Abstract
Oxolinic acid (OA), a quinolone, inhibits the activity of DNA gyrase composed of GyrA and GyrB and shows antibacterial activity against Burkholderia glumae. Since B. glumae causes bacterial seedling rot and grain rot of rice, both of which are devastating diseases, the emergence of OA-resistant bacteria has important implications on rice cultivation in Japan. Based on the MIC of OA, 35 B. glumae field isolates isolated from rice seedlings grown from OA-treated seeds in Japan were divided into sensitive isolates (OSs; 0.5 μg/ml), moderately resistant isolates (MRs; 50 μg/ml), and highly resistant isolates (HRs; ≥100 μg/ml). Recombination with gyrA of an OS, Pg-10, led MRs and HRs to become OA susceptible, suggesting that gyrA mutations are involved in the OA resistance of field isolates. The amino acid at position 83 in the GyrA of all OSs was Ser, but in all MRs and HRs it was Arg and Ile, respectively. Ser83Arg and Ser83Ile substitutions in the GyrA of an OS, Pg-10, resulted in moderate and high OA resistance, respectively. Moreover, Arg83Ser and Ile83Ser substitutions in the GyrA of MRs and HRs, respectively, resulted in susceptibility to OA. These results suggest that Ser83Arg and Ser83Ile substitutions in GyrA are commonly responsible for resistance to OA in B. glumae field isolates.
Burkholderia glumae-induced bacterial seedling rot and grain rot are devastating diseases that affect rice cultivation in Southeast Asian countries such as Japan and Korea. In growing seedlings, bacterial seedling rot is caused by a rapid increase in B. glumae populations in the epidermis of the plumules (14). After the rice seedlings are transplanted into paddy fields, the pathogen is usually isolated from the lower leaf sheaths (15). It can be found in the upper leaf sheaths, including the flag leaf sheaths before the heading stage, although symptoms do not appear on the leaf blades or leaf sheaths. It then invades the flowering spikelets, multiplies rapidly, and finally causes bacterial grain rot (19).
Oxolinic acid (OA) treatment of rice seeds and spray applications to heading rice plants have a high efficacy in the control of bacterial seedling rot and grain rot, respectively (14, 15). OA is a quinolone derivative with antibacterial activity against gram-negative bacteria, including B. glumae (13, 22, 37), and it inhibits the proliferation of bacteria in the plumules of seedlings and the spikelets of heading rice plants (14, 19). OA control involving seed treatment and spray applications to heading rice plants when combined with seed selection with salt solutions has a significant influence on B. glumae infection cycles and is highly efficacious in the control of these diseases (16).
Quinolones act by inhibiting the action of type II topoisomerase, DNA gyrase, and topoisomerase IV (25). DNA gyrase is a tetrameric enzyme composed of two A subunits and two B subunits, encoded by gyrA and gyrB, respectively (11, 35). Topoisomerase IV is also an A2B2 enzyme and is composed of ParC and ParE (23, 24), whose amino acid sequences share similarity to those of GyrA and GyrB, respectively. The main function of DNA gyrase is to catalyze the negative supercoiling of DNA (21), whereas topoisomerase IV seems to be associated with decatenation of the daughter replicons (25). The targets of quinolones in gram-negative bacteria are different from those in gram-positive bacteria. In gram-negative bacteria the target is DNA gyrase, whereas in gram-positive bacteria the target is topoisomerase IV (32). Therefore, either gyrA or gyrB mutations are responsible for the resistance of gram-negative bacteria. In addition, topoisomerase IV mutations can further increase this level of resistance. Decreased quinolone uptake is also involved in quinolone resistance and might be associated with two factors, an increased impermeability of bacteria to quinolones or the overexpression of efflux pumps (32). In gram-negative bacteria, such as Escherichia coli, Salmonella enterica serovar Typhimurium, and Neisseria gonorrhoeae, the major target of mutations is the gyrA gene (32). Most of the mutations identified thus far are located in a small region in GyrA, the quinolone resistance-determining region (QRDR) (40).
Recently, OA-resistant B. glumae was isolated from rice seedlings grown from OA-treated seeds (17). Because the conditions for raising seedlings are suitable for the growth of seed-borne bacteria, the growth of OA-resistant bacteria on rice plants can eventually lead to bacterial seedling rot and grain rot and significant production losses. OA-resistant B. glumae isolates obtained in vitro have been characterized (17), and these isolates also have a concomitant cross-resistance to other quinolones. OA-resistant B. glumae isolates grow rapidly in seedlings and rot the plants only when seeds are infested with a high density of OA-resistant bacteria and then treated with OA. However, the mechanisms of resistance to OA in B. glumae remain unclear.
In this study, the mechanisms of resistance to OA in B. glumae field isolates were investigated. The results suggest that an amino acid substitution at position 83 of GyrA is implicated in OA resistance in field isolates, because among various OA-resistant bacteria only those with a substitution in GyrA might retain the ability to survive on rice plants.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. B. glumae strains were routinely grown in peptone-yeast medium (peptone, 5 g; yeast extract, 2 g; deionized water, 1 liter; PY medium) at 30°C (20). E. coli strains were grown in LM medium (12) at 37°C. The following antibiotics were used in the selective media: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | recA1 endA1 gyrA96 thi-1 hsdR17supE44 Δ(lac)U169(φ80lacΔM15) | 12 |
| Pg-10-GyrA | Holding pGYRA10 | This study |
| B. glumae | ||
| Pg-10 | Wild type | 17 |
| FU13 | Wild type | This study |
| M3 | Wild type | This study |
| Pg-7 | Wild type | 17 |
| Pg-15 | Wild type | 17 |
| Pg-10G | Recombinant of Pg-10 with 3.9-kb DNA fragment of pGYRA-S; Kmr | This study |
| FU13G | Recombinant of FU13 with 3.9-kb DNA fragment of pGYRA-S; Kmr | This study |
| M3G | Recombinant of M3 with 3.9-kb DNA fragment of pGYRA-S; Kmr | This study |
| Pg-7G | Recombinant of Pg-7 with 3.9-kb DNA fragment of pGYRA-S; Kmr | This study |
| Pg-15G | Recombinant of Pg-15 with 3.9-kb DNA fragment of pGYRA-S; Kmr | This study |
| Pg-10R | Recombinant of Pg-10 with Ser83Arg substitution in GyrA; Kmr | This study |
| Pg-10I | Recombinant of Pg-10 with Ser83Ile substitution in GyrA; Kmr | This study |
| FU13RS | Recombinant of FU13 with Arg83Ser substitution in GyrA; Kmr | This study |
| M3IS | Recombinant of M3 with Ile83Ser substitution in GyrA; Kmr | This study |
| Pg-7RS | Recombinant of Pg-7 with Arg83Ser substitution in GyrA; Kmr | This study |
| Pg-15IS | Recombinant of Pg-15 with Ile83Ser substitution in GyrA; Kmr | This study |
| Plasmids | ||
| pBluescript KS | Ampr | Stratagene |
| pGEM-T | Ampr | Promega |
| pHSG398 | Cmr | TaKaRa |
| pUCK191 | pUC19 derivative containing the Kmr gene | 36 |
| pCUD800 | sacB; Kmr | 10 |
| pHSG398RI | pHSG398 derivative | This study |
| pGYRA-QRDR | 338-bp DNA fragment including gyrA-QRDR from Pg-10 in pGEM-T | This study |
| pGYRA10 | 8.7-kb DNA fragment including gyrA from Pg-10 in pBluescript KS | |
| pGYRA-1 | 5.1-kb ApaI/BamHI fragment of pGYRA10 in pBluescript KS | This study |
| pGYRA-2 | 2.5-kb BamHI/KpnI fragment of pGYRA-1 in pHSG398RI | This study |
| pGYRA-3 | 1.4-kb Kmr gene in pGYRA-2 | This study |
| pGYRAKm | 2.6-kb KpnI fragment of pGYRA-1 in pGYRA-3 | This study |
| pGYRA-S | 2.6-kb sacB of pGYRAKm | This study |
| pGY-R2 | 1.2-kb RI-KpnI-Arg fragment substituted for the EcoRI/KpnI fragment in pGYRA-2 | This study |
| This study | ||
| pGY-I2 | 1.2-kb RI-KpnI-Ile fragment substituted for the EcoRI/KpnI fragment in pGYRA-2 | This study |
| pGY-R2Km | 1.4-kb Kmr gene in pGY-R2 | This study |
| pGY-I2Km | 1.4-kb Kmr gene in pGY-I2 | This study |
| pGYRA-R | 2.6-kb sacB in pGY-R2Km | This study |
| pGYRA-1 | 2.6-kb sacB in pGY-I2Km | This study |
| pGYRA-7 | 2.6-kb sacB and 1.4-kb Kmr gene and 1.2-kb DNA fragment amplified by recombinant PCR from Pg-7 in pGYRA-2 | This study |
| pGYRA-13 | 2.6-kb sacB and 1.4-kb Kmr gene and 1.2-kb DNA fragment amplified by recombinant PCR from FU13 in pGYRA-2 | This study |
| pGYRA-15 | 2.6-kb sacB and 1.4-kb Kmr gene and 1.2-kb DNA fragment amplified by recombinant PCR from Pg-15 in pGYRA-2 | This study |
| pGYRA-3 | 2.6-kb sacB and 1.4-kb Kmr gene and 1.2-kb DNA fragment amplified by recombinant PCR from M3 in pGYRA-2 | This study |
Antibiotic susceptibility assay.
The MIC of OA was determined by using the sequential dilution method. A total of 102 freshly grown cells of the tested B. glumae strain were incubated on an OA-containing PY agar medium at 30°C for 2 days. The lowest concentration of OA, which completely inhibited growth, was defined as the MIC.
DNA manipulations.
Plasmid and chromosomal DNA isolation, restriction mapping, cloning, subcloning, Southern blot hybridization, and PCR were performed according to standard procedures (33). Restriction enzymes were obtained from TaKaRa, Otsu, Japan. B. glumae was transformed by electroporation as described by Allen et al. (1). Double-stranded DNA sequencing templates were prepared with the GenElute plasmid miniprep kit (Sigma, St. Louis, Mo.). DNA sequences were determined by using an Automated DNA sequencer model 373 (Applied Biosystems, Tokyo, Japan), and analyzed by using DNASIS-Mac software (Hitachi Software Engineering, Yokohama, Japan).
Cloning of the gyrA gene.
To create a B. glumae Pg-10 genomic library, the Pg-10 DNA genome was digested with BamHI, and 8 to 10-kb DNA fragments were collected with sucrose density gradient centrifugation. The DNA fragments were ligated into the BamHI site of pBluescript KS (Stratagene, La Jolla, Calif.) and transformed into E. coli DH5α to create a Pg-10 genomic library.
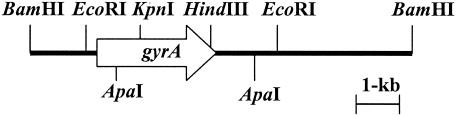
To probe the gyrA gene from the Pg-10 genomic library, the following primers were designed based on putative conserved sequences of gyrA from N. gonorrhoeae, N. meningitidis, and Ralstonia solanacearum to amplify an internal 338-bp gyrA DNA fragment: 5′-TACAAGAAGTCGGCGCG-3′ (gyrA1) and 5′-GCGATCCCGGACGAGCC-3′ (gyrA2). PCR amplification was performed with 1 cycle of 94°C for 2 min, 25 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 30 s by using a thermal cycler (TaKaRa). The resulting amplified 338-bp DNA fragment was cloned into pGEM-T (Promega, Madison, Wis.) to create pGYRA-QRDR. The plasmid was used as a template in the following PCR. To probe gyrA from the Pg-10 genomic library, Southern blot hybridization was performed by using a digoxigenin-labeled DNA probe (Roche Molecular Biochemicals, Indianapolis, Ind.) PCR amplified with gyrA1 and gyrA2 according to the manufacturer's protocol. The resultant positive transformant, Pg-10-GyrA, held a plasmid pGYRA10 containing an 8.7-kb insert (Fig. 1), which was used to sequence gyrA.
FIG. 1.
Physical map of pGYRA10, including gyrA from the genomic DNA of B. glumae Pg-10. The arrow represents the localization and orientation of the gyrA open reading frame.
Recombination of OA-resistant B. glumae field isolates with gyrA from Pg-10.
To create recombinants from the OA-resistant B. glumae field isolates with gyrA from Pg-10, pGYRA10 was partially digested with ApaI and a resulting 8.1-kb fragment, including gyrA, was self-ligated to create pGYRA-1. Plasmid pHSG398 (TaKaRa) digested with EcoRI was blunt ended with a T4 DNA polymerase (TaKaRa) and self-ligated to create an EcoRI site-deficient pHSG398RI. A 2.5-kb BamHI/KpnI fragment was subcloned from pGYRA-1 into pHSG398RI to create pGYRA-2. A 1.4-kb KpnI-DNA fragment containing a kanamycin resistance (Kmr) gene from pUCK191 (36) was blunt ended by T4 DNA polymerase and inserted into a blunt-ended pGYRA-2 EcoRI site to create pGYRA-3. A 2.6-kb KpnI fragment from pGYRA-1 was ligated into KpnI site of pGYRA-3 to create pGYRAKm. A 2.6-kb BamHI/PstI fragment containing sacB from pUCD800 (10) was blunt ended by T4 DNA polymerase and inserted into the blunt-ended pGYRAKm BamHI site to create pGYRA-S. The plasmid was electroporated into the cells of 25 field isolates, and the resultant kanamycin and sucrose-resistant recombinants were selected. Southern blot analysis was performed to verify correct insertion of 3.9-kb DNA fragment containing the Kmr gene in the gyrA locus in the isolates genetic backgrounds (data not shown).
DNA gyrA sequencing.
To determine the DNA sequences of gyrA from B. glumae field isolates FU13, M3, Pg-7, and Pg-15, a 2,708-bp gyrA-containing DNA fragment was PCR amplified from chromosomal FU13, M3, Pg-7, and Pg-15 DNA by using gyrA+2 and gyrA−2 (Table 2), designed based on the nucleotide sequences of Pg-10 gyrA. PCR amplification was performed with 1 cycle of 94°C for 2 min, 25 cycles of 94°C for 1 min, 63°C for 1 min, and 72°C for 3 min. The DNA sequences of the amplified products were analyzed by using primers designed based on the DNA sequences of gyrA from Pg-10 (Table 2).
TABLE 2.
Primers used to obtain the nucleotide sequences of gyrA
| Primer | Sequence |
|---|---|
| gyrA+2 | 5′-GCGGTCCCGTCTTCGCTCCG-3′ |
| gyrA−2 | 5′-GTTGCACCGCCGGGCGCGAC-3′ |
| gyrA−2+ | 5′-ATGCTGGTCGACGGTCAGGG-3′ |
| gyrA−3 | 5′-CACCATCTGCTCGCGAATCC-3′ |
| gyrA−4 | 5′-GACATCCGCGACGAGTCCGA-3′ |
| gyrA−5 | 5′-ATGCGTCTGCAGCGCCTGAC-3′ |
| gyrA2−1+ | 5′-CACGATCTACGAACTGCG-3′ |
| gyrA2−3+ | 5′-CAGATGAAGGAAGACGACTG-3′ |
| gyrA2−4+ | 5′-ATGACGGCGACTTCCTGATCGG-3′ |
| gyrA2−5+ | 5′-TCACGGAATACACGCGTCACGG-3′ |
DNA sequencing of the QRDR in gyrA.
To analyze the DNA sequences of the QRDRs in B. glumae field isolate gyrA, a 338-bp DNA fragment was PCR amplified from the chromosomal DNA of field isolates by using gyrA1 and gyrA2 primers. The DNA sequences of the PCR products were analyzed by using gyrA1 and gyrA2.
Ser83Arg and Ser83Ile substitutions in the GyrA of Pg-10.
The single point mutations corresponding to Ser83Arg and Ser83Ile substitutions in the GyrA of Pg-10 were introduced into a 1.2-kb DNA fragment containing gyrA-QRDR from chromosomal Pg-10 DNA by using recombinant PCR as follows. DNA fragments, RI-Kpn-Arg and RI-Kpn-Ile fragments with Ser83Arg and Ser83Ile GyrA substitutions, respectively, were synthesized by PCR with two pairs of primers: (i) a forward primer 5′-GCGAATTCGGGGAGGGCGCG-3′ (RI-gyrA+) and a reverse primer X and (ii) a forward primer Y and a reverse primer 5′-GGGGTACCTCGCCGCGCTTG-3′ (Kpn-gyrA−). The X and Y primers are presented in Table 3. PCR amplification was performed with 1 cycle of 94°C for 2 min, followed by 5 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min; and finally with 20 cycles of 94°C for 1 min, 61°C for 1 min, and 72°C for 1 min. Each PCR product purified by agarose gel electrophoresis was mixed and then PCR amplified with the RI-gyrA+ and Kpn-gyrA− primers. PCR amplification was performed with 1 cycle of 94°C for 2 min, followed by 25 cycles of 94°C for 1 min, 61°C for 1 min, and 72°C for 1 min. RI-Kpn-Arg and RI-Kpn-Ile fragments were digested with EcoRI and KpnI and then substituted into EcoRI/KpnI-digested pGYRA-2 fragments to create pGY-R2 and pGY-I2, respectively. A 1.4-kb KpnI-DNA fragment containing the Kmr gene from pUCK191 was blunt ended by T4 DNA polymerase and inserted into the blunt-ended pGY-R2 and pGY-I2 EcoRI sites to create pGY-R2Km and pGY-I2Km, respectively. A 2.6-kb PstI- and BamHI-digested DNA fragment containing sacB from pUCD800 was blunt ended by T4 DNA polymerase and ligated into the blunt-ended pGY-R2Km and pGY-I2Km KpnI sites, as described above, to create pGYRA-R and pGYRA-I, respectively. These plasmids were electroporated into Pg-10 cells, and the resultant Kmr and sucrose-resistant recombinants, Pg-10R and Pg-10I, were selected. DNA sequencing of PCR-amplified DNA fragments by using gyrA+2 and gyrA−2 as primers were performed to verify correct substitution of gyrA in Pg-10R and Pg-10I (data not shown).
TABLE 3.
Templates and primers used for recombinant PCR to introduce amino acid substitutions at position 83 in the GyrA of B. glumae field isolates
| Amino acid substitution at GyrA83 | Genomic DNA used as a template | Primera | Sequenceb |
|---|---|---|---|
| Ser83Arg | Pg-10 | X | 5′-CGTAGACCGCTCTGTCGCCGTG-3′ |
| Y | 5′-CACGGCGACAGAGCGGTCTACG-3′ | ||
| Ser83Ile | Pg-10 | X | 5′-CGTAGACCGCGATGTCGCCGTG-3′ |
| Y | 5′-CACGGCGACATCGCGGTCTACG-3′ | ||
| Arg83Ser | Pg-7, Fu13 | X | 5′-CGTAGACCGCGCTGTCGCCGTG-3′ |
| Y | 5′-CACGGCGACAGCGCGGTCTACG-3′ | ||
| Ile83Ser | Pg-15, M3 | X | 5′-CGTAGACCGCGCTGTCGCCGTG-3′ |
| Y | 5′-CACGGCGACAGCGCGGTCTACG-3′ |
Primers X and Y are complementary and correspond to the gyrA sequence of isolates from which the genomic DNA used as template was isolated.
Position of the sequences was at 238 to 259 in the gyrA of each isolates. Underlined letters indicate nucleotide changes.
Arg83Ser and Ile83Ser substitutions in the GyrA of OA-resistant field isolates.
To introduce Arg83Ser and Ile83Ser substitutions into the GyrA of Pg-7 and FU13 and of Pg-15 and M3, single point mutations corresponding to these amino acid substitutions were introduced into 1.2-kb DNA fragments containing gyrA-QRDR from isolate chromosomal DNA by using recombinant PCR with Ser-Recom−- and Ser-Recom+ (Table. 3), as described previously for Ser83Arg and Ser83Ile substitutions in Pg-10 GyrA, to create pGYRA-7 and pGYRA-13 and to create pGYRA-15 and pGYRA-3, respectively. These plasmids were electroporated into Pg-7, FU13, Pg-15, and M3 cells, and the resultant Kmr and sucrose-resistant recombinants—Pg-7S, FU13S, Pg-15S, and M3S, respectively—were selected. DNA sequences of PCR-amplified DNA fragments with gyrA+2 and gyrA−2 as primers were performed to verify correct substitution of gyrA in Pg-7S, FU13S, Pg-15S, and M3S (data not shown).
Spontaneous in vitro mutants resistant to OA from Pg-10.
To isolate spontaneous OA-resistant mutants from Pg-10, Pg-10 was incubated in a PY medium for 24 h and washed three times with sterilized water by centrifugation at 10,000 × g for 30 s. The pellet was suspended in sterilized water, and the bacterial density was adjusted to 1.0 × 108 CFU/ml. The suspension was spread onto a PY agar medium containing 1.0 μg of OA/ml, followed by incubation at 30°C for 3 days.
Determination of GyrA83 by using mismatch amplification mutation assay PCR.
To detect GyrA83 substitutions of the spontaneous OA-resistant mutants obtained in vitro, the forward primers 5′-CATCCGCACGGCGACAGC-3′ (gyrA-Ser), 5′-CATCCGCACGGCGACAGR-3′ (gyrA-Arg), and 5′-CATCCGCACGGCGACAT-3′ (gyrA-Ile) and a reverse primer R4 (5′-GCGATCCCGGACGAGCC-3′) were used in the MAMA PCR (27). PCR amplification was performed with 1 cycle of 94°C for 4 min, followed by 25 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 30 sec. Then, 9-μl aliquots of each PCR product were loaded onto horizontal, 2.0% TAE agarose gels and stained with ethidium bromide for detection of 293-bp DNA fragments after electrophoresis.
Nucleotide sequence accession numbers.
Pg-10, Pg-7, Pg-15, FU13, and M3 gyrA nucleotide sequences were assigned DDBJ accession numbers AB121693, AB121694, AB121695, AB121693-6, and AB121697, respectively.
RESULTS
MIC of OA in B. glumae field isolates.
Thirty-five B. glumae field isolates obtained from diseased seedlings grown from OA-treated seeds were collected from five prefectures in Japan. Based on the OA MICs, these isolates were divided into three groups (Table 4): OA-sensitive isolates (OSs; MIC = 0.5 μg/ml), moderately resistant isolates (MRs; MIC = 50 μg/ml) and high resistant isolates (HRs; MIC ≥ 100 μg/ml).
TABLE 4.
MICs of OA in B. glumae field isolates and recombinants of the isolates with gyrA from B. glumae Pg-10 and deduced amino acid residues at position 83 in the GyrAs of these isolates
| MIC of OA (μg/ml) | Deduced amino acid residue at position 83 in GyrA (codon) | Isolatesa |
|---|---|---|
| 0.5 | Ser (AGC) | Pg-10, Pg-12, 0112 |
| 50 | Arg (AGA or AGG) | Pg-5, Pg-6, Pg-7, Pg-8, Pg-9, Pg-17, FU11, FU13, FU15, FU22 |
| ≥100 | Ile (ATC) | 0101, Pg-14, Pg-15, M1, M3, M6, M8, M9, M10, IZUMI, TONDA, H91, H92, H93, H94, H95, H101, H102 |
The MICs of OA in recombinants of underlined and italic isolates are 0.5 and 5.0 μg/ml, respectively.
Cloning of the gyrA gene.
It has been reported that gyrA mutations confer quinolone resistance in most clinical isolates of gram-negative bacteria, such as E. coli, Methylovorus sp., and N. gonorrhoeae (43). Moreover, many quinolone-resistant gram-negative bacteria, whose resistance to chemicals results from gyrA mutations, can be complemented by quinolone sensitivity with the introduction of the wild-type allele (31). To determine whether resistance to OA in B. glumae field isolates results from gyrA mutations, gyrA was cloned from the genomic DNA of an OS, B. glumae Pg-10, and recombinants of the isolates with gyrA from Pg-10 were created with marker exchange.
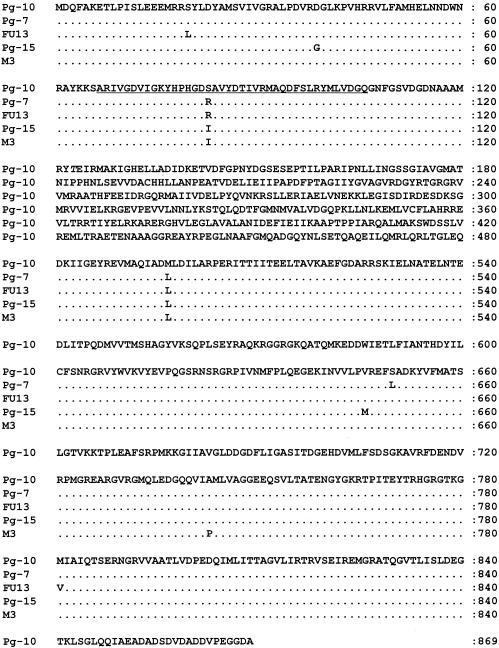
Using conserved primers, gyrA1 and gyrA2, based on the conserved sequences of gyrA in the β-subclasses of proteobacteria, N. gonorrhoeae MS11, N. meningitidis Z2491, Methylovorus sp. strain SS1, and R. solanacearum GMI1000, a 338-bp DNA fragment was PCR amplified from Pg-10. A clone containing a pGYRA10 (Fig. 1) was selected from the Pg-10 genomic library with Southern hybridization by using the amplified DNA fragment as a probe, and nucleotide sequences of the 8.7-kb insert were determined (Fig. 2A). The open reading frame of Pg-10 gyrA consisted of 2,610 nucleotides, with 56.2, 56.4, 65.9, and 78.1% identities to the gyrA of N. gonorrhoeae MS11, N. meningitidis Z2491, Methylovorus sp. strain SS1, and R. solanacearum GMI1000, respectively. The deduced amino acid sequence of GyrA showed 50.2, 50.1, 65.7, and 81.4% identities to GyrAs of N. gonorrhoeae MS11, N. meningitidis Z2491, Methylovorus sp. strain SS1, and R. solanacearum GMI1000, respectively, and had a homologous region consisting of 40 amino acids with 95.0, 90.0, and 97.5% identities to the QRDRs of E. coli K-12, N. gonorrhoeae MS11, and Methylovorus sp. strain SS1, respectively (Fig. 2B).
FIG. 2.
Deduced amino acid sequence of GyrA in B. glumae Pg-10 (A) and comparisons of the deduced amino acid sequences of the QRDR of GyrA (B). The underlined sequences are involved in QRDR.
Recombination of OA-resistant B. glumae field isolates with gyrA from Pg-10.
To examine the dependency of field isolate OA resistance on gyrA mutations, recombinants from 24 OA-resistant field isolates (6 MRs and 18 HRs) were created with marker exchange with gyrA-containing pGYRA-S from Pg-10. The insertion specificity was verified by Southern blot analysis (data not shown). The MICs of OA in the 12 field isolate recombinants (three MRs and nine HRs, S-type) and the other 12 field isolate recombinants (three MRs and nine HRs, MS-type) were 0.5 and 5 μg/ml, respectively (Table 4), suggesting that recombination with gyrA from Pg-10 resulted in any of the field isolates becoming OA susceptible. In particular, only gyrA mutations were responsible for the OA resistance of S-type isolates. On the other hand, MS-type isolates might have not only gyrA mutations but other mechanisms that are involved in OA resistance. The MIC of OA in the Pg-10 recombinants with pGYRA-S was 0.5 μg/ml.
DNA sequences of gyrA from B. glumae field isolates.
To elucidate the relationship between OA resistance and gyrA mutations in B. glumae field isolates, the DNA sequences of gyrA from an OS (Pg-10), MRs (Pg-7 and FU13), and HRs (Pg-15 and M3) were compared. The gyrA of each isolate was composed of 2,610 nucleotides. An amino acid at position 497 in the GyrAs of Pg-7, FU13, Pg-15, and M3 was Leu, although that of Pg-10 was Met (Fig. 3). In addition, the amino acid at position 83 of GyrA (GyrA83) in Pg-10 was Ser with anAGC codon; however, in all MRs and all HRs it was Arg and Ile, with AGA or AGG and ATC codons, respectively. These results suggest that substitutions of GyrA83 located in QRDR might be involved in OA resistance (Table 4).
FIG. 3.
Deduced amino acid sequences of GyrA from B. glumae Pg-10, Pg-7, FU13, Pg-15, and M3. The underlined sequences are involved in the QRDR.
GyrA83 involvement in the OA resistance of B. glumae field isolates.
To determine whether Ser83Arg and Ser83Ile substitutions result in OA-resistant bacteria, a point mutation was introduced into gyrA in Pg-10 with marker exchange. Pg-10R and Pg-10I containing the GyrA83 substitution for Arg and an Ile, respectively, were then created. Ser83Arg and Ser83Ile substitution specificity in GyrA83 of Pg-10R and Pg-10I was verified by DNA sequences of PCR-amplified DNA fragments with gyrA+2 and gyrA−2 as primers (data not shown). The MICs of OA for Pg-10R and Pg-10I were 50 and 100 μg/ml, respectively (Table 5), suggesting that Ser83Arg and Ser83Ile substitutions result in moderate and high resistance to OA, respectively.
TABLE 5.
MICs of OA in recombinants at position 83 in the GyrAs of B. glumae Pg-10, Pg-7, Pg-15, M3, and FU13
| B. glumae straina | Amino acid residue at position 83 in GyrA | MIC of OA (μg/ml) |
|---|---|---|
| Pg-10 | Ser | 0.5 |
| Pg-7 | Arg | 50 |
| FU13 | Arg | 50 |
| Pg-15 | Ile | 100 |
| M3 | Ile | 500 |
| Pg-10R | Arg | 50 |
| Pg-10I | Ile | 100 |
| Pg-7S | Ser | 0.5 |
| FU13S | Ser | 5.0 |
| Pg-15S | Ser | 0.5 |
| M3S | Ser | 5.0 |
Pg-10R and Pg-10I are recombinants of Pg-10 in which the amino acid residues at position 83 of GyrA were substituted by Arg and Ile, respectively, via marker exchange. Pg-7S, FU13S, Pg-15S, and M3S are recombinants of Pg-10 in which the amino acid residue at position 83 of GyrA was substituted by Ser via marker exchange.
To determine whether GyrA83 mutations in GyrA are involved in B. glumae field isolate OA resistance, a point mutation was introduced into gyrA in the S-type isolates (Pg-7 and Pg-15) and MS-type isolates (FU13 and M3) with marker exchange. Then, Pg-5RS and FU13RS, or Pg-15IS and M3IS containing an Arg83Ser or Ile83Ser GyrA83 substitution, respectively, were created. GyrA83 substitution specificity in Pg-5RS, FU13RS, Pg-15IS and M3IS was verified by DNA sequences of PCR-amplified DNA fragments with gyrA+2 and gyrA−2 as primers (data not shown). The MICs of OA for the S-type recombinants (Pg-7RS and Pg-15IS) and MS-type recombinants (FU13RS and M3IS) were 0.5 and 5.0 μg/ml, which are similar to those for Pg-7S and Pg-15S, and FU13S and M3S, respectively (Table 5). These results suggest that GyrA83 mutations are commonly responsible for OA resistance in B. glumae field isolates, and Leu497Met substitution in GyrA may not be involved in the resistance to OA of field isolates.
GyrA83 in spontaneous OA-resistant mutants from Pg-10.
To determine whether factors other than GyrA83 mutations might be involved in OA resistance, 89 spontaneous OA-resistant mutants (59 MRs and 30 HRs) of Pg-10 were isolated from Pg-10 grown on medium containing 1.0 μg of OA/ml. Analysis of single nucleotide polymorphisms with gyrA-Ser, gyrA-Arg, or gyrA-Ile and R4 primers showed that GyrA83 in 54 MRs and 24 HRs was Ser (Table 6). In five MRs and four HRs it was Arg, and in two HRs it was Ile, suggesting that the OA resistance of these spontaneous mutants does not necessarily depend only on GyrA83 mutations.
TABLE 6.
Amino acid residues of GyrA at position 83 in spontaneous OA-resistant mutants from B. glumae Pg-10
| MIC of OA (μg/ml) | Amino acid residue at position 83 of GyrA | No. of isolates |
|---|---|---|
| 50 | Ser | 54 |
| Arg | 5 | |
| ≥100 | Ser | 24 |
| Arg | 4 | |
| Ile | 5 |
DISCUSSION
Two main mechanisms of quinolone resistance in gram-negative bacteria were established: alterations in the quinolone targets, and decreased bacterial quinolone uptake (32). Amino acid substitutions involved in the development of quinolone resistance in E. coli have been described for GyrA/GyrB and ParC/ParE. A similar proportion of gyrA and gyrB mutations was observed for the presence of DNA gyrase mutations in quinolone-resistant E. coli strains obtained in vitro (31), whereas clinical isolate studies showed an exclusive prevalence of gyrA mutations (9, 31, 38). In the present study, amino acid analysis of QRDR in GyrA suggests that the GyrA83 of OA-resistant B. glumae field isolates is responsible for the level of OA resistance. Ser83Arg-Ser83Ile and Arg83Ser-Ile83Ser GyrA substitutions in OA-sensitive and OA-resistant isolates, respectively, show that bacterial OA resistance depends on GyrA83 substitutions. Therefore, GyrA83 substitutions are commonly responsible for OA resistance in B. glumae field isolates. On the other hand, resistance to OA in Pg-10 mutants in vitro did not necessarily depend on GyrA83 substitutions. This evidence suggests that only OA-resistant bacteria with Ser83Arg and Ser83Ile substitution might retain their ability to survive on rice plants grown in paddy fields and cause diseases on rice plants applied with OA.
Each GyrA dimmer subunit contains a short recognition helix on its presumed DNA-binding surface (10). The major quinolone resistance mutations map on the exposed surface of this helix at amino acids 83 and 87 in E. coli protein (6, 31, 32). Studies with purified DNA gyrase indicate that such mutations reduce drug binding (4, 39, 42). Since two recognition helices occur per GyrA dimmer (30) and biochemical studies indicate a stoichiometry of two quinolones per complex (5), it is likely that one quinolone binds to each recognition helix. Inspection of the amino acid changes conferring resistance at these positions reveals that the electronegative character of Ser at position 83 and Asn at position 87 is lost, mainly by mutations to the hydrophobic residues (4, 34). Substitution at Ser-83 to Ile, which is a hydrophobic residue, in the GyrA of B. glumae Pg-10 resulted in strong OA resistance, supporting data from GyrA substitutions of E. coli. Since Arg is a basic residue and tends to be located on the surface of protein molecules, Ser83Arg substitutions might cause weak changes in the structure of the GyrA-GyrA surface and reduce the binding activity of the GyrA dimmer to OA, resulting in moderate OA resistance. The codon at Ser-83 in the GyrA of OA-sensitive field isolates is AGC, and changes in this codon to AGA or AGG and ATC resulted in Arg-83 and Ile-83, respectively. A change into AGT results in Ser, and changes into AAC and ACC result in Asn and Thr, respectively. Since these amino acid residues are electronegative residues like Ser, these codon changes might not result in changes in GyrA-GyrA structure or OA resistance.
Baquero et al. suggested that a dangerous concentration range exists in which mutants are most frequently selected (2, 3). Furthermore, Drlica et al. (8) demonstrated that a threshold (mutant prevention concentration [MPC]) could be defined for restricting resistance development when the selection of resistant mutants is controlled by quinolone concentrations. A wide variety of mutants, including both target and putative efflux variants (7, 43), are selectively amplified in the concentration gap (the mutant selection window) between the MPC and the lowest concentration that inhibits growth of the majority of susceptible bacteria (MIC). At concentrations greater than the MPC, two resistance mutations are expected to be necessary for growth. According to this theory, the MPC and MIC of OA for B. glumae are 10 μg/ml (data not shown) and 0.5 μg/ml (Table 4), respectively. Ser83Arg and Ser83Ile substitutions in the GyrA of Pg-10 result in moderate and high resistance to OA, respectively, and the MICs of OA in the recombinants are 50 and 100 μg/ml. Analysis with recombinants with gyrA from Pg-10 shows that OA resistance in 12 of the 24 isolates depended only on GyrA mutations. Moreover, Arg83Ser and Ile83Ser substitutions in the GyrAs of Pg-7 and Pg-15, respectively, show that the OA resistance of these isolates depends only on the GyrA83 substitution. Most rice farmers in Japan apply 200 μg of OA/ml to heading rice plants by foliar application and immerse rice seeds in a 1,000-μg/ml solution of OA for 24 h or a 10,000-μg/ml solution for 20 min, according to protocols described in the literature (18). Since the solubility of OA in water is 3.2 μg/ml (18), B. glumae on rice plants grown in paddy fields might theoretically be exposed to OA solutions at concentrations lower than the MPC. Therefore, the OA resistance of these B. glumae field isolates depends on a single amino acid mutation at position 83 in GyrA.
On the other hand, the resistance to OA of the other 12 isolates might involve other mechanisms in addition to the GyrA83 substitution. There is no difference in the deduced amino acid sequences of QRDR in parC of all field isolates (Y. Maeda, unpublished data). In addition, the role of amino acid substitutions in ParE, which result in the development of quinolone resistance in clinical isolates of gram-negative bacteria, appear to be irrelevant (9), and alterations in the GyrB of E. coli, which result in quinolone resistance, have been described at positions 426 and 447 (41). However, it remains unclear whether ParE and GyrB alterations in B. glumae are involved in the resistance conferred by other mechanisms. On the other hand, although the MIC of OA for recombinants with Ser83Ile in the GyrA of Pg-10 was 100 μg/ml, that for Pg-14, of which GyrA83 was Ile, was >1,000 μg/ml. These results suggest that mechanisms other than GyrA83 substitution might be involved in the development of increased OA resistance in Pg-14. In our previous study of highly OA resistant B. glumae with an MIC of OA of >1,000 μg/ml, a DNA fragment containing one open reading frame involved in the development of increased OA resistance conferred by GyrA alterations was isolated (20). This DNA fragment does not confer resistance to OA on OA-sensitive B. glumae. These findings suggest that other mechanisms might be involved in the development of increased OA resistance in isolates with GyrA83 substitutions.
Although antibiotic pressure usually reduces bacterial diversity by favoring those that are better adapted (29), Low et al. (26) demonstrated that, in the long term, antibiotic treatment might provoke the diversification of antibiotic resistance. In Japan, since its registration 15 years ago, OA has been used for disease control three times per rice cultivation season. Recently, it was reported that an OA-resistant Erwinia amylovora was isolated from pear orchards in Israel, where since 1998 the number of OA applications was one to three sprays per season (28). Manulis et al. (28) demonstrated that the use of an at-risk compound where large bacterial populations exist should be avoided and that the number of risk compound applications should be reduced. B. glumae naturally exists saprophytically on roots, stem bases, and the lower leaf sheaths of rice plants (15). Bacteria existing on the flag leaf sheaths invade the flowering spikelets and utilize intermediate sugars in the biosynthesis of grain starch in these spikelets to remarkably multiply during 5 days after flowering (19). Therefore, OA applications to heading rice plants have shown a high efficacy in the control of bacterial grain rot of rice (15). However, applications to rice plants after heading results in reduced efficacy, since the bacteria in the spikelets have already proliferated vigorously. Moreover, OA applications to high populations of bacteria might result in the development of OA-resistant bacteria. The results of the present study suggest that several mechanisms are involved in increased resistance to OA in B. glumae field isolates with GyrA83 substitution. These evidences suggest that high resistance to OA might develop among MRs and HRs with GyrA83 substitutions, thus infecting rice plants, through selection by multiple OA applications.
Acknowledgments
We thank T. Okuno, Kyoto University, for critical reading of the manuscript.
This study was supported by a grant from Sumitomo Chemical Co., Ltd., and a Grant-in-Aid for Scientific Research (16658020) to Y.H. from the Japanese Society for Promotion of Sciences.
REFERENCES
- 1.Allen, C., Y. Huang, and L. Sequeira. 1991. Cloning of genes affecting polygalacturonase production in Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:147-154. [Google Scholar]
- 2.Baquero, F. 1990. Resistance to quinolones in gram-negative microorganisms: mechanisms and prevention. Eur. Urol. 17:3-12. [DOI] [PubMed] [Google Scholar]
- 3.Baquero, F., and M. C. Negri. 1997. Strategies to minimize the development of antibiotic resistance. J. Chemother. 9:29-37. [PubMed] [Google Scholar]
- 4.Barnard, F., and A. Maxwell. 2001. Interaction between DNA gyrase and quinolones: the effect of alanine mutations at A subunit residues Ser-83 and Asp-87. Antimicrob. Agents Chemother. 45:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Critchlow, S. E., and A. Maxwell. 1996. DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase-DNA complex. Biochemistry 35:7387-7393. [DOI] [PubMed] [Google Scholar]
- 6.Cullen, M., A. Wyke, R. Kuroda, and L. Fisher. 1989. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob. Agents Chemother. 33:886-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, Y., X. Zhao, J. Domagala, and K. Drlica. 1999. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1756-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drlica, K., and M. Malik. 2003. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 3:249-282. [DOI] [PubMed] [Google Scholar]
- 9.Everett, M. J., Y. F. Jin, V. Ricci, and L. J. V. Piddock. 1996. Contribution of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Hikichi, Y. 1993. Antibacterial activity of oxolinic acid on Pseudomonas glumae. Ann. Phytopathol. Soc. Jpn. 59:369-374. [Google Scholar]
- 14.Hikichi, Y. 1993. Mode of action of oxolinic acid against bacterial seedling rot of rice caused by Pseudomonas glumae 1: relationship between population dynamics of P. glumae on seedlings of rice and disease severity of bacterial seedling rot of rice. Ann. Phytopathol. Soc. Jpn. 59:441-446. [Google Scholar]
- 15.Hikichi, Y. 1993. Relationship between population dynamics of Pseudomonas glumae on rice plants and disease severity of bacterial grain rot of rice. J. Pesticide Sci. 18:319-324. [Google Scholar]
- 16.Hikichi, Y., and H. Egami. 1995. Control system for bacterial grain rot of rice with oxolinic acid and seed selection with salt solution. Ann. Phytopathol. Sci. Jpn. 61:405-409. [Google Scholar]
- 17.Hikichi, Y., H. Egami, Y. Oguri, and T. Okuno. 1998. Fitness for survival of Burkholderia glumae resistant to oxolinic acid in rice plants. Ann. Phytopathol. Soc. Jpn. 64:147-152. [Google Scholar]
- 18.Hikichi, Y., C. Noda, and K. Shimizu. 1989. Oxolinic acid. Jpn. Pestic. Infect. 55:21-23. [Google Scholar]
- 19.Hikichi, Y., T. Okuno, and I. Furusawa. 1994. Susceptibility of rice spikelets to infection with Pseudomonas glumae and its population dynamics. J. Pestic. Sci. 19:11-17. [Google Scholar]
- 20.Hikichi, Y., K. Tsujiguchi, Y. Maeda, and T. Okuno. 2001. Development of increased oxolinic acid resistance in Burkholderia glumae. J. Gen. Plant Pathol. 67:58-62. [Google Scholar]
- 21.Horowitz, D. S., and J. C. Wang. 1987. Mapping the activity site tyrosine of Escherichia coli DNA gyrase. J. Biol. Chem. 262:5339-5344. [PubMed] [Google Scholar]
- 22.Kaminsky, D., and R. I. Melzer. 1968. Quinolone antibacterial agents: oxolinic acid and related compounds. J. Med. Chem. 11:160-163. [DOI] [PubMed] [Google Scholar]
- 23.Kato, J.-I., Y. Nishimura, R. Imamura, H. Niki, S. Hiraga, and H. Suzuki. 1990. New topoisomerase essential for chromosome segregation in Escherichia coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 24.Khodursky, A. B., E. L. Zechiedrich, and N. R. Cozzarelli. 1995. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:11801-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine, C., H. Hiasa, and K. Marians. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400:29-43. [DOI] [PubMed] [Google Scholar]
- 26.Low, A. S., F. M. MacKenzie, I. M. Gould, and I. R. Booth. 2001. Protected environments allow parallel evolution of a bacterial pathogen in a patient subjected to long-term antibiotic therapy. Mol. Microbiol. 42:619-630. [DOI] [PubMed] [Google Scholar]
- 27.Maeda, Y., A., Kiba, K. Ohnishi, and Y. Hikichi. A new method to detect oxolinic acid-resistant Burkholderia glumae infesting rice seeds using a mismatch amplification mutation assay polymerase chain reaction. J. Gen. Plant Pathol., in press.
- 28.Manulis, S., F. Kleitman, D. Shtienberg, and H. Shwartz. 2003. Changes in the sensitivity of Erwinia amylovora populations to streptomycin and oxolinic acid in Israel. Plant Dis. 87:650-654. [DOI] [PubMed] [Google Scholar]
- 29.Matic, I. 2002. Selective compartments and antibiotic resistance diversity. Trends Microbiol. 10:66. [Google Scholar]
- 30.Morais-Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura, S., M. Nakamura, T. Kojima, and H. Yoshida. 1989. gyrA and gyrB mutants in quinolone-resistant strains of Escherichia coli. Antimicrob. Agents Chemother. 33:254-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz, J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation, and DNA gyrase protection. J. Antimicrob. Chemother. 51:1109-1117. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sreedharan, S., M. Oram, B. Jesen, L. Peterson, and L. Fisher. 1990. DNA gyrase gyrA mutations in ciprofloxacin-resistant strains of Staphylococcus aureus: close similarity with quinolone resistance mutations in Escherichia coli. J. Bacteriol. 172:7260-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugino, A., C. I. Peebles, K. N. Krenzer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuge, S., A. Furutani, R. Fukunaka, Y. Kubo, and O. Horino. 2001. Growth complementation of hrpXo mutants of Xanthomonas oryzae pv. oryzae by virulent strains in rice cultivars resistant and susceptible to the parental strain. J. Gen. Plant Pathol. 67:51-57. [Google Scholar]
- 37.Turner, F. J., S. M. Ringel, J. F. Martin, P. J. Storino, J. M. Daly, and B. S. Schwartz. 1967. Oxolinic acid, a new synthetic antimicrobial agent. I. In vitro and in vivo activity. Antimicrob. Agents Chemother. 7:475-479. [PubMed] [Google Scholar]
- 38.Vila, J., J. Ruiz, F. Marco, A. Barcelo, P. Goni, E. Giralt, and T. Jimenez de Anta. 1994. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob. Agents Chemother. 38:2477-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willmott, C. J. R., and A. Maxwell. 1993. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida, H., M. Nakamura, M. Bogaki, H. Ito, T. Kojima, H. Hattori, and S. Nakamura. 1993. Mechanism of action of quinolones against Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 37:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, J.-F., Y. Dong, X. Zhao, S. Lee, A. Amin, S. Ramaswamy, J. Domagala, J. M. Musser, and K. Drlica. 2000. Selection of antibiotic resistance: allelic diversity among fluoroquinolone-resistant mutation. J. Infect. Dis. 182:517-525. [DOI] [PubMed] [Google Scholar]