Abstract
Artisanal and industrial sausages were analyzed for their aerobic, heat-resistant microflora to assess whether new emerging pathogens could be present among Bacillus strains naturally contaminating cured meat products. Sixty-four isolates were characterized by randomly amplified polymorphic DNA (RAPD)-PCR and fluorescent amplified fragment length polymorphism (fAFLP). The biotypes, identified by partial 16S rRNA gene sequence analysis, belonged to Bacillus subtilis, Bacillus pumilus, and Bacillus amyloliquefaciens species. Both RAPD-PCR and fAFLP analyses demonstrated that a high genetic heterogeneity is present in the B. subtilis group even in strains harvested from the same source, making it possible to isolate 56 different biotypes. Moreover, fAFLP analysis made it possible to distinguish B. subtilis from B. pumilus strains. The strains were characterized for their toxigenic potential by molecular, physiological, and immunological techniques. Specific PCR analyses revealed the absence of DNA sequences related to HBL, BcET, NHE, and entFM Bacillus cereus enterotoxins and the enzymes sphingomyelinase Sph and phospholipase PI-PLC in all strains; also, the immunological analyses showed that Bacillus strains did not react with NHE- and HBL-specific antibodies. However, some isolates were found to be positive for hemolytic and lecithinase activity. The absence of toxigenic potential in Bacillus strains from the sausages analyzed indicates that these products can be considered safe under the processing conditions they were produced; however, great care should be taken when the ripening time is shortened, particularly in the case of traditional sausages, which could contain high amounts of Bacillus strains and possibly some B. cereus cells.
Bacillus spp. are common soil inhabitants and may frequently contaminate foods, including dairy products, meats, baby food, rice dishes, vegetables, spices, and cereals. The spore-forming ability of Bacillus spp. promotes the survival of members of this genus during food processing treatments, such as milk or juice pasteurization, and in many instances, in home-cooked meals (12). At least one member of this genus, Bacillus cereus, is a well-known food-poisoning organism that may cause illness due to the fact that it produces either heat-stable emetic toxin (cereulide) usually found in fried and cooked rice and pasta or heat-sensitive diarrheal enterotoxins (HBL, NHE, and BcET) most frequently found in milk and meat products (21). In Asia, the emetic form of B. cereus food poisoning is more commonly reported than the diarrheal type, which is the most frequent one in Europe and North America (20).
Although few papers report the presence of B. cereus in cured meats (10, 37, 50), Bacillus spp. strains are widespread in meat and meat products (5, 43) and in traditional European sausages (17, 18, 39). Bacilli such as Bacillus licheniformis, Bacillus subtilis, Bacillus pumilus, and Bacillus thuringiensis have rarely been linked to incidents of food-borne illnesses (16, 33), and isolates of Bacillus mycoides, B. thuringiensis, Bacillus circulans, Bacillus lentus, B. pumilus, Bacillus polymyxa, and Bacillus carotarum have been found to produce active B. cereus toxins (23). Beattie and Williams (7) demonstrated that most isolates of B. mycoides, B. thuringiensis, B. subtilis, B. lentus, B. circulans, B. licheniformis, and B. laterosporus were positive with the cytotoxicity assay, but only 36% gave positive results with immunological tests, too. Contradictory results were reported for cytotoxicity for B. subtilis and B. pumilus strains (44, 45).
The data regarding dry cured sausages show that Germany, Italy, Spain, and France rank as the biggest producers and consumers of sausage (34). In Italy, the sector of meat-based product processing is represented by more than 3,500 companies, including those turning out artisanal products which continue to hold their ground in the production of traditional Italian foods. In 2001, Italian consumption of charcuterie products was on average 18.5 kg per person (+0.3% versus 2000) according to data available at the Istituto per la Valorizzazione dei Salumi Italiani website (http://www.salumi-italiani.it/ivsi_eng/default.asp).
Although few reports have been made on outbreaks of Bacillus spp. associated with the consumption of fermented meat products (11, 49, 52), the shortening of ripening time, the presence of different levels of gram-positive endospore-forming aerobic bacteria in dry sausages, and the production of toxins by non-B. cereus strains led us to investigate whether new emerging pathogenic Bacillus strains were eventually present in cured sausages.
In this study randomly amplified polymorphic DNA (RAPD)-PCR and fluorescent amplified fragment length polymorphism (fAFLP) analyses were carried out on Bacillus isolates from cured sausages to evaluate the genetic heterogeneity in the Bacillus genus, mainly among known Bacillus anthracis, B. cereus, and B. thuringiensis species (4, 13, 29, 35, 51).
The combined use of molecular, immunological, and physiological techniques for the analysis of genetic determinants and the presence and expression of toxins and virulence factors made it possible to evaluate the potential risk related to the consumption of artisanal and industrial cured sausages containing Bacillus strains.
MATERIALS AND METHODS
Sausage sampling.
Eight samples of cured ready-to-eat sausages, artisanally or industrially processed, were collected from a local market. The industrial sausages were Salame Felino (sample F; Parma, Italy), Salame Ungherese (sample U; Mantua, Italy), Salame Milano (sample M; Milan, Italy), and Spanish Tunel sausage (sample Spa; Girona, Spain). The artisanal sausages Spianata Piccante (sample SP), Salame Lucano (sample SL), Salame Campagnolo (sample C), and Soppressata Calabra (sample SOP) were produced in three regions of Southern Italy (Apulia, Basilicata, and Calabria). Seven sausages contained only minced pork meat, whereas sausage M could contain up to 25% beef. The sausages were coarse grain (samples SOP, SP, C, and SL), medium grain (samples F and SPA), or fine grain (samples M and U). Black pepper was present in six sausages, hot chili pepper was the main spice in sausage SP, and powder from a local pepper cultivar (Capsicum annuum cv. Senise) was present in sausage SL. The curing time varied from 25 days (sample SL) to 3 months (samples SOP, M, and U).
The pHs of the sausages were determined by direct insertion of a solid pH meter. Water activity (aw) was measured with the aw recorder AquaLab, series 3, model TE (Decagon Devices, Inc., Pullman, Wash.).
Microbiological analysis of sausage samples.
The sausage casing was removed aseptically. A 20-g sample from the central portion of each sausage was added to 180 ml of sterile 0.1% (wt/vol) peptone saline solution (0.9% NaCl) and homogenized in a blender (Lab Blender; Seward, London, United Kingdom) at room temperature for 1 to 3 min, depending on the consistency of each sample. All samples were stored at −80°C.
The frozen samples were thawed and serially diluted in peptonate saline solution. The appropriate dilutions were plated in triplicate on Rogosa SL agar for the isolation of lactobacilli, mannitol salt agar (MSA) to make a count of the staphylococci present, and tryptic soy agar (TSA) to isolate the bacilli. In addition, a selective isolation of B. cereus strains was carried out with mannitol-egg yolk-polymyxin (MYP) agar. MSA, MYP, and TSA plates were incubated under aerobic conditions, whereas Rogosa plates underwent incubation under anaerobic conditions (AnaeroGen; Oxoid S.p.A., Garbagnate, Milan, Italy). Colonies were counted after 48 h of growth at 30°C. The counts of heat-resistant cells on MYP and TSA plates were obtained after pasteurization (10 min at 80°C) of 5 ml of cell suspension. After heat treatment, the samples were immediately placed on ice to prevent spore germination.
For each sample, 15 to 20 randomly selected colonies isolated from TSA were analyzed for Gram staining, cell morphology, presence of endospores, and catalase reaction. Sixty-four gram-positive, catalase-positive bacilli endowed with endospores were inoculated in brain heart infusion (BHI) broth, incubated overnight with shaking at 30°C, frozen at −80°C, and used for further characterization. The strains from sample SL were also isolated from raw meat. The strains analyzed in this work are listed in Table 1.
TABLE 1.
Bacterial strains analyzed
| Source | Strain(s) | Species |
|---|---|---|
| Salame Felino | fel55 | B. pumilus |
| fel56 | B. subtilis | |
| Salame Ungherese | ung16, ung17, ung19, ung20, ung21, ung22 | B. pumilus |
| ung18 | B. subtilis | |
| Salame Milano | mil37, mil38, mil42, mil43, mil46, mil47 | B. pumilus |
| mil36, mil39, mil40, mil41, mil44, mil45 | B. subtilis | |
| Spanish sausage | spa1, spa2, spa3, spa4, spa5, spa7, spa8, spa9, spa10, spa11, spa12, spa13, spa14 | B. subtilis |
| Spianata Piccante | spia48 | B. pumilus |
| spia49, spia51, spia53 | B. subtilis | |
| Salame Lucano | tr56a | B. amyloliquefaciens |
| tr51,a tr44a | B. pumilus | |
| tr53,a tr54, trf22, tr50 | B. subtilis | |
| Soppressata Calabra | sop24, sop25, sop26, sop27, sop28, sop29, sop30, sop31, sop33, sop34, sop35 | B. subtilis |
| Dairy isolate | BAC 1 | B. cereus |
| Type strain | DSM 7 | B. amyloliquefaciens |
| DSM 10 | B. subtilis | |
| DSM 12 | B. firmus | |
| DSM 13 | B. licheniformis | |
| DSM 27 | B. pumilus | |
| DSM 32 | B. megaterium | |
| DSM 2046 | B. thuringiensis | |
| DSM 4312 (emetic type) | B. cereus | |
| DSM 4313 (diarrheal type) | B. cereus |
These strains were recovered from raw meat at the beginning of the processing and were not isolated in the ripened sausage.
Rogosa, TSA, and MSA media were obtained from Oxoid, whereas MYP was purchased from Merck S.p.A. (Milan, Italy).
DNA isolation and purification.
Five milliliters of BHI broth was inoculated with a single Bacillus isolate colony, and the cultures were incubated overnight with shaking at 30°C. Bacterial cells were harvested by centrifugation at 1,000 × g for 5 min, and bacterial pellets were rinsed with sterile water. DNA was isolated by using the Wizard genomic DNA purification kit (Promega, Madison, Wis.) according to the protocol provided by the manufacturer. DNA quantity and quality were determined by electrophoresis with known amounts of lambda DNA, marker VI (Roche S.p.a., Milan, Italy), as a standard.
Strain typing analysis. (i) RAPD-PCR.
RAPD-PCR analysis was performed on 64 randomly selected colonies of gram-positive, catalase-positive endospore-forming bacilli by using the two-step RAPD-PCR protocol, as described by Baruzzi et al. (6). Amplification consisted of an initial denaturation at 94°C for 1 min, followed by 20 cycles of 94°C for 30 s, 29°C for 1 min, and 72°C for 1 min and 40 additional cycles of 94°C for 30 s, 55°C for 40 s, and 72°C for 30 s; the annealing and extension of the last 40 cycles were elongated for 1 and 2 s per cycle, respectively. A final extension at 72°C for 5 min was added. Control reaction mixtures lacking DNA template were also included in each experiment. PCR amplification was carried out in a Thermal Cycler 9700 (Perkin-Elmer, Alameda, Calif.). Taq polymerase, deoxynucleoside triphosphates, and DNA molecular weight markers were purchased from Sigma (Milan, Italy), and high-purity salt-free purified oligonucleotides were obtained from MWG Biotech S.r.L. (Florence, Italy). The amplified fragments, separated by electrophoresis on a 2.0% agarose gel, were sized by using Quantity One, version 4.3.1, software (Bio-Rad Laboratories S.r.L., Milan, Italy). Fingerprints from sausage isolates were subjected to similarity analysis, calculating the Dice coefficient, and a dendrogram was obtained after UPGMA (unweighted pair group method with arithmetic means) analysis. The bands subjected to cluster analysis included PCR products ranging from 300 to 2,500 bp in size.
(ii) fAFLP.
fAFLP analysis was carried out by using the AFLP microbial fingerprinting protocol of PE Applied Biosystems (Perkin-Elmer) on the biotypes differentiated by RAPD-PCR analysis and on Bacillus amyloliquefaciens, Bacillus firmus, B. licheniformis, Bacillus megaterium, B. subtilis, B. thuringiensis, and B. cereus reference strains. Briefly, 10 ng of genomic DNA was digested with 5 U of EcoRI and 1 U of MseI and simultaneously ligated (1 U of T4 ligase) with EcoRI and MseI site-specific adapters (55) overnight at room temperature. The enzymes were obtained from New England BioLabs, Inc. (Beverly, Mass.). The preselective PCR was carried out in 20 μl (final volume) with 4 μl of restriction ligation mixture. Five couples of primers with different fluorescent labeled nucleotides at the 3′ end were tested to choose the pair that displayed the highest discriminatory power after selective PCR. Selective PCR was performed in 10 μl (final volume) with 1.5 μl of diluted (1:20 in Tris-EDTA) preselective amplification product, 0.5 pmol of EcoRI adaptor-specific primer (EcoRI+C), and 2.5 pmol of MseI adaptor-specific primer (MseI+CA) fluorescently labeled and 7.5 μl of AFLP core amplification mix. The preselective and selective PCR amplifications were carried out following the AFLP microbial fingerprinting protocol of PE Applied Biosystems in a Thermal Cycler 9700. One microliter of the diluted and denatured amplicons from selective PCR was analyzed in an ABI PRISM 310 genetic analyzer (PE Applied Biosystems, Inc.). To each sample, 1 μl of Genescan-500 (ROX) (PE Applied Biosystems) internal lane standard ranging in size from 35 to 500 bp was added. Fragments were sized by using ABI Genescan, version 2.1, software (PE Applied Biosystems). Electropherograms were visually inspected for polymorphisms. The binary matrix obtained by detection for the presence or absence of peaks, with Genotyper, version 2.1, software, converted into Excel format, was used for the output of a TREE cluster with NTSYS PC software. The Dice coefficient of similarity was computed, and cluster analysis was performed by using the UPGMA algorithm.
For both RAPD-PCR and fAFLP analyses, some strains were replicated to assess the threshold within which two strains were closely related.
Taxonomic identification of strains.
All strains were identified by amplification and sequencing of 16S rDNA, as already described (32); furthermore, three random strains per sample were analyzed for the 16S-23S rRNA spacer region, as described by Berthier and Ehrlich (9).
The DNA sequences were obtained by using an ABI PRISM Big Dye Terminator cycle sequencing kit (PE Applied Biosystems), and both the forward and reverse primers for 16S rDNA were used. The reaction products were analyzed with an ABI PRISM 310 genetic analyzer. Taxonomic strain identification was performed by comparing the sequences from sausage isolates with those present in a Basic BLAST search (2).
Moreover, to confirm B. subtilis and B. pumilus molecular taxonomic identification, the ability of Bacillus isolates to hydrolyze starch was investigated as follows. Strains were streaked onto plates containing agar (15 g liter−1) and wheat flour (8 g liter−1), and the plates were incubated under aerobic conditions at 30°C for 5 days. Then an 8% iodine solution was poured on the plates to detect the presence of a halo around the colonies. After taxonomic identification, isolated strains were stored at −80°C in the Istituto di Scienze delle Produzioni Alimentari bacterial collection and are available on request.
PCR screening of B. cereus genes encoding toxins.
The major B. cereus enterotoxins and virulence factors were investigated by means of PCR analysis in Bacillus strains isolated from sausages.
The enterotoxins searched for were HBL, a three-component hemolysin (L1, L2 [hbl-D/A], and B), NHE, a three-component nonhemolytic enterotoxin (nheB), B. cereus enterotoxin T (bceT), and enterotoxin FM (entFM). In addition to these toxins, two enzymes, sphingomyelinase (sph) and phosphatidylinositol- and phosphatidylcholine-specific phospholipase (piplc) were also investigated. The primers and PCR fragments obtained from the sph, bceT, entFM/S, piplc, nheB, and hbl-D/A genes are listed in Table 2. Primers for HBL were designed along the hbl-D/A gene sequence (GenBank accession number AJ007794) from the B. cereus ATCC 14579 reference strain by using Primer3 online software (46). PCR mixtures of 25 μl contained 20 to 30 ng of genomic DNA, 1 μM concentrations of each primer, 0.2 mM deoxynucleoside triphosphates, and 2.5 U of Taq polymerase (Sigma) in the supplied buffer. All primers were purchased from MWG Biotech S.r.L. Amplification consisted of an initial denaturation at 94°C for 3 min, followed by 40 cycles of 94°C for 25 s, 55°C for 45 s, and 72°C for 2 min and a final extension at 72°C for 5 min. PCR products were analyzed on a 1% (wt/vol) agarose gel stained with 5 μg of ethidium bromide (Sigma) ml−1 and visualized by UV. PCR fragments were sequenced by following the method described above for 16S rRNA sequencing.
TABLE 2.
PCR primers used and virulence factors
| Target gene | Primer name | Primer sequence (5′-3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| hbl-D/A | hblD-f | GGAGCGGTCGTTATTGTTGT | 623 | This study |
| hblA-r | GCCGTATCTCCATTGTTCGT | |||
| nheB | nheB 1500 S | CTATCAGCACTTATGGCAG | 769 | 22 |
| nheB 2269 A | ACTCCTAGCGGTGTTCC | |||
| bceT | ETF | TTACATTACCAGGACGTGCTT | 428 | 1 |
| ETR | TGTTTGTGATTGTAATTCAGG | |||
| entFM | EntA | ATGAAAAAAGTAATTTGCAGG | 1,269 | 3 |
| EntB | TTAGTATGCTTTTGTGTAACC | |||
| sph | Ph1 | CGTGCCGATTTAATTGGGGC | 558 | 25 |
| Ph2 | CAATGTTTTAAACATGGATGCG | |||
| piplc | PC105 | CGCTATCAATGGACCATGG | 569 | 14 |
| PC106 | GGACTATTCCATGCTGTACC |
Immunological assays for B. cereus HBL and NHE toxins.
In vitro enterotoxin detection was assessed for all Bacillus strains isolated by using the Bacillus diarrheal enterotoxin visual immunoassay BDE kit (TECRA International Pty. Ltd., Frenchs Forest, New South Wales, Australia) and the B. cereus enterotoxin-reversed passive latex agglutination kit according to the manufacturer's instructions (Oxoid). The BDE kit makes it possible to detect the 41-kDa subunit of NHE, whereas the B. cereus enterotoxin-reversed passive latex agglutination kit contains antisera for the L2 subunit of HBL.
Determination of hemolytic and lecithinase activities.
Hemolytic activity was determined at 30°C on blood agar plates (Merck) containing 5% sheep blood. B. subtilis DSM 10 and B. pumilus DSM 27 were used as negative controls, and B. cereus DSM 4312 and B. cereus DSM 4313 were used as positive controls. Five microliters of cells grown overnight in BHI broth at 30°C were placed on blood agar plates. The cell-free supernatants of positive strains were also tested for hemolytic activity.
Lecithinase (phosphatidylinositol-specific phospholipase C) activity was determined at 30°C on nutrient agar supplemented with 8% egg yolk emulsion (Oxoid): lecithinase-positive strains produced a halo around the colonies.
Nucleotide sequence accession number.
The partial sequence (1,481 bp) of the 16S rRNA gene of type strain B. pumilus DSM 27 has been submitted to GenBank under accession number AY456263.
RESULTS
Microbiological analysis.
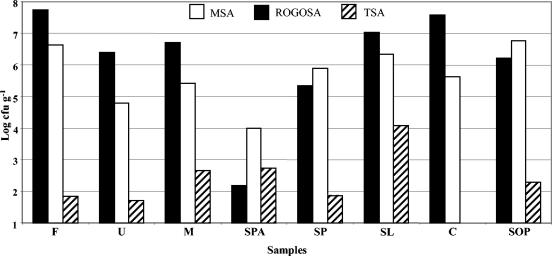
The viable counts of bacteria from Rogosa, MSA, and TSA plates are shown in Fig. 1.
FIG. 1.
Distribution of main bacterial groups in sausage samples. Bacterial counts on Rogosa agar (black bars), MSA (white bars), and after heat treatment (10 min at 80°C), on TSA (striped bars). Industrial samples: F, Salame Felino; U, Salame Ungherese; M, Salame Milano; SPA, Spanish Tunel sausage. Artisanal samples: SP, Spianata Piccante; SL, Salame Lucano; C, Salame Campagnolo; SOP, Soppressata Calabra.
Lactobacillus counts were found to range between 1.50 × 102 and 5.32 × 107 CFU g−1, whereas Staphylococcus counts ranged from 6.15 × 104 to 5.70 × 106 CFU g−1. The presumptive coagulase-negative Staphylococcus colonies counted on MSA plates were all surrounded by reddish purple zones.
After pasteurization (10 min at 80°C), gram-positive aerobic rods, endowed with endospores, were recovered from seven samples. Their amounts varied from 0.50 × 102 to 1.20 × 104 CFU g−1. No Bacillus strains were recovered from sample C. On MYP agar plates, no rough, pink-purple presumptive B. cereus colonies surrounded by dense precipitate were seen, but many yellow colonies were found. Lactobacilli were the dominant microorganisms in all samples except in samples SPA, SP, and SOP, where staphylococci dominated. The lowest number of microorganisms was given by Bacillus isolates in all samples.
The pH values in sausage samples ranged from 4.77 to 5.65, whereas the aw ranged from 0.877 to 0.911.
Strain typing analyses.
RAPD-PCR analysis, performed on 64 gram-positive endospore-forming isolates from TSA plates, demonstrated the presence of 56 different patterns, revealing a wide heterogeneity of fingerprints. The biotypes in each sample ranged from 2 to 13. The RAPD-PCR patterns recovered from different sausage samples were never identical. The reproducibility of the developed RAPD-PCR protocol was tested by separately growing the same strains twice and subjecting the replicates to RAPD-PCR analysis. The replicates of each strain exhibited a Dice similarity coefficient value between 0.85 and 0.93 on the dendrogram.
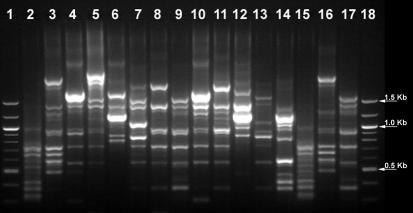
The band pattern varied little between different experiments with the same strain, but individual bands varied greatly in intensity. Faint PCR bands that may lead to faulty cluster analysis were therefore discarded. Each RAPD-PCR pattern contained 3 to 15 bands, varying in size between 250 and 2,500 bp. Figure 2 shows the RAPD-PCR fingerprints of Bacillus biotypes from sample SPA.
FIG. 2.
RAPD-PCR patterns of B. subtilis isolates from sausage SPA. Lanes: 1 and 18, molecular size marker (100-bp DNA ladder; New England Biolabs, Inc.); 2 and 15, spa1; 3 and 16, spa2; 4, spa3; 5, spa4; 6, spa5; 7, spa7; 8, spa8; 9 and 17, spa9; 10, spa10; 11, spa11; 12, spa12; 13, spa13; 14, spa14.
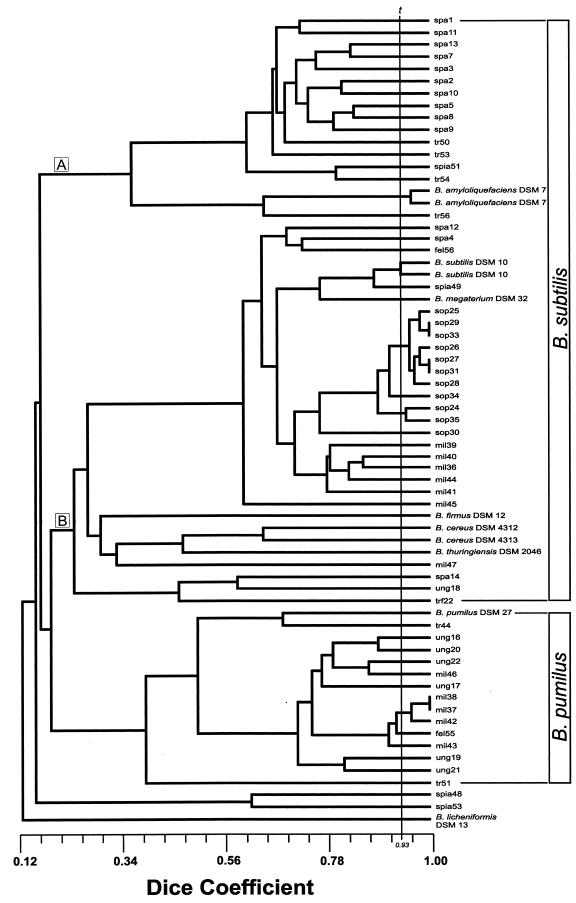
fAFLP analysis, performed on 56 RAPD-PCR biotypes demonstrated the presence of 47 different fingerprints. No identical fAFLP fingerprints were recovered from different sausage samples. fAFLP analysis generated over 30 fragments, sized within 1 bp, and ranging in size from 35 to 500 bp. The result of fAFLP analysis is shown as a dendrogram in Fig. 3. fAFLP analysis made it possible to distinguish between the closely related species B. subtilis and B. pumilus. The B. subtilis cluster displayed two distinct clades. The first group (A) included B. subtilis and B. amyloliquefaciens strains; the second clade (B) included many B. subtilis isolates and the B. subtilis, B. cereus, and B. thuringiensis type strains. Only the B. pumilus mil47 was included in B. subtilis cluster B. B. licheniformis DSM 13, B. subtilis spia53, and B. pumilus spia48 fell outside both the B. subtilis and B. pumilus clusters. Replicates of data generated by B. amyloliquefaciens DSM 7 and B. subtilis DSM 10 type strains exhibited a 0.93 Dice similarity coefficient (Fig. 3, line t). Two or more strains displaying this value or a higher one were considered indistinguishable.
FIG.3.
Dendrogram of Bacillus sausage isolates and type strains based on fAFLP analysis with EcoRI-C and MseI-AC primers based on Dice coefficient and UPGMA analysis. The B. subtilis cluster displayed the distinct A and B clusters. The line labeled “t ” shows the threshold similarity Dice coefficient value based on replicate analysis of B. subtilis strain DSM 10.
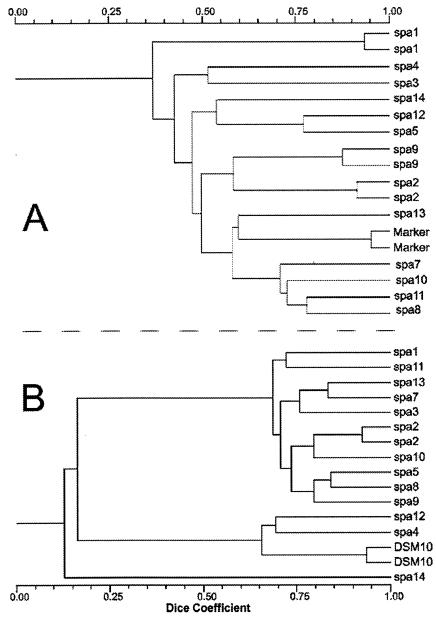
In Fig. 4, the dendrograms from RAPD-PCR and fAFLP analyses of isolates from sample SPA are shown. In this sausage and in the majority of samples, both analyses made it possible to distinguish the same number of biotypes. For sausage sample SOP, seven different RAPD-PCR biotypes showed a Dice similarity coefficient of more than 0.93 by fAFLP analysis, clustering them as a single strain.
FIG. 4.
Comparison of dendrograms of RAPD-PCR (A) and fAFLP (B) fingerprint cluster analysis of isolates from SPA sausage.
Taxonomic identification of strains.
The strains analyzed in this work are listed in Table 1. The taxonomic position of all biotypes was achieved by means of sequence analysis of at least 500 bp of the 5′ region of the 16S rRNA gene.
The analysis of the 16S-23S rDNA spacer region carried out on 20 randomly selected strains from the seven sausage samples confirmed the results obtained by 16S rDNA sequence comparison. The agreement between data from rDNA and spacer region sequence analysis of the 20 strains was considered sufficient to confirm the taxonomic identification of all strains. The percentage of similarity between the rRNA gene sequences of Bacillus isolates and those in the database ranged from 95 to 99%. Also, the assay of starch hydrolysis confirmed that the positive strains belonged to B. subtilis, whereas the negative ones were B. pumilus.
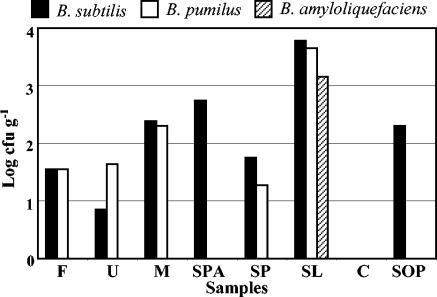
The distribution of Bacillus strains in the sausages is shown in Fig. 5. No Bacillus strains were found in sample C. Strains belonging to B. subtilis were present in the remaining samples. No B. pumilus strains were isolated from samples SPA and SOP. B. amyloliquefaciens was isolated only from sample SL.
FIG. 5.
Relative amounts of B. subtilis, B. pumilus, and B. amyloliquefaciens in industrial (F, U, M, and SPA) and artisanal (SP, SL, C, and SOP) sausage samples.
PCR screening of B. cereus genes encoding toxins.
All 56 strains were assayed for toxins and virulence factors usually present in B. cereus. The presence of genes of the main important toxins responsible for diarrheal food poisoning (HBL, NHE, and BcET) and the entFM toxin were investigated by means of PCR analysis. In addition, two B. cereus enzymes (Sph and PI-PLC), whose role in food-borne disease is not well known, were also studied. The sequence analyses of DNA fragments amplified from the positive control strains (B. cereus type strains DSM 4312 and DSM 4313 and B. cereus BAC1 dairy isolate) showed that they exhibited a high degree of identity (97 to 100%) with the B. cereus virulence genes. Molecular analysis on the B. subtilis, B. pumilus, and B. amyloliquefaciens strains isolated from traditional and industrial cured sausages, performed with primers from B. cereus virulence determinants, showed that all strains were devoid of these enterotoxin- and enzyme-encoding genes.
Immunological assays for B. cereus HBL and NHE toxins.
To complete the toxigenic characterization of the isolated Bacillus strains, even though no positive PCR results were obtained, the presence of HBL and NHE toxins was assessed. B. cereus DSM 4313 (diarrheal type) and B. thuringiensis DSM 2046 were found to be positive for HBL toxins, whereas B. cereus DSM 4312 (emetic type), B. subtilis DSM 10, B. pumilus DSM 27, and B. amyloliquefaciens DSM 7 proved to be negative under the same assay conditions. All strains isolated from sausages were found to be negative for HBL toxins. When strains were analyzed for NHE, the type strains B. cereus DSM 4313 and B. thuringiensis DSM 2046 proved to be positive, whereas all sausage strains were found to be negative.
Physiological tests.
Some sausage strains exhibited hemolytic activity on 5% sheep blood agar plates. In particular, 2 B. subtilis strains and 15 of 16 B. pumilus strains showed weak halos after 48 h of growth at 30°C compared with the halos produced by B. cereus strains, already evident after 12 h of growth. The halos from B. cereus were large and clear, whereas the halos from B. subtilis and B. pumilus were small and greenish, suggesting alpha-hemolysis (results not shown). The isolated strains, which were weakly positive, were further analyzed for the hemolytic activities of their cell-free supernatants. The cell-free supernatants from B. cereus type strains exhibited a hemolytic activity weaker than those developed by B. cereus cells. Among the 17 weakly positive isolated strains, only cell-free supernatants from B. subtilis ung18 and B. subtilis tr50 still showed a weak hemolytic activity. Lecithinase activity was developed by B. pumilus mil47 and 26 B. subtilis strains (67%).
DISCUSSION
The risk of food-borne diseases due to bacilli other than B. cereus can be considered low, although some papers report the presence of B. cereus virulence-like factors in strains belonging to several Bacillus spp. (33, 42). Toxins from B. thuringiensis similar to those from B. cereus were frequently found in foods (15) and in the environment (41). This could, however, be due to the fact that B. thuringiensis is a member of the B. cereus group. To our knowledge, no information is available on the risk related to the consumption of cured meat products containing Bacillus spp. other than B. cereus.
In many European countries, traditional dry sausages are still produced without selected starters, using different spices and ingredients, and cured for at least 2 months, following traditional processing methods without taking any hygienic measures. Some authors focused on the survival of pathogenic bacteria in meat products cured for long or brief periods of time (18, 28), reaching the conclusion that long ripening helps to lower the risk of food-borne infections caused by cured sausages. In the last few years, a shortening of ripening time was introduced in sausage production with the aim of increasing the profit margin and the competitiveness of the end product. The studies carried out to reach this goal included ripening at elevated temperatures, the use of genetically modified starter bacteria, and the addition of lipase and/or protease to meats (19).
Although limited information is available on outbreaks of Bacillus spp. associated with the consumption of fermented meat products (11, 49, 52), the shortening of ripening time, more and more often adopted in both industrial and artisanal products, prompted us to carry out studies on the toxigenic potential of bacilli naturally contaminating sausages.
In the sausages analyzed, no B. cereus strains were isolated and only one product (Salame Lucano) presented more than 103 CFU of gram-positive aerobic heat-resistant cells g−1. The isolated strains belonged mainly to B. pumilus and B. subtilis. These species are usually considered safe, and in some cases, they are used as probiotics in foods or in pharmaceutical preparations (26, 47). However, Turnbull (53) reported that B. subtilis, B. pumilus, and B. licheniformis have been associated with a range of clinical conditions, food spoilage, and incidents of food-borne gastroenteritis, although these infections were found in patients in debilitated immune states according to data available at the U.S. Environmental Protection Agency website (http://www.epa.gov/biotech_rule/pdf/fra009.pdf) (54).
Recent studies deal with the genetic diversity of strains of B. subtilis (30, 36, 38, 48, 56), and the two closely related species B. licheniformis and B. pumilus have been placed in the B. subtilis group.
To our knowledge, the present study is the first to investigate the genetic heterogeneity of B. subtilis group strains from cured meat products. The analyses carried out by means of RAPD-PCR and fAFLP indicated that there was a high degree of genetic heterogeneity in the Bacillus strains from sausages. In some samples, more than 10 different strains were isolated, picking up no more than 15 distinct colonies. The high numbers of Bacillus strains found may be due to the different sources of contamination (fats, meats, salt, and spices, etc.). The results of fAFLP analyses substantially agreed with those obtained by RAPD analyses. In the case of the SOP sample, seven RAPD biotypes were clustered together in fAFLP analysis. For these isolates, RAPD fingerprints were constituted by 4 to 5 bands similar in size, and thus, the calculation of similarity with the Dice coefficient could be misleading (data not shown). fAFLP analysis made it possible to cluster strains belonging to the same species within the B. subtilis group in accordance with similar results obtained with strains belonging to the B. cereus group (31, 35, 51).
The lack of any positive results from the PCR analyses of B. cereus virulence factors and from the enzyme-linked immunosorbent assays of HBL and NHE toxins indicates that B. subtilis, B. pumilus, and B. amyloliquefaciens strains isolated from the sausages analyzed do not code for the known toxins of B. cereus.
When hemolytic activity was assayed, only rinsed cells from some strains produced weak halos, and only the cell-free supernatants from B. subtilis ung18 and B. subtilis tr50 still proved positive, as did the cell-free supernatants from B. cereus type strains.
Lecithinase activity was also developed by many B. subtilis strains, although PCR fragments related to phosphatidylinositol- and phosphatidylcholine-specific phospholipase (pipIC gene) were not amplified.
It was previously demonstrated that some hemolytic activities of Bacillus strains could probably be due to the proteolytic activity usually present in B. subtilis strains (40).
Lacking other results, these hemolytic and lecithinase activities could be considered normal metabolisms in cells from the food matrix, although more in-depth studies should be carried out to understand whether new potential virulence factors are expressed in B. pumilus and B. subtilis.
The preliminary results reported in this paper are reassuring with regard to the potential risk due to the presence of Bacillus strains in both industrial and traditional cured sausages. In any case, some considerations need to be made. It should be noted that Bacillus strains are usually counted after pasteurization. In this way, only spores are counted, and therefore, the total amount of Bacillus cells is unknown and their potential risk could be underestimated. Some information is available about the ability of Bacillus spores to germinate during sausage ripening (8, 24, 27); therefore, further studies are needed to understand which processing parameters can stimulate or repress spore germination and cell replication.
Since B. cereus and other toxigenic bacilli could derive from different sources (fats, meats, and spices, etc.) and their presence cannot be ruled out, thorough control measures should be applied for Bacillus strains when new food technologies or new ingredients are adopted.
Great care should be taken when the ripening time of traditional sausages is shortened, particularly when other sanitary precautions, such as the addition of starter cultures and bacteriocins, are not taken, because they are considered foreign to their nature. In addition, further in-depth investigations are required to gain an insight into the behavior of cells and spores under different chemical and physical conditions present during sausage curing and to understand the role played by bacilli in these artisanal meat products.
The data obtained and the above considerations lead one to think that the potential risk from Bacillus in cured meat products could derive from gross errors in the processing and curing of sausages, which may have particularly harmful effects on children or elderly people and patients in a state of immune deficiency.
Acknowledgments
We thank G. Stea for technical assistance in DNA sequencing and fAFLP analysis.
REFERENCES
- 1.Agata, N., M. Ohta, Y. Arakawa, and M. Mori. 1995. The bceT gene of Bacillus cereus encodes an enterotoxic protein. Microbiology 141:983-988. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano, S.-I., Y. Nukumizu, H. Bando, T. Hzuka, and T. Yamamoto. 1997. Cloning of novel enterotoxin genes from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 63:1054-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ash, C., J. A. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 5.Asplund, K., E. Nurmi, P. Hill, and J. Hirn. 1988. The inhibition of the growth of Bacillus cereus in liver sausage. Int. J. Food Microbiol. 7:349-352. [DOI] [PubMed] [Google Scholar]
- 6.Baruzzi, F., M. Morea, A. Matarante, and P. S. Cocconcelli. 2000. Changes in the Lactobacillus community during Ricotta forte cheese natural fermentation. J. Appl. Microbiol. 89:807-814. [DOI] [PubMed] [Google Scholar]
- 7.Beattie, S. H., and A. G. Williams. 1999. Detection of toxigenic strains of Bacillus cereus and other Bacillus spp. with an improved cytotoxicity assay. Lett. Appl. Microbiol. 28:221-225. [DOI] [PubMed] [Google Scholar]
- 8.Bell, R. G., and K. M. de Lacy. 1984. Influence of NaCl, NaNO2 and oxygen on the germination and growth of Bacillus licheniformis, a spoilage organism of chub-packed luncheon meat. J. Appl. Bacteriol. 57:523-530. [DOI] [PubMed] [Google Scholar]
- 9.Berthier, F., and S. D. Ehrlich. 1998. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett. 161:97-106. [DOI] [PubMed] [Google Scholar]
- 10.Borch, E., and P. Arinder. 2002. Bacteriological safety issues in red meat and ready-to-eat meat products, as well as control measures. Meat Sci. 62:381-390. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2000. Human ingestion of Bacillus anthracis-contaminated meat. Morb. Mortal. Wkly. Rep. 49:813-816. [Google Scholar]
- 12.Christiansson, A., J. Bertilsson, and B. Svensson. 1999. Bacillus cereus spores in raw milk: factors affecting the contamination of milk during the grazing period. J. Dairy Sci. 82:305-314. [DOI] [PubMed] [Google Scholar]
- 13.Daffonchio, D., A. Cherif, L. Brusetti, A. Rizzi, D. Mora, A. Boudabous, and S. Borin. 2003. Nature of polymorphisms in 16S-23S rRNA gene intergenic transcribed spacer fingerprinting of Bacillus and related genera. Appl. Environ. Microbiol. 69:5128-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damgaard, P. H., C. S. Jacobson, and J. Sorensen. 1996. Development and application of a primer set for specific detection of Bacillus thuringiensis and Bacillus cereus in soil using magnetic capture hybridization and PCR amplification. Syst. Appl. Microbiol. 19:436-441. [Google Scholar]
- 15.Damgaard, P. H., H. D. Larsen, B. M. Hansen, J. Bresciani, and K. Jorgensen. 1996. Enterotoxin-producing strains of Bacillus thuringiensis isolated from food. Lett. Appl. Microbiol. 23:146-150. [DOI] [PubMed] [Google Scholar]
- 16.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Encinas, J. P., J. Sanz-Gomez, M. L. García-López, M. R. García-Armesto, and A. Otero. 1996. Evaluation of different systems for the identification of Bacillus strains isolated from Spanish fermented sausages. Meat Sci. 42:127-131. [DOI] [PubMed] [Google Scholar]
- 18.Encinas, J. P., J. J. Sanz, M. L. García-Lopez, and A. Otero. 1999. Behaviour of Listeria spp. in naturally contaminated chorizo (Spanish fermented sausage). Int. J. Food Microbiol. 46:167-171. [DOI] [PubMed] [Google Scholar]
- 19.Fernández, M., J. A. Ordóñez, J. M. Bruna, B. Herranz, and L. de la Hoz. 2000. Accelerated ripening of dry fermented sausages. Trends Food Sci. Tech. 11:201-209. [Google Scholar]
- 20.Granum, P. E. 2001. Bacillus cereus, p. 373-381. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. ASM Press, Washington, D.C.
- 21.Granum, P. E., and T. Lund. 1997. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157:223-228. [DOI] [PubMed] [Google Scholar]
- 22.Granum, P. E., K. O'Sullivan, and T. Lund. 1999. The sequence of the non-hemolytic enterotoxin operon from Bacillus cereus. FEMS Microbiol. Lett. 177:225-229. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths, M. W. 1990. Toxin production by psychrotrophic Bacillus spp. present in milk. J. Food Prot. 53:790-792. [DOI] [PubMed] [Google Scholar]
- 24.Grohs, B. M., and B. Kunz. 1999. Antimicrobial effect of spices on sausage-spoiling microorganisms using a model medium for sausage type frankfurter. Adv. Food Sci. 21:128-135. [Google Scholar]
- 25.Hisieh, Y. M., S. J. Sheu, Y. L. Chen, and H. Y. Tsen. 1999. Enterotoxigenic profiles and polymerase chain reaction detection of Bacillus cereus group cells and B. cereus strains from food-borne outbreaks. J. Appl. Microbiol. 87:481-490. [DOI] [PubMed] [Google Scholar]
- 26.Hosoi, T., R. Hirose, S. Saegusa, A. Ametani, K. Kiuchi, and S. Kaminogawa. 2003. Cytokine responses of human intestinal epithelial-like Caco-2 cells to the nonpathogenic bacterium Bacillus subtilis (natto). Int. J. Food Microbiol. 82:255-264. [DOI] [PubMed] [Google Scholar]
- 27.Houben, J. H., and B. Krol. 1989. Effect of citric acid, citrate and slight aw decreases on the bacteriological stability of Hague liver sausage. Meat Sci. 24:163-176. [DOI] [PubMed] [Google Scholar]
- 28.Incze, K. 1998. Dry fermented sausages. Meat Sci. 49:169-177. [PubMed] [Google Scholar]
- 29.Jackson, P. J., K. K. Hill, M. T. Laker, L. O. Ticknor, and P. Keim. 1999. Genetic comparison of Bacillus anthracis and its close relatives using amplified fragment length polymorphism and polymerase chain reaction analysis. J. Appl. Microbiol. 87:263-269. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, Y. A., M. Nagpal, M. T. Krahmer, K. F. Fox, and A. Fox. 2001. Precise molecular weight determination of PCR products of the rRNA intergenic spacer region using electrospray quadrupole mass spectrometry for differentiation of B. subtilis and B. atrophaeus, closely related species of bacilli. J. Microbiol. Methods 40:241-254. [DOI] [PubMed] [Google Scholar]
- 31.Keim, P., A. Kalif, J. Schupp, K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. Hugh-Jones, C. R. Kuske, and P. J. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klijn, N., A. H. Weerkamp, and W. M. de Vos. 1995. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl. Environ. Microbiol. 61:788-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer, J. M., and R. J. Gilbert. 1989. Bacillus cereus and other Bacillus species, p. 21-70. In M. P. Doyle (ed.), Foodborne bacteria pathogens. Marcel Dekker, New York, N.Y.
- 34.Lücke, F. K. 1998. Fermented sausages, p. 441-483. In B. J. B. Wood (ed.), Microbiology of fermented foods, 2nd ed., vol. 2. Blackie Academic & Professional, London, United Kingdom.
- 35.Manzano, M., L. Cocolin, C. Cantoni, and G. Comi. 2003. Bacillus cereus, Bacillus thuringiensis and Bacillus mycoides differentiation using a PCR-RE technique. Int. J. Food Microbiol. 81:249-254. [DOI] [PubMed] [Google Scholar]
- 36.Marten, P., K. Smalla, and G. Berg. 2000. Genotypic and phenotypic differentiation of an antifungal biocontrol strain belonging to Bacillus subtilis. J. Appl. Microbiol. 89:463-471. [DOI] [PubMed] [Google Scholar]
- 37.Nortje, G. L., S. M. Vorster, R. P. Greebe, and P. L. Steyn. 1999. Occurrence of Bacillus cereus and Yersinia enterocolitica in South African retail meats. Food Microbiol. 16:213-217. [Google Scholar]
- 38.Ouoba, L. I., B. Diawara, W. Amoa-Awua, A. S. Traore, and P. L. Moller. 2004. Genotyping of starter cultures of Bacillus subtilis and Bacillus pumilus for fermentation of African locust bean (Parkia biglobosa) to produce Soumbala. Int. J. Food Microbiol. 90:197-205. [DOI] [PubMed] [Google Scholar]
- 39.Palumbo, S. A., A. I. Rivenburgh, J. L. Smith, and J. C. Kissinger. 1975. Identification of Bacillus subtilis from sausage products and spices. J. Appl. Bacteriol. 38:99-105. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen, P. B., M. E. Bjøornvad, M. D. Rasmussen, and J. N. Petersen. 2002. Cytotoxic potential of industrial strains of Bacillus sp. Regul. Toxicol. Pharm. 36:155-161. [DOI] [PubMed] [Google Scholar]
- 41.Perani, M., A. H. Bishop, and A. Vaid. 1998. Prevalence of β-exotoxin, diarrheal toxin and specific δ-toxin in natural isolates of Bacillus thuringiensis. FEMS Microbiol. Lett. 160:55-60. [DOI] [PubMed] [Google Scholar]
- 42.Phelps, R. J., and J. L. McKillip. 2002. Enterotoxin production in natural isolates of Bacillaceae outside the Bacillus cereus group. Appl. Environ. Microbiol. 68:3147-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodel, W., and F. K. Lucke. 1990. Effect of redox potential on Bacillus subtilis and Bacillus licheniformis in broth and in pasteurized sausage mixtures. Int. J. Food Microbiol. 10:291-301. [DOI] [PubMed] [Google Scholar]
- 44.Rowan, N. J., K. Deans, J. G. Anderson, C. G. Gemmel, I. S. Hunter, and T. Chaithong. 2001. Putative virulence factor expression by clinical and food isolates of Bacillus spp. after growth in reconstituted infant milk formulae. Appl. Environ. Microbiol. 67:3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowan, N. J., G. Caldow, C. G. Gemmell, and I. S. Hunter. 2003. Production of diarrheal enterotoxins and other potential virulence factors by veterinary isolates of Bacillus species associated with nongastrointestinal infections. Appl. Environ. Microbiol. 69:2372-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 47.Sanders, M. E., L. Morelli, and T. A. Tompkins. 2003. Sporeformers as human probiotics: Bacillus, Sporolactobacillus, and Brevibacillus. Compr. Rev. Food Sci. Food Safety 2:101-110. [DOI] [PubMed] [Google Scholar]
- 48.Shaver, Y. J., M. L. Nagpal, K. F. Fox, R. Rudner, and A. Fox. 2001. Variation in 16S-23S rRNA intergenic spacer regions among Bacillus subtilis 168 isolates. Mol. Microbiol. 42:101-109. [DOI] [PubMed] [Google Scholar]
- 49.Singh, D. K., K. G. Narayan, and M. K. Gupta. 1992. Mechanisms of Bacillus cereus enteropathy. Indian J. Exp. Biol. 30:324-326. [PubMed] [Google Scholar]
- 50.Te Giffel, M. C., R. R. Beumer, S. Leijendekkers, and F. M. Rombouts. 1996. Incidence of Bacillus cereus and Bacillus subtilis in foods in The Netherlands. Food Microbiol. 13:53-58. [Google Scholar]
- 51.Ticknor, L. O., A.-B. Kolstø, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trinchese, N. M., L. Carraciuolo, E. Giffoni, and M. G. Panico. 2000. An outbreak of food poisoning caused by Bacillus cereus in the province of Naples: epidemiological investigation and case-control study. Ig. Mod. 113:335-346. [Google Scholar]
- 53.Turnbull, P. 1997. The role of Bacillus genus in infection. Culture 18:5-8. [Google Scholar]
- 54.U.S. Environmental Protection Agency. 1997. Final risk assessment of Bacillus subtilis. Item no. 3177. U.S. Environmental Protection Agency, Washington, D.C.
- 55.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu, D., and J. C. Côté. 2003. Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 3′ end 16S rDNA and 5′ end 16S-23S ITS nucleotide sequences. Int. J. Syst. Evol. Microbiol. 53:695-704. [DOI] [PubMed] [Google Scholar]