Abstract
The bacterium Xenorhabdus nematophila is a mutualist of Steinernema carpocapsae nematodes and a pathogen of insects. Presently, it is not known what nutrients the bacterium uses to thrive in these host environments. In other symbiotic bacteria, oligopeptide permeases have been shown to be important in host interactions, and we therefore sought to determine if oligopeptide uptake is essential for growth or symbiotic functions of X. nematophila in laboratory or host environments. We identified an X. nematophila oligopeptide permease (opp) operon of two sequential oppA genes, predicted to encode oligopeptide-binding proteins, and putative permease-encoding genes oppB, oppC, oppD, and oppF. Peptide-feeding studies indicated that this opp operon encodes a functional oligopeptide permease. We constructed strains with mutations in oppA1, oppA2, or oppB and examined the ability of each mutant strain to grow in a peptide-rich laboratory medium and to interact with the two hosts. We found that the opp mutant strains had altered growth phenotypes in the laboratory medium and in hemolymph isolated from larval insects. However, the opp mutant strains were capable of initiating and maintaining both mutualistic and pathogenic host interactions. These data demonstrate that the opp genes allow X. nematophila to utilize peptides as a nutrient source but that this function is not essential for the existence of X. nematophila in either of its host niches. To our knowledge, this study represents the first experimental analysis of the role of oligopeptide transport in mediating a mutualistic invertebrate-bacterium interaction.
The gram-negative bacterium Xenorhabdus nematophila has two types of eukaryotic hosts: the nematode Steinernema carpocapsae and various species of larval-stage insects (for a review of X. nematophila and its host interactions, see reference 11). The nematode acts as a mutualistic vector to allow X. nematophila access to the hemocoel (circulatory system) of insects. Once there, the bacterium is expelled from the nematode, the two organisms collaborate to kill the insect, and the invaders multiply and consume the insect corpse. Inside the insect, the bacterium is thought to produce exoenzymes including proteases, lipases, and lecithinases (11), and X. nematophila-secreted protease II has been shown to promote the digestion of some proteins of insect hemolymph (circulatory fluid) (8). The identities of the breakdown products of the protease activities are not yet known, but some bacteria, such as Salmonella enterica, are able to utilize peptides or free amino acids as nitrogen and carbon sources (15). Thus, oligopeptides could be a source of nutrients for X. nematophila in insect hosts.
As the food supply within the insect runs low, the bacterium and the nematode reassociate, the nematode develops into its vector stage (called the infective juvenile [IJ]), and the bacterium-nematode pair leaves the spent insect carcass in search of a new host. The bacterium resides in a specialized compartment of the IJ nematode intestine called the vesicle, within which X. nematophila multiplies to a population of 30 to 200 cells after an initial colonization by 1 to 2 cells (22). Even though IJ nematodes are nonfeeding, X. nematophila bacteria can survive within them for over 4 months in laboratory storage (21). At present, it is not known what nutrients support bacterial growth in the vesicle or how X. nematophila survives nutritionally during long-term IJ nematode quiescence. It is possible that X. nematophila bacteria residing in the nematode vesicle use nutrients released from dying bacterial siblings, which could include peptides. Alternatively, or perhaps additionally, the nonfeeding nematode may provide X. nematophila bacteria with nutrients to its own detriment. Consistent with the latter possibility, uncolonized nematodes survive longer in storage than X. nematophila-colonized nematodes (25).
Bacterial ABC (ATP-binding complex)-type oligopeptide permeases bring short peptides (two to six amino acid residues in enteric bacteria) into the bacterial cell. The oligopeptides to be transported are first bound by a periplasmic (in gram-negative bacteria) oligopeptide-binding protein (e.g., OppA), which directs the oligopeptides to the permease. The major known functions of oligopeptide transport in bacteria are the salvaging of cell wall components (using MppA instead of OppA) (26) and the uptake of nutrients (14, 27). Additionally, the peptide-binding proteins of some bacteria play important roles in host interactions, participating in intracellular survival (6) and binding to host cells (9), host proteins (10), or other bacterial cells in the host environment (18, 19, 23). Consistent with a specific role for oligopeptide-binding proteins in host interactions, several pathogens have been shown to have multiple oppA gene homologues. For example, Borrelia burgdorferi, which has two types of eukaryotic hosts (ticks, which act as its vector, and mammals), has five oppA homologues—three immediately upstream of the permease-encoding genes, and two individually carried on separate plasmids (5, 20). It was proposed that B. burgdorferi utilizes multiple oligopeptide-binding proteins to broaden substrate recognition for different oligopeptides encountered in each host environment (20). Indeed, the various B. burgdorferi oppA genes are differentially expressed as the bacterium traverses between host environments (32). However, to our knowledge, this hypothesis has not been tested further with B. burgdorferi.
In this work, we identify an X. nematophila opp locus consisting of two oppA homologues and putative permease-encoding genes oppB, oppC, oppD, and oppF. We constructed strains with mutations in the opp locus, which allowed us to directly examine the role of these genes in bacterial uptake of peptides, growth, and host interactions. We demonstrate that the permease is not essential for X. nematophila-host interactions, even though loss of the permease resulted in an altered growth phenotype in a peptide-rich laboratory medium and in hemolymph isolated from Manduca sexta insects.
MATERIALS AND METHODS
Bacterial strains and standard growth conditions.
Table 1 shows a list of bacterial strains used. Cultures were grown in a tube roller at 30°C in Luria-Bertani (LB) broth (24) that had been stored in the dark (34). LB agar (20 g liter−1) plates were supplemented with 1 g of sodium pyruvate liter−1 (34) and, when appropriate, kanamycin (KAN; 20 mg liter−1 for X. nematophila or 50 mg liter−1 for Escherichia coli) or chloramphenicol (CHL; 20 mg liter−1). Solid lipid agar (LA) medium was prepared as described elsewhere (30). For peptide-feeding experiments, defined medium consisted of 22 mM KH2PO4, 40.2 mM K2HPO4, 15.1 mM (NH4)2SO4, 0.41 mM nicotinic acid, 9.1 mM sodium pyruvate, 50 mM glucose, 10 ml of SL4 salts liter−1 (3), 50 mg each of all amino acids except leucine (the amino acid being tested) liter−1, and glutamate, which was added at 250 mg liter−1 (50 mg of glutamate liter−1 was subsequently determined to be sufficient for growth of X. nematophila), and 15 g of noble agar liter−1. Leucine or peptides (Leu-Leu, Tyr-Leu, Leu-Leu-Leu, and Tyr-Tyr-Tyr; Sigma, St. Louis, Mo.) suspended in water were spread on defined medium plates to give a final concentration of 70 μM (17). Permanent stocks of the cultures were stored at −80°C in LB broth supplemented with 10% dimethyl sulfoxide (Sigma, St. Louis, Mo.).
TABLE 1.
Strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| X. nematophila | ||
| HGB007 | ATCC 19061, wild type | American Type Culture Collection |
| HGB081 | AN6/1 Rifr | S. Forst, University of Wisconsin-Milwaukee |
| HGB585 | HGB007 oppA1::Tn5-12 Kanr | This study |
| HGB586 | HGB007 oppA2::Tn5-15 Kanr | This study |
| HGB717 | HGB007 oppB::Tn5-20-4 Kanr | This study |
| HBG731 | HGB081 leuB::Tn5 Camr | This study |
| HBG732 | HGB081 leuB::Tn5 CamroppA1::Tn5-12 Kanr | This study |
| HBG733 | HGB081 leuB::Tn5 CamroppA2::Tn5-15 Kanr | This study |
| HGB734 | HGB081 leuB::Tn5 CamroppB::Tn5-20-4 Kanr | This study |
| E. coli | ||
| S17-1 λpir | E. coli general cloning and conjugation donor strain | 29 |
Conjugation of plasmids into X. nematophila.
E. coli S17-1 λpir was used as the donor strain to conjugate plasmids for gene knockout constructions into X. nematophila as described previously (13), except that each strain was washed and resuspended in phosphate-buffered saline (PBS; pH 7.5) (28), and the conjugation plates were incubated at room temperature to allow X. nematophila to kill and/or outcompete the E. coli donor strain. Following selective growth, exconjugant cells were verified to be X. nematophila by their negative catalase reaction when hydrogen peroxide was applied and by their characteristic smell.
DNA manipulations and sequence analysis.
DNA manipulations and transformation of E. coli were performed using standard protocols (reference 4 and product literature). Integrated DNA Technologies (Coralville, Iowa) synthesized all oligonucleotide primers. All compared sequences were obtained from the GenBank database (2) through the National Center for Biotechnology Information.
Identification of the opp locus.
oppA1 and part of oppA2 were identified on an X. nematophila chromosomal fragment used in unrelated studies on tdk (S. S. Orchard and H. Goodrich-Blair, unpublished data). PCR with arbitrary primers (7) was used to sequence downstream of the oppA2 gene in its chromosomal location, leading to the discovery of an oppB homologue. To determine if the oppC, oppD, and oppF genes were further downstream, we designed a degenerate primer (oppFdeg1; NARNGGRTCNACYTTNARRCA, where N is A+C+T+G, R is A+G, and Y is C+T) based on the C-terminal OppF amino acid sequence, CLKVDPL, of E. coli K12, S. enterica serovar Typhimurium LT2, and Yersinia pestis. This degenerate primer was used in a PCR with primer oppXnrev1 (AAAACAGATGAAGAGCGTGC) to amplify and clone a 4.1-kb fragment from the X. nematophila chromosome, which was sequenced to reveal oppB, oppC, oppD, and oppF homologues.
Identification of an X. nematophila leucine auxotroph.
HGB081 was conjugated with S17-1 λpir containing mini-Tn5 (CHLr) donor plasmid pBSL203 (1). CHLr exconjugants were tested for leucine auxotrophy on defined medium lacking leucine. The transposon insertion site of one leucine auxotroph, designated HGB731, was sequenced using primer P6 (16). The sequence appears to encode the first 34 amino acids of a LeuB homologue with 71% identity to E. coli LeuB (3-isopropylmalate dehydrogenase), the enzyme responsible for the penultimate step of leucine biosynthesis.
Construction of X. nematophila opp and leuB opp mutant strains.
To construct oppA1 and oppA2 mutants, a 4.7-kb fragment of the X. nematophila chromosome containing oppA1, oppA2, and part of oppB was PCR amplified using primers KpnIoppA (GGTACCATTGGCCCTTTAGTGATTCC) and SacIoppB (GAGCTCAGTCCCATTTGGTATTCTGG). To create an oppB mutant strain, a 1.6-kb fragment of the X. nematophila chromosome including most of oppB (lacking the 5′ 89 bp) and oppC (lacking the 3′ 287 bp) was PCR amplified using primers KpnIopp5for (GGTACCACCTGAATGACCCGATG) and SacIoppAflIIrev (GAGCTCCTTAAGAAAGATTCAAACAG). These PCR products were cloned independently into pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.) to create plasmids poppA-B′ and popp′B-C′, respectively. Each insert was subcloned into pKR100 (Camr oriR6K suicide vector; K. Visick, Loyola University) using KpnI and SacI digests to create plasmids pKRoppA-B′ and pKRopp′B-C′, respectively. The GeneJumper primer insertion kit (Invitrogen) was used to insert GeneJumper KANr transposons into these plasmids, and the mutagenized plasmids were transformed into E. coli S17-1 λpir. Strains containing transposed pKRoppA-B′ or pKRopp′B-C′ were selected on the basis of KANr and CHLr. pKRoppA1::Tn5-12, pKRoppA2::Tn5-15, and pKRoppB::Tn5-20-4, chosen for suitable transposon location, were conjugated into X. nematophila, and exconjugants were selected on LB plates containing KAN. Strains that had undergone double recombination events with concurrent loss of the plasmid (KANr CHLs) were chosen for further study: X. nematophila oppA1::Tn5-12 (HGB585), oppA2::Tn5-15 (HGB586), and oppB::Tn5-20-4 (HGB717). The insertion in oppA1 disrupts the predicted encoded protein at amino acid 169 of 546. oppA2 is disrupted at predicted encoded amino acid 192 of 548, and oppB is disrupted at predicted encoded amino acid 235 of 306. To construct X. nematophila leuB oppA1, leuB oppA2, and leuB oppB mutant strains, HGB731 (leucine auxotroph) was conjugated with E. coli S17-1 λpir containing plasmid pKRoppA1::Tn5-12, pKRoppA2::Tn5-15, or pKRoppB::Tn5-20-4 as described for making the oppA1, oppA2, and oppB mutants, respectively. KANr exconjugants were screened for double recombination by PCR analysis.
Nematode maintenance and cocultivations with bacteria (noncompetitive).
S. carpocapsae All (Weiser) was propagated by passage through Galleria mellonella larvae (Vanderhorst Wholesale, Inc., St. Marys, Ohio), harvested in White traps (33), and stored in water. Culture of nematodes on bacterial lawns for colonization assays was performed on LA plates as described previously (30). Axenic nematode eggs were isolated from gravid adult females as described previously (31), except that the eggs were washed and resuspended in LB broth. The eggs were left in LB broth at room temperature for 1 to 2 days to assess sterility and to enable hatching into larval-stage nematodes, which were used to inoculate coculture plates. IJ nematodes from the cocultures were harvested by allowing their migration to water. The IJ nematodes were surface sterilized by using a 3-min incubation in a 2% HOCl solution followed by three rinses with sterile distilled water. To release the bacteria from the nematode vesicles, a suspension of 104 nematodes in 0.5× LB broth was sonicated in a water bath sonicator (Branson Ultrasonics, Danbury, Conn.) for 1 min. The sonicated nematode suspension was vortexed for 3 s and then diluted and plated on LB agar to determine average IJ CFU counts.
Competitive bacterial colonization assays.
Overnight cultures of mutant and wild-type strains (200 μl each) were mixed and inoculated onto 6-cm-diameter LA plates. After overnight incubation, each plate was inoculated with 2,000 to 4,000 early-larval-stage nematodes and incubated for 8 days at 25°C. KAN resistance of individual colonies was used to assess the proportion of opp mutant strains in the bacterial lawn at day 9 (plate input) and in the resulting nematode progeny (plate output). The competitive index was calculated with the ratio proportionplate output/proportionplate input. To determine if the mutant strains are competitive during a natural infection process, these IJ nematodes were then used to infect G. mellonella insects (insect input). G. mellonella was selected for use due to the ease of recovering IJ nematodes. The insects were infected with 100, 500, or 1,000 IJ nematodes, and all insects died within 72 h. After 2 weeks, the proportion of colonization by the opp mutant strains of the IJ nematode progeny from the insect infections (insect output) was determined, and the competitive index during infection was calculated with the ratio proportioninsect output/proportioninsect input. A ratio of 1 in either assay would indicate that the mutant strain competes equally as well as the wild-type strain for colonization of nematodes, and a ratio of <1 would indicate that the mutant has a defect in competitive colonization. Six independent experiments were conducted for all competition assays, except that only three experiments were performed for the insect infections with the oppB mutant strain. The oppA2 mutant strain was outcompeted by the wild-type strain on plates in four of the six experiments, and competitive indices for those experiments could not be reported. Competitive indices were calculated for each experiment, and the values were averaged and used to calculate standard errors of the means (SEM).
M. sexta insect rearing and injection assays.
M. sexta (tobacco hornworm) insect eggs were obtained from W. Goodman (University of Wisconsin—Madison), and larvae were reared as described previously (30). After 18 to 24 h of growth in LB broth from −80°C freezer stocks, strains were subcultured at a concentration of 1:100 into fresh LB broth and grown for an additional 24 h. The strains were washed and resuspended in PBS, serially diluted, and plated on LB agar for subsequent CFU determination. One to one thousand CFU in 10 μl of PBS were injected behind the left first proleg of fourth-instar M. sexta. The insects were fed and monitored, and deaths in each set of 10 insects were recorded to provide a value for percent mortality at 72 h postinjection. Previous observations indicated that only rarely do additional insects die in response to injection beyond 72 h postinjection.
Analysis of growth profile in hemolymph.
M. sexta larvae were chilled on ice for 30 min and cleaned with 95% ethanol. An incision was made in the left first proleg, and hemolymph was drained into a microcentrifuge tube. Extracted hemolymph was centrifuged at 10,000 × g at 4°C for 10 min, and glutathione (Sigma, St. Louis, Mo.) was added to a final concentration of 5 mM to retard the phenoloxidase reaction. Hemolymph, typically from ∼10 to 20 insects per experiment, was filtered through a 0.8-μm-pore-size filter and then through a 0.2-μm-pore-size filter (Millipore, Bedford, Mass.) and used the day of harvest or following a single freeze and less than 1 week of storage at −20°C. Bacteria from isolated colonies were cultured overnight in LB broth, washed, and resuspended in PBS and diluted 1:100 in hemolymph. Samples were placed in non-tissue-culture-treated, 96-well, flat-bottom polystyrene plates (Becton Dickinson Labware, Franklin Lakes, N.J.) and incubated at 30°C with shaking in a Molecular Devices SPECTRAmax 190 plate reader. Absorbance data was collected using SOFTMax PRO 3.1.2 software (Molecular Devices, Sunnyvale, Calif.).
Growth profile in LB broth and relative oppA1 and oppA2 transcript analyses.
Bacteria from isolated colonies were cultured overnight in LB broth, and 400 μl of the cultures was used to inoculate 40 ml of fresh LB broth into 125-ml flasks. Because overnight cultures of the mutant and wild-type strains did not differ in viable cell counts (see Results), no adjustment was made for the difference in optical density of the starter strains. Three independent cultures for each strain were shaken at 250 rpm in a water bath at 30°C. Samples of the triplicate wild-type cultures were harvested to analyze relative levels of oppA1 and oppA2 transcripts. Total RNA from the wild-type strain was isolated, DNase treated, and used to make cDNA with random hexamer primers (Integrated DNA Technologies, Coralville, Iowa) and avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.). As a control to detect DNA contamination of the DNase-treated RNA, samples with no added avian myeloblastosis virus reverse transcriptase were analyzed by PCR for amplification of oppF, and, as expected, none showed a product. Reactions (25 μl) for real-time PCR were performed in duplicate with iQ SYBR Green Supermix (Bio-Rad, Hercules, Calif.), cDNA template, appropriate primers, and a three-step cycling protocol on a Bio-Rad iCycler and analyzed with Bio-Rad iCycler iQ software. The amount of oppA1 and oppA2 transcript was measured by amplification with primers oppAIprfor (ATGTGCAGAACGCCGATG) and opp12rev (AGTTTTCCGGCAACGTCC) for oppA1 and primers oppfor8 (CAACATCGCCCGTGATTTG) and oppAIIprrev1 (TATGGACAAATAACTTGACC) for oppA2. As a negative control, water was used in place of cDNA template. Cycle threshold results for each sample were adjusted according to recA levels (amplified with the following primers designed from the X. nematophila recA sequence published under GenBank accession no. AF127333: recaminfor, TGTCCGTTTGGATATCCGCC, and recaminrev, CCCAGAGTATTAATACCTTCCCCAT) and then converted to arbitrary units, factoring in a twofold change in PCR product per cycle. The X. nematophila recA gene is not expressed at constant levels over a growth curve (K. N. Cowles and H. Goodrich-Blair, unpublished data), and we therefore chose to present the data as oppA1 transcript level as a proportion of the oppA2 transcript level at each sampling point.
Statistical analysis.
One-way analysis of variance (ANOVA) with Dunnett's posttest was used to analyze the colonization and pathogenesis data. It was used to compare each mutant strain to the wild-type strain, or to a value of 1 for the competitive nematode colonization assays, at the 95% confidence interval. One-way ANOVA with Tukey's posttest was used to analyze the relative oppA1 and oppA2 transcript data. All statistical analyses were performed using Prism version 3.0a for Macintosh (GraphPad Software, San Diego, Calif.).
Nucleotide sequence accession numbers.
All opp genes in the described X. nematophila locus have been sequenced from PCR-amplified fragments, and a 15.08-kb sequence including the opp genes has been deposited in GenBank (accession no. AY363171). A region of DNA including a 3′ portion of the putative leuA gene and a 5′ portion of the putative leuB gene has been sequenced from a cloned copy, and the 734-nucleotide sequence has been deposited in GenBank (accession no. AY443349).
RESULTS
Construction of X. nematophila oppA1, oppA2, and oppB mutant strains.
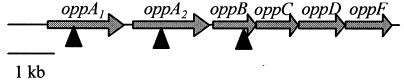
We identified an opp locus putatively encoding an oligopeptide permease (OppB, OppC, OppD, and OppF) and two OppA peptide-binding protein homologues, OppA1 and OppA2 (see Materials and Methods). The entire operon was sequenced, and the genetic organization of the opp operon is presented in Fig. 1. The amino acid sequences of the predicted X. nematophila peptide-binding proteins, OppA1 and OppA2, had 58% identity to each other and 57 and 67% identity, respectively, to OppA of E. coli. The predicted permease proteins, OppB, OppC, OppD, and OppF, had 83 to 88% identity to the corresponding Opp permease proteins in E. coli. Insertions were made in oppA1, oppA2, and oppB to examine the role of the putative oligopeptide permease and associated peptide-binding proteins in the host interactions of X. nematophila. The mutations were confirmed by PCR and Southern hybridization analysis (data not shown). Insertions in oppA2 and oppB reduced transcription of oppF to levels similar to that of the negative control (no template) as measured by real-time PCR (data not shown), indicating that these insertions completely abolish transcription of the downstream opp genes under the tested conditions. However, the oppA2-oppF genes were still expressed in the oppA1 mutant strain, suggesting the presence of a promoter downstream of the insertion in oppA1 (data not shown).
FIG. 1.
opp operon organization in X. nematophila and locations of Tn5 insertions. Triangles indicate approximate locations of insertions in opp mutant strains used in these studies. Gene designations are indicated above the arrows, which represent open reading frames. The direction of the arrows indicates the direction of transcription. This region spans 8.35 kb.
The X. nematophila opp operon encodes a functional oligopeptide permease.
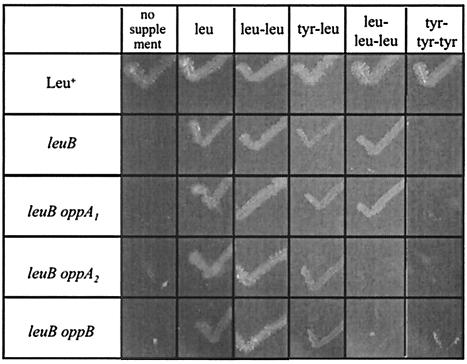
To determine if the opp homologues encode a functional oligopeptide permease, we tested the ability of an X. nematophila leucine auxotroph to acquire leucine through peptide uptake. Insertions were made in oppA1, oppA2, and oppB in a leuB background. These double mutant strains were tested for growth on the dipeptides Tyr-Leu and Leu-Leu and the tripeptides Leu-Leu-Leu and Tyr-Tyr-Tyr (Fig. 2). Tyr-Tyr-Tyr was included as a negative control because it contains no leucine, and, as expected, none of the leuB strains grew on this tripeptide. The leuB (Opp+) auxotroph grew on all tested peptides that contain a leucine residue, while the leuB oppA2 and leuB oppB strains only grew on leucine-containing dipeptides. In contrast, the leuB oppA1 strain, like the leuB (Opp+) strain, grew on Leu-Leu-Leu, indicating that OppA2 and the permease are sufficient to transport the Leu tripeptide.
FIG. 2.
Growth of X. nematophila leuB opp mutant strains on peptides. Peptides and leucine (Leu), where indicated, were spread on defined medium plates at a final concentration of 70 μM. The plates were inoculated by scoring with a toothpick coated with the appropriate strain. Three independent colonies each of HGB081 (Leu+), HGB731 (leuB), HGB732 (leuB oppA1), HGB733 (leuB oppA2), and HGB734 (leuB oppB) were tested; a representative result is shown for each condition. Growth at 96 h after inoculation is shown.
X. nematophila opp mutant strains colonize and persist in S. carpocapsae nematodes.
X. nematophila opp mutant strains were assayed for their ability to colonize and survive in S. carpocapsae nematodes following cocultivation on LA plates. All mutant strains colonized the nematode to levels similar to that of the wild-type strain after a 2- to 3-week cocultivation (Table 2). To test if X. nematophila opp mutant strains could recolonize nematodes in the natural habitat of an insect corpse, X. nematophila oppA1- and oppA2-colonized IJ nematodes were allowed to infect and kill G. mellonella insects. IJ nematode progeny were tested for level of colonization by bacteria, and again the X. nematophila opp mutant strains were able to colonize progeny IJ nematodes to an extent similar to that of the wild-type strain (data not shown).
TABLE 2.
Host interactions of X. nematophila opp mutant strains
| Strain | No. of CFU per IJa (SEM) | Competitive index (SEM)
|
% Mortalityd (SEM) | |
|---|---|---|---|---|
| Platesb | Insectsc | |||
| HGB007 (Opp+) | 43.8 (4.1) | NAe | NAe | 48.1 (6.1) |
| HGB585 (oppA1) | 44.1 (5.9) | 1.0 (0.16) | 0.83 (0.17) | 60 (13.9) |
| HGB586 (oppA2) | 49.2 (6.8) | 0.76 (0.04) | 1.17 (0.44) | 62.9 (9.7) |
| HGB717 (oppB) | 47.3 (3.1) | 1.74f (0.27) | 0.57 (0.16) | 42 (4.7) |
Average bacterial colonization level of S. carpocapsae nematodes.
Competitiveness with wild-type strain for colonization of S. carpocapsae nematodes as calculated by proportionplate output/proportionplate input.
Competitiveness with wild-type strain for colonization of S. carpocapsae nematodes as calculated by proportioninsect output/proportioninsect input.
Average percent mortality of sets of 10 M. sexta larvae following injection with 1 to 1,000 CFU of the appropriate strain.
NA, not applicable.
Significantly different from 1 (P < 0.001) as determined by one-way ANOVA with Dunnett's posttest.
X. nematophila opp mutant strains are competitive for nematode colonization.
Because competitive assays can sometimes reveal subtle phenotypes, the X. nematophila opp mutant strains were tested for their ability to compete with wild-type X. nematophila in colonizing nematodes. On LA plates, the oppA1 and oppA2 mutant strains were as competitive as the wild-type strain (P was >0.05 for difference from 1), and the oppB mutant strain was significantly (P < 0.001) more competitive than the wild-type strain for nematode colonization (Table 2). The oppA2 mutant strain was outcompeted by the wild-type strain for growth on LA plates in four of the six experiments performed (data not shown). However, because the oppA2 mutant strain was able to compete with the wild-type strain for nematode colonization in the remaining two experiments and for competitive colonization of the nematode and growth in the insect infection assays (see below), this phenotype was not explored further.
To determine if the mutant strains are as competitive as the wild-type strain during the natural infection of insects, populations of nematodes carrying both the wild-type and opp mutant strains were allowed to infect G. mellonella insects. The calculated proportioninsect output/proportioninsect input ratios for the opp mutant strains (Table 2) are not significantly different from 1 (P > 0.05). Thus, X. nematophila opp mutant strains competed as well as the wild-type strain during growth and nematode recolonization inside the insect.
X. nematophila opp mutant strains kill and grow in insects normally.
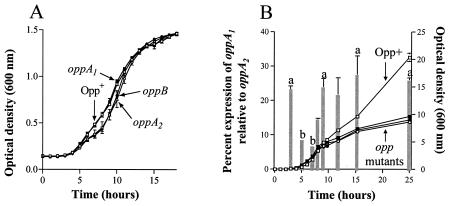
The X. nematophila opp mutant strains were tested for their ability to kill fourth-instar M. sexta larvae. Wild-type X. nematophila kills ∼50% of insects in 72 h when 1 to 1,000 CFU are injected per insect. All mutant strains were as virulent as the wild-type strain towards M. sexta, killing ∼40 to 60% of insects by 72 h after injection at 1 to 1,000 CFU per insect (Table 2). The majority of bacterial growth in the insect appears to occur after the insect's death (12). Thus, we reasoned that oligopeptide uptake might be essential for bacterial outgrowth in insects. The ability of the X. nematophila opp mutant strains to recolonize nematodes in G. mellonella carcasses (see above) suggested that they were able to grow in G. mellonella insects. However, to more easily measure bacterial growth rates, we studied the in vitro growth of X. nematophila in insect hemolymph. G. mellonella larvae are more susceptible to X. nematophila infection than M. sexta larvae are, and we thus chose to use hemolymph extracted from M. sexta insects for the growth analysis to subject the bacteria to a possibly more stringent environment for growth. A small but reproducible difference in growth rate (doubling times were 1.9 h for wild-type and oppA1 mutant strains and 2.1 and 2.5 h for oppA2 and oppB mutant strains, respectively) was observed for the oppA2 and oppB strains relative to the wild-type and oppA1 strains (Fig. 3A). However, all strains ultimately reached the same optical density.
FIG. 3.
Growth of X. nematophila opp mutant strains in hemolymph (A) and LB (B) liquid culture and analysis of relative oppA1 transcript levels in the wild-type strain in LB broth (B). HGB007 (Opp+, open squares), HGB585 (oppA1, filled squares), HGB586 (oppA2, open circles), and HGB717 (oppB, filled circles) were grown in 100 μl of hemolymph plus 5 mM glutathione in microtiter plates (A) or in 40 ml of LB broth in 125-ml flasks (B) at 30°C with shaking. (B) oppA1 transcript levels (gray columns) are presented as percentages of oppA2 transcript levels. Error bars indicate SEM of three independent cultures of each strain, except for the 8-h time point, which shows duplicate results for the transcript analysis due to the loss of an RNA sample. Columns with different letters are significantly different from each other (P < 0.05) as determined by one-way ANOVA with Tukey's posttest analysis, except that the 3-h column is not significantly different from the 5-h column.
X. nematophila opp mutant strains achieve a lower final optical density in LB liquid culture.
To determine if the inability of the X. nematophila opp mutant strains to use oligopeptides as a nutrient source hinders their growth in a peptide-rich laboratory medium, their growth in LB broth was measured. The opp mutant strains consistently did not achieve as high a final optical density as the wild-type strain in flasks (Fig. 3B), microtiter plates, or 5-ml culture in tubes (data not shown). This difference was not due to a difference in final viable cell counts, proportion of dead cells (by propidium iodide staining and fluorescence-activated cell sorter analysis), a difference in the size of the bacteria, or a difference in absorbance of the culture supernatant (data not shown). In addition, it was not an effect of the transposon per se: an unrelated mutant in the laboratory collection, X. nematophila cobB::Tn5 (KANr) (GeneJumper transposon) does not display a reduced final optical density (data not shown). Attempts at complementation for this phenotype were hindered by the fact that all tested constructs of the oppA1 gene in a single copy or the entire operon in a low copy number inhibited growth of both the wild-type and mutant strains (data not shown). However, the same phenotype was seen in both the HGB007 (Fig. 3B) and HGB731 (HGB081 leuB) backgrounds (data not shown), suggesting that the phenotype occurs regardless of strain background. Furthermore, the growth phenotype was observed in X. nematophila oppB::Tn5-13-7, a CHLr oppB mutant strain (data not shown). Therefore, the aberrant optical density phenotype appears to be specific to the opp disruption. We analyzed the level of oppA1 transcript relative to the level of oppA2 transcript and found that it was low during log-phase growth and increased over twofold during stationary phase (Fig. 3B). The 3-h time point was in lag phase and thus likely reflects transcript levels from the stationary-phase starter culture. This same trend was observed when oppA1 transcript levels were compared to oppF transcript levels (data not shown). In contrast, oppF transcript levels remained at the same level proportional to oppA2 transcript levels over the entire growth curve (data not shown). Consistent with the idea that normal transcription from a promoter upstream of oppA1 contributes to obtaining normal late-stage optical density, relative oppA1 transcript levels increased at a point in the growth curve when the opp mutant growth profile began to taper off (Fig. 3B).
DISCUSSION
In this work, we describe an oligopeptide permease operon in the nematode mutualist and insect pathogen X. nematophila and directly test its role in X. nematophila growth and host interactions. The operon comprises six genes whose products are homologous to oligopeptide permease proteins of gram-negative and -positive bacteria and includes two OppA homologues (Fig. 1). It had previously been suggested that B. burgdorferi utilizes multiple OppA homologues to bind different peptides in its various host niches (20). To genetically address the contribution of oligopeptide uptake to the interactions between a microbe and its invertebrate host, we constructed X. nematophila oppA1, oppA2, and oppB mutant strains and tested their ability to grow in laboratory culture and to associate with the nematode and insect hosts.
Small-peptide growth assays demonstrated that the X. nematophila opp locus encodes a functional oligopeptide permease (Fig. 2). The oppA2 and oppB mutant strains also had a reduced initial growth rate in hemolymph, whereas the oppA1 mutant had normal growth in this medium (Fig. 3A). Taken together, these data suggest that hemolymph contains an oligopeptide that is utilized early in the X. nematophila growth cycle and that OppA2 and the permease are sufficient and essential for normal utilization of this nutrient. However, an inability to utilize this nutrient does not prevent the population from achieving maximum optical density in hemolymph.
Analysis of oppA1, oppA2, and oppF transcript levels in the wild-type strain indicated that oppA1 transcript levels increased relative to those of oppA2 (Fig. 3B) and oppF (data not shown) as the cultures entered stationary phase of growth in LB broth. These results are consistent with the finding that B. burgdorferi oppA homologues are differentially expressed in different environments (e.g., laboratory growth, ticks, or mice) (32). Although differential expression of oppA homologues can occur in X. nematophila, this regulation is not required for any tested host interaction or successful transition between hosts. Intriguingly, the X. nematophila opp mutant strains consistently achieved a lower final optical density in LB broth than the wild-type strain (Fig. 3B). Thus, the altered final optical density of the X. nematophila opp mutant strains in stationary phase in LB broth could indicate that this increased transcription of the opp operon from the promoter upstream of oppA1 is required to reach maximum optical density in LB broth. The cause of the difference in optical density is unclear, and to our knowledge this phenotype has not been previously observed during growth analyses of opp mutant strains of other bacterial species. Technical difficulties in complementation analyses prevented examination of the causality of the opp gene mutations for this growth phenotype. However, we believe it is associated with mutation of the opp locus because the phenotype was always correlated with the mutation, regardless of strain background or type of insertion, and was concurrent with a change in operon transcription that would be precluded in the mutant strains.
The X. nematophila opp mutant strains colonized the nematode to an extent similar to that of the wild-type bacterium soon after the colonization event (Table 2) and after long-term storage. They were also competitive with the wild-type strain for nematode colonization and killed experimental groups of M. sexta insects to the same extent and within the same time frame as the wild-type strain (Table 2). While these data do not exclude the possibility that X. nematophila uses oligopeptides as a nutrient source in the nematode or insect hosts (indeed, the hemolymph growth profiles suggest that they may), they do demonstrate that oligopeptide transport by the identified permease is not an essential function for success in these host environments. It should be noted, however, that colonization of individual nematodes by X. nematophila is a clonal or occasionally near-clonal event (22). Therefore, in the competition assays presented here, the X. nematophila opp mutant strains did not likely have to compete directly with the wild-type strain following the initial colonization event, except in the ∼30% biclonally colonized nematodes. The oppB mutation provided an apparent competitive advantage in nematode colonization in plate assays but did not provide a competitive advantage in the insect infection assays. Because the insect infection assays simulate a more natural environment for the bacteria, we are discounting the observed competitive advantage on plates as an experimental artifact.
Little is known of the nutrients available to the bacterium in either the nematode or the insect host; taken together, the data presented here indicate that transport of oligopeptides by the identified permease is not essential for nutrient acquisition by X. nematophila in host environments. The selective pressure for maintenance of the opp operon, including a second oppA gene, thus remains to be determined.
Acknowledgments
We gratefully acknowledge the assistance of David Majewski for help with early opp operon sequencing and cloning, Walter Goodman for supplying M. sexta eggs, and Eric Martens for developing a defined medium for X. nematophila and for isolating the leucine auxotroph used in the peptide-feeding experiments.
This work was supported by National Institutes of Health RO1 grant GM59776, by the Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Foundation, and by USDA/CREES grant CRHF-0-6055 (awarded to H.G.-B. and used to support S.S.O. in part). S.S.O. received additional support through National Institutes of Health predoctoral training grant T32 GM07215 in Molecular Biosciences and through National Science Foundation Graduate Teaching Fellows in K-12 Education award DUE-9979628 to the K-Through-Infinity program.
REFERENCES
- 1.Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene 160:59-62. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atlas, R. M. 1997. Handbook of microbiological media, 2nd ed. CRC Press, Boca Raton, Fla.
- 4.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. Wiley and Sons, New York, N.Y.
- 5.Bono, J. L., K. Tilly, B. Stevenson, D. Hogan, and P. Rosa. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144:1033-1044. [DOI] [PubMed] [Google Scholar]
- 6.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caetano-Annolles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-92. [DOI] [PubMed] [Google Scholar]
- 8.Caldas, C., A. Cherqui, A. Pereira, and N. Simões. 2002. Purification and characterization of an extracellular protease from Xenorhabdus nematophila involved in insect immunosuppression. Appl. Environ. Microbiol. 68:1297-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cundell, D., B. Pearce, J. Sandros, A. Naughton, and H. Masure. 1995. Peptide permeases from Streptococcus pneumoniae affect adherence to eukaryotic cells. Infect. Immun. 63:2493-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenno, J. C., M. Tamura, P. M. Hannam, G. W. K. Wong, R. A. Chan, and B. C. McBride. 2000. Identification of a Treponema denticola OppA homologue that binds host proteins present in the subgingival environment. Infect. Immun. 68:1884-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forst, S., and D. Clarke. 2002. Bacteria-nematode symbioses, p. 57-77. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, United Kingdom.
- 12.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 13.Forst, S. A., and N. Tabatabai. 1997. Role of the histidine kinase, EnvZ, in the production of outer membrane proteins in the symbiotic-pathogenic bacterium Xenorhabdus nematophilus. Appl. Environ. Microbiol. 63:962-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodell, E. W., and C. F. Higgins. 1987. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J. Bacteriol. 199:3861-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutnick, D., J. M. Calvo, T. Klopotowski, and B. N. Ames. 1969. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 100:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 17.Jenkinson, H., R. Baker, and G. Tannock. 1996. A binding-lipoprotein-dependent oligopeptide transport system in Streptococcus gordonii essential for uptake of hexa- and heptapeptides. J. Bacteriol. 178:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkinson, H. F. 1992. Adherence, coaggregation, and hydrophobicity of Streptococcus gordonii associated with expression of cell surface lipoproteins. Infect. Immun. 60:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkinson, H. F., and R. A. Easingwood. 1990. Insertional inactivation of the gene encoding a 76-kilodalton cell surface polypeptide in Streptococcus gordonii Challis has a pleiotrophic effect on cell surface composition and properties. Infect. Immun. 58:3689-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornacki, J. A., and D. B. Oliver. 1998. Lyme disease-causing Borrelia species encode multiple lipoproteins homologous to peptide-binding proteins of ABC-type transporters. Infect. Immun. 66:4115-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, E. E., S. Selvan, J. F. Campbell, and R. Gaugler. 1995. Changes in foraging behaviour during the infective stage of entomopathogenic nematodes. Parasitology 110:585-590. [Google Scholar]
- 22.Martens, E. C., K. Heungens, and H. Goodrich-Blair. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J. Bacteriol. 185:3147-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNab, R., and H. Jenkinson. 1998. Altered adherence properties of a Streptococcus gordonii hppA (oligopeptide permease) mutant result from transcriptional effects on cshA adhesin gene expression. Microbiology 144:127-136. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Mitani, D. K., H. K. Kaya, and H. Goodrich-Blair. 2004. Comparative study of the entomopathogenic nematode, Steinernema carpocapsae, reared on mutant and wild-type Xenorhabdus nematophila. Biol. Control 29:382-391. [Google Scholar]
- 26.Park, J. T., D. Raychaudhuri, H. Li, S. Normark, and D. Mengin-Lecreulx. 1998. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate. J. Bacteriol. 180:1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payne, J. W., and C. Gilvarg. 1968. Size restriction on peptide utilization in Escherichia coli. J. Biol. Chem. 243:6291-6299. [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 30.Vivas, E. I., and H. Goodrich-Blair. 2001. Xenorhabdus nematophilus as a model for host-bacterium interactions: rpoS is necessary for mutualism with nematodes. J. Bacteriol. 183:4687-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volgyi, A., A. Fodor, A. Szentirmai, and S. Forst. 1998. Phase variation in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 64:1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, X.-G., B. Lin, J. M. Kidder, S. Telford, and L. T. Hu. 2002. Effects of environmental changes on expression of the oligopeptide permease (opp) genes of Borrelia burgdorferi. J. Bacteriol. 184:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodring, J. L., and H. K. Kaya. 1988. Steinernematid and Heterorhabditid nematodes: a handbook of biology and techniques. Southern cooperative series bulletin 331. Arkansas Agricultural Experiment Station, Fayetteville.
- 34.Xu, J., and R. E. Hurlbert. 1990. Toxicity of irradiated media for Xenorhabdus spp. Appl. Environ. Microbiol. 56:815-818. [DOI] [PMC free article] [PubMed] [Google Scholar]