a silicon nitride AFM tip (Fig. 1c). Examples of force-distance curves generated from data measured over the top of a trapped and physically adsorbed bacterium as well as between the bacterium-coated tip and a silicon nitride sheet are presented below the corresponding images.
FIG. 1.
AFM deflection images of K. terrigena ATCC 33527 trapped in an Isopore polycarbonate membrane (image size, 2 μm by 2 μm; z-range, 75 nm) (a) and physically adsorbed onto poly-l-lysine-coated glass (image size, 3 μm by 3 μm; z-range, 150 nm) (b), together with a scanning micrograph of an AFM tip coated with K. terrigena cells (c). Examples of force-distance curves for interactions between K. terrigena and silicon nitride for each immobilization method are presented below the corresponding images. Solid lines, approach curves; dashed lines, retraction curves.
At first sight, similarities between the force-distance curves associated with mechanically trapped and physically adsorbed bacteria (Fig. 1a and b) can be observed. Upon approach of the tip, a long-range (about 500 to 800 nm) repulsive force was encountered, while in the examples given no adhesion was recorded upon retraction of the tip from the bacterial cell surface. In contrast, as the bacterium-coated tip approached a silicon nitride sheet, repulsion began at a much shorter separation distance (about 15 nm), and a single adhesion peak was always present upon retraction.
Quantitative features of the force-distance curves are summarized in Table 1. It is remarkable that intimate contact between the interacting surfaces was achieved for an applied force F0 that varied from 2.6 to 12 nN depending on the bacterial immobilization method used, whereas the decay length λ of the repulsive force upon approach ranged from 2.0 to 111 nm. As the AFM tip was retracted, adhesion forces were found in 13 to 15% of all cases, with average attractive forces of −0.26 and −0.5 nN at separation distances of 60 and 102 nm for mechanically trapped and physically adsorbed bacteria, respectively. However, the retraction of bacteria immobilized onto an AFM tip away from a silicon nitride sheet always showed adhesion, with an average attractive force of −35 nN at a 78-nm separation distance.
TABLE 1.
Characteristics of force-distance curves between K. terrigena ATCC 33527 and silicon nitride for three different bacterial immobilization methodsa
| Immobilization method | F0 (nN) | λ (nm) | % Adhesion | Fadh (nN) | Dadh (nm) |
|---|---|---|---|---|---|
| Mechanical trapping | 2.6 ± 1.7 | 59 ± 52 | 15 | −0.26 ± 0.05 | 60 ± 8 |
| Physical adsorption | 12 ± 4 | 111 ± 57 | 13 | −0.5 ± 0.2 | 102 ± 35 |
| Bacterium-coated AFM tip | 3.7 ± 0.5 | 2.0 ± 0.5 | 100 | −35 ± 2 | 78 ± 13 |
F0 is the repulsive force at zero separation distance and λ is the decay length of this repulsive force upon approach, while Fadh is the average adhesion force recorded upon retraction and Dadh is the separation distance at which the adhesion force occurred. The percentages of force-distance curves for which adhesion upon retraction occurred are given, since not all force-distance curves showed adhesion upon retraction. All data are average values ± standard deviations of 150 force-distance curves taken over 5 to 10 different organisms at 10 different locations per organism.
DISCUSSION
A proper interpretation of the force-distance curves generated for interacting surfaces in AFM requires bacterial immobilization that fully preserves the chemical and structural integrity of the cell surface. For this paper, we compared the interaction forces between K. terrigena and silicon nitride for three immobilization methods. Force-distance curves were different when bacteria were attached by fixation to the tip (Fig. 1c) from those obtained for mechanically trapped or physically adsorbed bacteria (Fig. 1a and b). For the last two methods, qualitative similarities were found in the force-distance curves, although for a bacterium immobilized by attachment to poly-l-lysine-treated glass, stronger repulsive forces occurring at larger separation distances were measured upon approach of the tip (Table 1).
Mechanical trapping of a single bacterium in a membrane filter with a pore size comparable to the dimensions of the cell does not require any chemical treatment or surface modification, and the highest part of a trapped organism protrudes through the holes of the filter. Therefore, it can be easily probed with an AFM under physiological conditions. In contrast, physical adsorption onto a positively charged surface may stimulate the secretion of excess EPS by K. terrigena. The surface of K. terrigena adsorbed onto a positively charged surface (Fig. 1b) shows a similar morphology to those of EPS substances previously scanned by Van der Aa et al. (24). The surface presents stretchable coil-like structures in the scanning direction. A thicker, negatively charged, and highly hydrated EPS layer could account for the higher repulsion forces operating over larger distances observed upon approach of the AFM tip to such physically adsorbed bacteria. This is in line with observations by Razatos et al. (20), who reported that an Escherichia coli mutant that overproduced colanic acid in buffer experienced more repulsion upon approach of the AFM tip than the parent strain, which was attributed to the higher negative charge density of the capsular material produced.
The interaction forces between a bacterium-coated AFM tip (Fig. 1c) and silicon nitride sheets yielded qualitatively and quantitatively distinct force-distance curves. Most notably, upon approach, the distance over which repulsion was probed was significantly reduced compared to that for both other methods. In addition, retraction of the bacterium-coated tip from the silicon nitride sheet always showed adhesion, whereas for the other two immobilization methods, very weak adhesion forces (<−0.5 nN) upon retraction were observed in only 13 to 15% of all force-distance curves recorded. The differences between adhesion forces could be readily attributed to the larger contact area probed by the bacterium-coated AFM tip. Assuming that five bacteria interact with the silicon nitride substratum, an average adhesion force per bacterium of 7 nN can be calculated. The contact area for the other two immobilization methods is generally estimated based on the effective AFM tip radius, which is thought to be ∼250 nm. Therefore, average adhesion forces per bacterium of ∼1 and 2 nN were found for mechanically trapped and physically adsorbed bacteria, respectively.
We envisage that glutaraldehyde fixation of bacteria to a tip stiffens the bacteria by cross-linking proteins and amino acids in the peptidoglycan layer, with an impact on adhesive properties. It has been found, for instance, that glutaraldehyde fixation causes Saccharomyces cerevisiae cells to become more hydrophobic (6). However, Razatos et al. (20) argued that glutaraldehyde treatment did not affect the adhesive properties of E. coli strains, because both contact angle and zeta potentials before and after glutaraldehyde treatment remained unchanged. In contrast, Burks et al. (8) found that the adhesion of these E. coli strains to glass was affected by a glutaraldehyde treatment. Furthermore, AFM-based results showed that the addition of glutaraldehyde consistently increased the rigidity of the E. coli strains studied.
Table 2 summarizes the advantages and disadvantages of each AFM immobilization method evaluated in this study. In general, trapping bacterial cells in filters guarantees the physical and chemical integrity of the bacterial cell surface, whereas the applied vacuum needed to pull the cells into the holes could induce changes in mechanical cell surface properties. Also, the adsorption of living cells onto positively charged surfaces may promote structural rearrangements in the bacterial cell surface structure. Glutaraldehyde fixation of bacteria to the AFM tip clearly affects the chemical and structural integrity of the bacterial cell surface, with a major impact on the interaction forces probed by AFM. Furthermore, complete coverage of the AFM tip by bacterial cells constitutes another problem. It is our experience that in three of five cases the coverage is incomplete, as shown in Fig. 2.
TABLE 2.
Summary of advantages and disadvantages associated with the bacterial immobilization methods employed in this study
| Immobilization method | Advantages | Disadvantages |
|---|---|---|
| Mechanical trapping | Simple preparation | Rod-shaped bacteria are difficult to trap |
| No chemical pretreatment of either the tip or bacteria | Cells may be compressed as a result of applied vacuum | |
| Physicochemical properties of bacterial cell remain unchanged | EPS may accumulate on the top of a trapped bacterium | |
| Exact positioning of the tip on the bacterial cell surface | ||
| Contact area can be estimated based on the dimensions of the tip | ||
| Physical adsorption | Simple preparation | Chemical treatment of the substratum is required |
| Bacteria with different shapes and dimensions can be studied | Physicochemical properties of bacterial cell surface are possibly affected by the surface modification | |
| Exact positioning of the tip on the bacterial cell surface | Immobilization not always adequate for different strains (11) | |
| Contact area can be estimated based on the dimensions of the tip | ||
| Bacterium-coated tip | Versatile choice of substratum | Long and difficult preparation procedure |
| Requires chemical treatment of both bacterial cell and substratum surface | ||
| Physicochemical and mechanical properties of the bacterial cell surface change | ||
| The number of interacting cells is unknown, as is their spatial orientation when interacting with the substratum | ||
| Bacterium-coated tips need to be checked regularly for full coverage by electron microscopy |
FIG. 2.
Electron micrographs of AFM silicon nitride tips after being coated with K. terrigena bacteria according to the procedure developed by Razatos et al. (20). Bar, 1 μm.
In conclusion, the results from this study indicate that different methods for the immobilization of bacteria for AFM affect the qualitative and quantitative features of the force-distance curves for interactions between K. terrigena and silicon nitride. Mechanical trapping of single cells in a membrane filter is inferred to be the most suitable technique, as the other two methods evaluated change the chemical and structural integrity of the bacterial cell surface.
Acknowledgments
Part of this study was funded by The Procter & Gamble Company, Mason, Ohio.
REFERENCES
- 1.Arnoldi, M., M. Fritz, E. Baurerlein, M. Radmacher, E. Sackmann, and A. Boulbitch. 2000. Bacterial turgor pressure can be measured by atomic force microscopy. Phys. Rev. E 62:1034-1043. [DOI] [PubMed] [Google Scholar]
- 2.Boland, T., and B. D. Ratner. 1995. Direct measurement of hydrogen bonding in DNA nucleotide bases by atomic force microscopy. Proc. Natl. Acad. Sci. USA 92:5297-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolshakova, A. V., O. I. Kiselyoava, A. S. Filonov, O. Y. Frolova, Y. L. Lyubchenko, and I. V. Yaminsky. 2001. Comparative studies of bacteria with an atomic force microscopy operating in different modes. Ultramicroscopy 86:121-128. [DOI] [PubMed] [Google Scholar]
- 4.Boonaert, C. J. P., V. Toniazzo, C. Mustin, Y. F. Dufrêne, and P. G. Rouxhet. 2002. Deformation of Lactococcus lactis surface in atomic force microscopy study. Colloids Surf. B 23:201-211. [Google Scholar]
- 5.Bottomley, L. A., J. E. Coury, and P. N. First. 1996. Scanning probe microscopy. Anal. Chem. 68:185-230. [Google Scholar]
- 6.Bowen, W. R., R. W. Lovitt, and C. J. Wright. 2001. Atomic force microscope study of the adhesion of Saccharomyces cerevisiae. J. Colloid Interface Sci. 237:54-61. [DOI] [PubMed] [Google Scholar]
- 7.Browing-Kelly, M. E., K. Wadu-Mesthrige, V. Hari, and G. Y. Liu. 1997. Atomic force microscopic study of specific antigen/antibody binding. Langmuir 13:343-350. [Google Scholar]
- 8.Burks, G. A., S. B. Velegol, E. Paramonova, B. E. Lindenmuth, J. D. Feick, and B. E. Logan. 2003. Macroscopic and nanoscale measurements of the adhesion of bacteria with varying outer layer surface composition. Langmuir 19:2366-2371. [Google Scholar]
- 9.Bustamante, C., and C. Rivetti. 1996. Visualizing protein-nucleic acid interactions on a large scale with the scanning force microscope. Annu. Rev. Biophys. Biomol. Struct. 25:395-429. [DOI] [PubMed] [Google Scholar]
- 10.Camesano, T., and B. E. Logan. 2000. Probing bacterial electrosteric interactions using atomic force microscopy. Environ. Sci. Technol. 34:3354-3362. [Google Scholar]
- 11.Camesano, T. A., M. J. Natan, and B. E. Logan. 2000. Observation of changes in bacterial cell morphology using tapping mode atomic force microscopy. Langmuir 16:4563-4572. [Google Scholar]
- 12.Chilkoti, A., T. Boland, B. D. Ratner, and P. S. Stayton. 1995. The relationship between ligand-binding thermodynamics and protein-ligand interaction forces measured by atomic force microscopy. Biophys. J. 69:2125-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dufrêne, Y. F. 2000. Direct characterization of the physicochemical properties of fungal spores using functionalized AFM probes. Biophys. J. 78:3286-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel, A., C.-A. Schoenenberger, and D. J. Mueller. 1997. High resolution imaging of native biological sample surfaces using scanning probe microscopy. Curr. Opin. Struct. Biol. 7:279-284. [DOI] [PubMed] [Google Scholar]
- 15.Florin, W.-L., V. T. Moy, and H. E. Gaub. 1994. Adhesive forces between individual ligand-receptor pairs. Science 264:415-417. [DOI] [PubMed] [Google Scholar]
- 16.Hamers, R. J. 1996. Scanned probe microscopies in chemistry. J. Phys. Chem. 100:13103-13120. [Google Scholar]
- 17.Kasas, S., and A. Ikai. 1995. A method for anchoring round shaped cells for atomic force microscope imaging. Biophys. J. 68:1678-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, G. U., L. A. Chrisy, and R. J. Colton. 1994. Direct measurement of the forces between complementary strands of DNA. Science 266:771-773. [DOI] [PubMed] [Google Scholar]
- 19.Lower, S. K., C. J. Tadamnier, and M. F. Hochella. 2000. Measuring interfacial and adhesion forces between bacteria and mineral surfaces with biological force microscopy. Geochim. Cosmochim. Acta 64:3133-3139. [Google Scholar]
- 20.Razatos, A., Y.-L. Ong, M. M. Sharma, and G. Georgiou. 1998. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl. Acad. Sci. USA 95:11059-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rief, M., F. Oesterhelt, B. Heymann, and H. E. Gaub. 1997. Single molecule force spectroscopy on polysaccharides by AFM. Science 275:1295-1297. [DOI] [PubMed] [Google Scholar]
- 22.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin Ig-domains by AFM. Science 276:1109-1112. [DOI] [PubMed] [Google Scholar]
- 23.Shao, Z., J. Mou, D. M. Czajkowsky, J. Yang, and J. Y. Yuan. 1996. Biological atomic force microscopy: what is achieved and what is needed. Adv. Phys. 45:1-86. [Google Scholar]
- 24.Van der Aa, B. C., and Y. F. Dufrêne. 2002. In situ characterization of bacterial extracellular polymeric substances by AFM. Colloids Surf. B 23:173-182. [Google Scholar]
- 25.van der Mei, H. C., H. J. Busscher, R. Bos, J. de Vires, C. J. P. Boonaert, and Y. F. Dufrêne. 2000. Direct probing by atomic force microscopy of the cell surface softness of a fibrillated and nonfibrillated oral streptococcal strain. Biophys. J. 78:2662-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velegol, S. B., and B. E. Logan. 2002. Contributions of bacterial surface polymers, electrostatics, and cell elasticity to the shape of AFM force curves. Langmuir 18:5256-5262. [Google Scholar]




