Abstract
Quorum sensing is a cell population-density dependent regulatory system which in gram-negative bacteria often involves the production and detection of N-acyl homoserine lactones (AHLs). Some Pseudomonas putida strains have been reported to produce AHLs, and one quorum-sensing locus has been identified. However, it appears that the majority of strains do not produce AHLs. In this study we report the identification and regulation of the AHL-dependent system of rhizosphere P. putida WCS358. This system is identical to the recently identified system of P. putida strain IsoF and very similar to the las system of Pseudomonas aeruginosa. It is composed of three genes, the luxI family member ppuI, the putative repressor rsaL, and the luxR family member ppuR. A genomic ppuR::Tn5 mutant of strain WCS358 was identified by its inability to produce AHLs when it was cross-streaked in close proximity to an AHL biosensor, whereas an rsaL::Tn5 genomic mutant was identified by its ability to overproduce AHL molecules. Using transcriptional promoter fusions, we studied expression profiles of the rsaL, ppuI, and ppuR promoters in various genetic backgrounds. At the onset of the stationary phase, the autoinducer synthase ppuI gene expression is under positive regulation by PpuR-AHL and under negative regulation by RsaL, indicating that the molecules could be in competition for binding at the ppuI promoter. In genomic rsaL::Tn5 mutants ppuI expression and production of AHL levels increased dramatically; however, both processes were still under growth phase regulation, indicating that RsaL is not involved in repressing AHL production at low cell densities. The roles of the global response regulator GacA and the stationary-phase sigma factor RpoS in the regulation of the AHL system at the onset of the stationary phase were also investigated. The P. putida WCS358 gacA gene was cloned and inactivated in the genome. It was determined that the three global regulatory systems are closely linked, with quorum sensing and RpoS regulating each other and GacA positively regulating ppuI expression. Studies of the regulation of AHL quorum-sensing systems have lagged behind other studies and are important for understanding how these systems are integrated into the overall growth phase and metabolic status of the cells.
Global regulation of gene expression in the fluorescent pseudomonads has recently been the subject of extensive investigation. Global regulatory systems are involved in regulation of important traits, in virulence in Pseudomonas aeruginosa, and in plant-bacterium interactions in beneficial strains of Pseudomonas putida and Pseudomonas fluorescens. Three important examples of such global regulatory systems are the GacA-GacS two-component system, the stress and stationary-phase sigma factor RpoS, and the cell population density-dependent system known as quorum sensing (reference 33 and references therein). The global two-component GacA-GacS system is well conserved in a variety of gram-negative bacteria and has been implicated in the regulation of secondary metabolites and virulence factors in Pseudomonas (15). The stationary-phase alternative sigma factor RpoS (also known as σs or σ38) has been shown to be a global regulator in P. aeruginosa, P. putida, and P. fluorescens and to be involved in stress adaptation to regulate virulence factors and secondary metabolites (references 27, and 33 and references therein). Regulation by quorum sensing relies on autoinducer molecules that accumulate in the medium and allows the cells to respond, via transcriptional regulation of target genes, to their own population density. In gram-negative bacteria, N-acyl homoserine lactone (AHL) autoinducers to date appear to be the most commonly used autoinducers, and they are most commonly produced by an autoinducer synthase belonging to the LuxI protein family. A transcriptional regulator belonging to the LuxR family then forms a complex with the cognate AHL at high threshold levels and affects the transcriptional activity of target genes. The AHLs produced by different bacterial species are believed to be freely diffusible across the cell envelope and to differ in the length and structure of the acyl chain. AHL-dependent quorum sensing has been extensively studied in P. aeruginosa, in which two hierarchically organized systems [the LasR/I and RhlR/I systems responsible for the production of and response to N-(3-oxododecanoyl)-AHL (3-oxo-C12-AHL) and N-butyryl-AHL, respectively] regulate biofilm formation and virulence gene expression (28, 29). AHL-dependent regulation has been reported to be present in some strains of P. fluorescens and P. putida; however, studies regarding their regulons have yet to be performed (11, 32).
From various studies with different Pseudomonas species there is evidence linking AHL-dependent quorum sensing, the GacA-GacS two-component system, and the RpoS response. Recently, the RpoS and quorum-sensing regulons in P. aeruginosa have been deciphered by transcriptome analysis, which revealed that both of these regulons control a very large set of genes and that there is considerable overlap (27, 28). Some of this overlap is thought to be due to RpoS and quorum sensing mildly cross-regulating each other, most probably resulting in indirect activation of genes in the stationary phase at high cell densities. Previous reports have shown that in P. aeruginosa, rpoS expression is positively regulated by the Las and Rhl quorum-sensing systems (19); however, a more recent study indicated that RpoS negatively regulates rhl expression (36). According to a model that has been proposed, both of these effects can contribute to the regulation of genes by RpoS and quorum sensing (27). Interestingly, a link between quorum sensing and RpoS has also been observed in Ralstonia solanacearum (13), indicating that the connection between the two global regulatory systems, which often regulate genes under the same environmental conditions, could be widespread in bacteria.
The GacA-GacS system is a global two-component system in which GacS is the sensor kinase, which perceives a signal that has not been identified yet, and GacA is the response regulator. This system has been found in numerous bacteria and has been shown to control an array of phenotypes, including plant growth-promoting ability, pathogenicity, production of secondary metabolites, biofilm formation, and secreted enzymes and proteins (15). In P. aeruginosa, GacA has been implicated in positive regulation of N-butyryl-AHL production through modulation of rhlI transcription and thus has an indirect influence on quorum-sensing-controlled loci (24, 25). A similar effect of GacA on quorum sensing also occurs in Pseudomonas syringae since in a gacA mutant there are lower levels of AHLs and ahlI/R mRNA transcripts (7). In P. fluorescens GacA has also been reported to positively influence the transcription of rpoS and σs accumulation (35). A similar positive effect on RpoS regulation by GacA has been observed in Azotobacter vinelandii (6), indicating that this regulatory cascade could be a general phenomenon in bacteria.
Evidence linking the GacA-GacS system, quorum sensing, and RpoS is accumulating; however, some of the data are contradictory, the precise molecular mechanisms still remain far from clear, and there have been no studies on the cross-regulation of the three global regulators in the same bacterial species. In this study we identified and studied the regulation of the AHL-quorum-sensing system of the rhizosphere-colonizing plant growth-promoting organism P. putida WCS358. This system is identical to the recently described system of P. putida strain IsoF (32). It is composed of the luxR family member ppuR, the luxI member ppuI, and the negative regulator rsaL. Here we present data showing that the quorum-sensing system of P. putida is under strong negative autoregulation by the putative RsaL repressor and under positive regulation by PpuR. In addition, we show that there is a link between the system with GacA and RpoS, demonstrating that the three global regulatory controls are intimately connected in response to the stationary phase. AHL-dependent quorum sensing has a positive effect on rpoS transcription; RpoS has a negative effect on quorum sensing, while GacA has a positive effect. A model of how these three global regulatory systems are organized is presented below.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The P. putida bacterial strains and plasmids used in this study are listed in Table 1. P. putida WCS358 is a plant growth-promoting strain isolated from the rhizosphere of potato roots. Chromobacterium violaceum CVO26 is a double mini-Tn5 mutant derived from ATCC 31532; this mutant is nonpigmented, and production of a purple pigment can be induced by providing exogenous AHL inducer molecules (21). Escherichia coli DH5α (26) and C. violaceum CVO26 were grown in Luria-Bertani (LB) medium (22) at 37 and 30°C, respectively; P. putida WCS358 was grown in LB medium or M9 minimal medium (26) supplemented with 0.2% citric acid as a carbon source at 30°C. The following antibiotic concentrations were used: ampicillin, 100 μg/ml; kanamycin, 100 μg/ml; nalidixic acid, 25 μg/ml; tetracycline, 10 μg/ml (E. coli) or 40 μg/ml (Pseudomonas); chloramphenicol, 25 μg/ml (E. coli) or 250 μg/ml (Pseudomonas); and gentamicin, 10 μg/ml (E. coli) or 40 μg/ml (Pseudomonas). Transcriptional fusion plasmids for the various gene promoters based on the pMP220 promoter probe vector were constructed as follows. The 732-bp fragment containing the gacA promoter region was amplified by PCR by using Vent DNA polymerase (New England Biolabs) according to the instructions of the supplier, genomic DNA of P. putida WCS358 as the template, and oligonucleotides GacK and GacX (Table 1), and it was cloned in pMOSBlue (Amersham Biosciences, Amersham, United Kingdom), yielding pMOP2, and verified by DNA sequencing (GATC Biotech, Constance, Germany). The gacA promoter was then removed as a KpnI-XbaI fragment and cloned in pMP220, yielding pGAC220. Similarly, the ppuR promoter was amplified as a 190-bp fragment by using oligonucleotides PRRE and PRRX and cloned in pMOSBlue, yielding pMOP3. The ppuR promoter was then removed as an EcoRI-XbaI fragment and cloned in pMP220, yielding pPUR220. The ppuI promoter was amplified as a 162-bp fragment by using oligonucleotides PRIE and PRIX and cloned in pMOSBlue, yielding pMOP1. The ppuI promoter was then removed as an EcoRI-XbaI fragment and cloned in pMP220, yielding pPUI220. The rsaL promoter was amplified as a 162-bp fragment by using oligonucleotides PRIX and PRAL and cloned in pMOSBlue, yielding pMOP4. The rsaL promoter was then removed as an SphI-XbaI fragment and cloned in pMP220, yielding pRSA220. The ppuI gene was cloned in expression vector pQE30 as follows: the ppuI gene was PCR amplified by using oligonucleotides QEIS and QEIE and pBQS1 as the DNA template and cloned as a BamHI-KpnI fragment in pQE30, yielding pQSH1.
TABLE 1.
Plasmids, P. putida strains, and oligonucleotides used
| Strain, plasmid(s), or oligonucleotide | Characteristics or sequence | Reference or source |
|---|---|---|
| Strains | ||
| P. putida WCS358 | Wild type | 14 |
| P. putida M17 | psrA178::Tn5 of WCS358, Kmr | 18 |
| P. putida MKO1 | rpoS880::Tn5 of WCS358, Kmr | 17 |
| P. putida IBE1 | gacA400::Tn5 of WCS358, Kmr | This study |
| P. putida IBE2 | ppuR1793::Tn5 of WCS358, Kmr | This study |
| P. putida IBE3 | rsaL1640::Tn5 of WCS358, Kmr | This study |
| Plasmids | ||
| pMOSBlue | Cloning vector, Ampr | Amersham-Pharmacia |
| pBluescriptKS | Cloning vector, Ampr | Stratagene |
| pQE30 | Expression vector | Qiagen |
| pRK2013 | Kmr Tra+ Mob+ ColE1 replicon | 12 |
| pPH1JI | IncP1, Gmr | 8 |
| pMP220 | Promoter probe vector, IncP1, Tcr | 30 |
| pMP190 | Promoter probe vector, IncQ, Cmr | 30 |
| pMP77 | Promoter probe vector, IncQ, Cmr | 30 |
| pH3.5 | rpoS gene of strain WCS358 in pBluescriptKS | 17 |
| pSCR1 | lacZ-based AHL biosensor based on cep system | 1 |
| pGAC210 | pMOSblue containing 210-bp P. putida WCS358 gacA | This study |
| pRPO220B | rpoS promoter in pMP220 | 17 |
| pCOS67 | pLAFR3 containing WCS358 DNA | This study |
| pBIB27 | pBluescript containing 2.4-kb KpnI fragment from pCOS67 | This study |
| pGAC27 | pMP220 containing 2.4-kb KpnI fragment from pCOS67 | This study |
| pGAC275 | pGAC27 with Tn5 insertion in gacA | This study |
| pMOS15, pMOS49, pMOS50, pMOS133 | pMOSblue containing Tn5 WCS358 DNA | This study |
| pMOS2B, pMOS3C | pMOSblue containing Tn5 WCS358 DNA | This study |
| pBQS1 | pBluescriptKS containing 2.6-kb KpnI-SalI fragment of WCS358 | This study |
| pMQS1 | pMP190 containing 2.6-kb KpnI-SalI fragment of WCS358 | This study |
| pQSH1 | ppul cloned in expression vector pQE30 | This study |
| pRPO770 | pMP77 containing 3.5-Kb HindIII fragment from pH3.5 | This study |
| pPUI220 | ppul promoter cloned in pMP220 | This study |
| pGAC220 | gacA promoter cloned in pMP220 | This study |
| pPUR220 | ppuR promoter cloned in pMP220 | This study |
| pRSA220 | rsaL promoter cloned in pMP220 | This study |
| pMOP1 | ppul promoter cloned in pMOSblue | This study |
| pMOP2 | gacA promoter cloned in pMOSblue | This study |
| pMOP3 | ppuR promoter cloned in pMOSblue | This study |
| pMOP4 | rsaL promoter cloned in pMOSblue | This study |
| Oligonucleotides | ||
| GAC1 | 5′-ATCGGCGGCCTGGAGGC-3′ | This study |
| GAC2 | 5′-TGCTGGGCGATCTGCGGGCT-3′ | |
| GACK | 5′-GGGTACCGAGGATGAATGCTTCGTGCG-3′ | This study |
| GACX | 5′-CTCTAGATGTCGGGATGGCTGCGCAAC-3′ | |
| PRRE | 5′-GGAATTCGTTGTGGCGGGTTGGGCA-3′ | This study |
| PRRX | 5′-CTCTAGATCATTCCCTTCCTCGTAG-3′ | |
| PRIE | 5′-GGAATTCAGCTTCATCGGGAATCAATC-3′ | This study |
| PRIX | 5′-CTCTAGACAATGGAGATAAGCATAGG-3′ | |
| PRAL | 5′-ATGCATGCAGCTTCATCGGGAATCAATC-3′ | This study |
| QEIS | 5′-CGGGATCCATGCTTATCCATTGATCGC-3′ | This study |
| QEIE | 5′-GGGGTACCTATTCAATGGCAGTTATTCTT-3′ |
Recombinant DNA techniques.
DNA manipulations, including digestion with restriction enzymes, agarose gel electrophoresis, purification of DNA fragments, ligation with T4 ligase, end filling with the Klenow enzyme, hybridization, radioactive labeling by random priming, and transformation of E. coli, were performed as described previously (26). Southern hybridizations were performed by using N+Hybond membranes (Amersham Biosciences); plasmids were purified by using Jet star columns (Genomed GmbH, Löhne, Germany) or by the alkaline lysis method (5); and total DNA from Pseudomonas was isolated by Sarkosyl-pronase lysis as described previously (4). Triparental matings between E. coli and P. putida were carried out with the helper strain E. coli DH5α(pRK2013). The DNA sequences flanking the transposon insertions in genomic mutants P. putida IBE2 and IBE3 were determined by using an arbitrary PCR technique as previously described (23).
Cloning and construction of the genomic gacA null mutant of P. putida WCS358.
Part of the gacA gene of P. putida WCS358 was amplified by using genomic DNA as the template and oligonucleotides GAC1 and GAC2 designed from conserved regions of closely related P. putida KT2440 and P. aeruginosa gacA sequences. This resulted in amplification of a 210-bp fragment, which was cloned in pMOSBlue, yielding pGAC210. This fragment was then used as a probe with a pLAFR3-based cosmid library of P. putida WCS358. One cosmid, designated pCOS67, was identified, and the gacA gene was localized in a 2.4-kb KpnI fragment, which was cloned in the corresponding sites in pBluescript KS, yielding pBIB27. The 2.4-kb KpnI fragment was also cloned in pMP220, yielding pGAC27, which was used in a Tn5 mutagenesis procedure in which HB101::Tn5 was used as the source of the transposon, as described previously (3, 20). Plasmid pGAC275 was then identified; this plasmid contained a Tn5 insertion in the gacA gene, as verified by DNA sequencing (Fig. 1). Plasmid pGAC275 was then homogenized with the corresponding target region of the genome of P. putida WCS358 by the marker exchange procedure as previously described (3, 8). pPH1JI was used as the incoming IncP1 incompatible plasmid, and selection was performed on LB medium containing nalidixic acid, ampicillin, gentamicin, and kanamycin. Putative marker-exchanged mutants were streaked onto LB medium containing tetracycline to confirm loss of the IncP1 recombinant plasmid. The marker exchange was further confirmed by restriction and Southern hybridization of the chromosomal DNA. This resulted in construction of a P. putida WCS358 null mutant which was designated P. putida IBE1 (gacA::Tn5).
FIG. 1.
(A) Gene map of the 2.456-kb KpnI fragment isolated in this study, which contained the gacA response regulatory gene. The ORFs spanning this region are shown, as is the position of the Tn5 insertion which was isolated in this study in the cloned gene and transferred by homologous recombination into the genome of P. putida WCS 358, creating IBE1 (see text for details). Tn5 was at position 1507 of this fragment, and gacA spanned from position 1177 to position 1707. The ORFs adjacent to gacA displayed high identity to pp4101 and pp4100 of P. putida KT2440, which encoded an acetyl transferase belonging to the GNAT family and a transcriptional regulator belonging to the Cro/Ci family, respectively. (B) Gene map of the 2.683-kb SalI-KpnI fragment isolated in this study, which contained the AHL-dependent system of P. putida WCS358. The locations within this fragment of the ppuI gene (spanning positions 1383 to 790), the rsaL gene (spanning positions 1523 to 1753), and the ppuR gene (spanning positions 2473 to 1757) are shown. Also shown are the positions of the Tn5 insertions in the different Tn5 genomic mutants of P. putida WCS358 isolated in this study. The positions of the genomic Tn5 insertions into the rsaL gene are labeled 3C (Tn5 inserted at position 1640) and 2B (Tn5 inserted at position 1672). The positions of the genomic Tn5 insertions into the ppuR gene are labeled 49 (Tn5 inserted at position 1793), 15 (Tn5 inserted at position 2073), 133 (Tn5 inserted at position 2133), and 50 (Tn5 inserted at position 2292). The work described here was performed by using Tn5 mutants 2B (IBE3) and 50 (IBE2); the sequences of these loci have been deposited in data banks under accession numbers AJ629219 and AJ629218, respectively.
Isolation of quorum-sensing mutants and cloning of the ppuI-rsaL-ppuR quorum-sensing locus of P. putida WCS358.
P. putida WCS358 induces β-galactosidase production on solid growth medium in E. coli DH5α(pSCR1) by providing an exogenous AHL autoinducer(s). Plasmid pSCR1 contains components of the cepIR quorum-sensing system, including the cepR gene and the cepI promoter transcriptionally fused to a promoterless lacZ gene. Thus, in solid media in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), exogenously provided AHL can be conveniently detected when it is streaked close to a test strain (2). In order to identify genomic mutants which did not produce an AHL molecule(s), 10,000 Tn5 genomic mutants (34) of strain WCS358 were independently streaked on LB media close to E. coli(pSCR1). Four Tn5 genomic mutants which were unable to induce β-galactosidase production were identified. The chromosomal DNA of all four mutants were purified, and arbitrary PCR (23) was performed in order to amplify, clone, and characterize short DNA fragments which were adjacent to Tn5 (the four PCR fragments corresponding to the four mutants were cloned in pMOSblue, yielding pMOS15, pMOS49, pMOS50, and pMOS133). All four mutants had Tn5 insertions at different positions within a gene, which had very high levels of identity with ppuR of P. putida IsoF (see below) (Fig. 1); the mutants were designated P. putida IBE2.
In order to identify P. putida WCS358 mutants that overproduce AHL, another type of mutant screening was performed. P. putida WCS358 induced only slight pigmentation when it was streaked close to C. violaceum strain CVO26. Consequently, the Tn5 genomic mutant bank was screened with CVO26 for AHL overproducers as described by Yao et al. (38). This led to identification of two Tn5 mutants which could induce very strong pigmentation of CVO26. The chromosomal DNA of both mutants were purified, and arbitrary PCR (23) was performed in order to amplify, clone, and characterize short DNA fragments which were adjacent to Tn5 (two PCR fragments corresponding to the two mutants were cloned in pMOSblue, yielding pMOS2B and pMOS3C). The two mutants had Tn5 insertions at different positions within a gene which had very high levels of identity with rsaL of P. putida IsoF and P. aeruginosa (see below) (Fig. 1); the mutants were designated P. putida IBE3. The quorum-sensing locus was then identified by using the arbitrary PCR fragment of pMOS50 containing part of the ppuR gene as the probe, and it was cloned from the chromosome of P. putida WCS358 as a 2.6-kb KpnI-SalI fragment in the corresponding sites in pBluescriptKS, yielding pBQS1. The same fragment was cloned from pBQS1 as a KpnI-BamHI fragment in broad-host-range plasmid pMP190 KpnI-BglII sites, yielding pMQS1.
Reporter gene fusion assay.
β-Galactosidase activities were determined during growth in LB medium essentially as described by Miller (22), with the modifications of Stachel et al. (31). All experiments were performed in triplicate, and the mean values are given below. β-Galactosidase activities were determined at various times after a 20-ml LB medium culture was started with an initial inoculum of 1.6 × 108 CFU. The growth curves of all mutants were comparable to the growth curve obtained for the parent strain (see Fig. 3 to 5).
FIG. 3.
ppuI, rsaL, and ppuR promoter activities in parent strain P. putida WCS358 and mutant derivatives. All experiments were performed in triplicate, and the mean values are indicated; the standard deviations are not shown and were all within 10% of the reported values. β-Galactosidase activities were determined at various times after a 20-ml LB medium culture was started with an inoculum of 1.6 × 108 CFU. The growth curves of all mutants were comparable to the curve obtained for the parent strain, as indicated in each graph. (A) ppuI promoter activities obtained by using plasmid pPUI220. Also shown are the promoter activities in P. putida IBE3(pPUI220) in the presence of pMQS1, which carried the intact rsaL gene; the values for these activities are indicated by black bars. (B) rsaL promoter activities obtained by using plasmid pRSA220. Also shown are the promoter activities in P. putida IBE3(pRSA220) in the presence of pMQS1, which carried the intact rsaL gene; the values for these activities are indicated by black bars. (C) ppuR promoter activities obtained by using pPUR220 in P. putida WCS358. See text for details. β-gal, β-galactosidase; OD600, optical density at 600 nm.
FIG. 5.
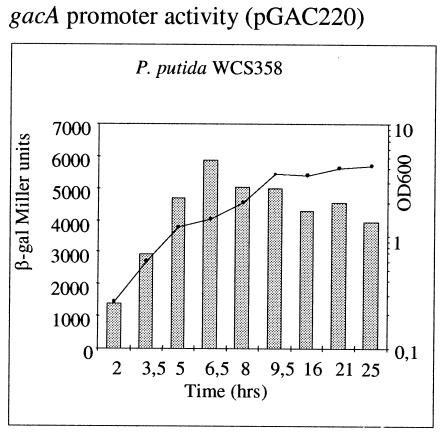
gacA promoter activities in parent strain P. putida WCS358 obtained by using plasmid pGAC220. All experiments were performed in triplicate, and the means are shown; the standard deviations are not shown and were all within 5% of the values reported. β-Galactosidase activities were determined at various times after a 20-ml LB medium culture was started an inoculum of 1.6 × 108 CFU. The growth curve was comparable to the curve obtained for the parent strain, as indicated in the graph. See text for details. β-gal, β-galactosidase; OD600, optical density at 600 nm.
Purification, detection, and characterization of AHLs.
Purification, detection, and characterization of AHLs were performed as previously described (21). P. putida WCS358 and the knockout mutants were grown overnight in M9 minimal medium supplemented with citric acid, and the optical density at 600 nm was measured. The supernatants of the cultures were extracted two times with the same culture volume of ethyl acetate-0.1% acetic acid. The extracts were then dried and resuspended in ethyl acetate by using an amount which resulted in 1 μl of final extract corresponding to 2 × 107 cells of the original culture. Synthetic AHLs, including 3-oxo-C6-AHL, 3-oxo-C8-AHL, 3-oxo-C10-AHL, and 3-oxo-C12-AHL, were used as standard molecules in a thin-layer chromatography (TLC) analysis (kindly provided by P. Williams, University of Nottingham, Nottingham, United Kingdom) performed with C18 reverse-phase chromatography plates by using 60% (vol/vol) methanol in water as the mobile phase. The AHL molecules on the TLC plate were detected by overlaying the TLC plate with a thin layer of LB top agar seeded with E. coli(pSB401) or E. coli(pSB1075) (37).
DNA sequence determination.
The complete sequence of the 2.4-kb KpnI insert containing gacA from pBIB27 and the complete sequence of the 2.6-kb KpnI-SalI insert containing ppuR, rsaL, and ppuI from pBQS1 were determined. These sequences were determined on both strands by GATC Biotech.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 2.4-kb KpnI insert containing gacA from pBIB27 and the 2.6-kb KpnI-SalI insert containing ppuR, rsaL, and ppuI from pBQS1 have been deposited in the GenBank/EMBL/DDBJ database under accession numbers AJ629219 and AJ629218, respectively.
RESULTS
Identification and characterization of AHL-dependent quorum-sensing mutants of P. putida WCS358.
The plant growth-promoting rhizosphere-colonizing organism P. putida WCS358 produces several AHL signal molecules (17). In order to identify mutants which were unable to synthesize AHL molecules, Tn5 genomic mutants were streaked for growth on solid LB media close to E. coli DH5α(pSCR1); this strain produces β-galactosidase when AHL molecules are provided exogenously. A total of 10,000 mutants were examined, and 4 of these mutants were unable to induce β-galactosidase production (as detected on solid media with the aid of X-Gal); it was postulated that these mutants had Tn5 insertions in loci important for AHL biosynthesis. Characterization of these four mutants revealed that they all contained an independent Tn5 insertion within an open reading frame (ORF) with a very level of high identity to the luxR regulatory family member ppuR of P. putida IsoF (Fig. 1) (32). These mutants were designated P. putida IBE2 (having ppuR::Tn5). Cloning and sequencing of a 2.6-kb KpnI-SalI chromosomal fragment in which the Tn5 was located revealed that adjacent to ppuR were two other ORFs related to AHL-dependent quorum sensing. As shown in Fig. 1, the region contained the 714-bp ppuR ORF encoding a protein with a predicted molecular mass of 27 kDa displaying 97% identity with PpuR of P. putida IsoF (32), 42% identity with LasR of P. aeruginosa PAO1 (accession number P25084), and 42% identity with MpuR of P. fluorescens NCIMB10586 (accession number Q9AH79). Adjacent to ppuR in the opposite orientation was a putative 228-bp ORF encoding a protein with a predicted molecular mass of 8.6 kDa displaying 100% identity to RsaL of P. putida IsoF (32) and 42% identity with RsaL of P. aeruginosa, a negative regulator of quorum sensing (9). Finally, an ORF representing a putative luxI family member was adjacent to rsaL in the opposite orientation; this 591-bp ORF encoded a protein with a predicted molecular mass of 22 kDa displaying 100% identity to PpuI of P. putida IsoF (32) and 58% identity to P. aeruginosa LasI (accession number P33883). We concluded that P. putida WCS358 contained an AHL-dependent quorum-sensing system identical to that of P. putida IsoF, consisting of ppuI, rsaL, and ppuR (Fig. 1); the ppuI gene encoded an AHL synthase, ppuR encoded the transcriptional regulator, and rsaL encoded a putative negative regulator. The AHL-dependent quorum-sensing system of P. putida WCS358 appeared to have characteristics identical to those of P. putida IsoF; thus, it was responsible for synthesizing 3-oxo-C12-AHL, 3-oxo-C10-AHL, 3-oxo-C8-AHL, and 3-oxo-C6-AHL, as observed when ppuI was expressed in E. coli DH5α (Fig. 2) (see below), and it was under positive feedback control (see below).
FIG. 2.
TLC analysis of AHLs produced by parent strain P. putida WCS358 and mutant derivatives. In panels A, B, and C the E. coli(pSB401) AHL biosensor was used for detection of the spots, whereas in panel D, E. coli(pSB1075) was used. In all experiments a volume corresponding to 5 × 108 CFU was loaded. Synthetic AHL compounds were used as reference compounds.
Identification and characterization of AHL-overproducing mutants of P. putida WCS358.
P. putida strain WCS358, unlike P. aeruginosa PAO1, was unable to promote strong pigment formation when it was grown on solid growth media side by side with the AHL biosensor C. violaceum CVO26. It was of interest to determine whether the quorum-sensing system was under negative regulation, and this led us to attempt to isolate P. putida WCS358 Tn5 mutants which hyperproduced AHL molecules. As described by Yao et al. (38), we spread between 1,000 and 2,000 C. violaceum CVO26 cells and 300 to 500 P. putida WCS358 Tn5 mutant cells from a Tn5 mutant bank on one plate and screened for strong purple loci. After 10,000 WCS358 Tn5 mutants were scored, 2 mutants were isolated which strongly induced pigment production in CVO26. Characterization of these two mutants revealed that they both contained an independent Tn5 insertion in the rsaL gene (Fig. 1), indicating that inactivation of this gene resulted in much higher levels of AHL production, as also revealed by analysis of spent culture supernatants (Fig. 2) (see below). These mutants were designated P. putida IBE3 and contained rsaL::Tn5.
Cloning and construction of a gacA null mutant of P. putida WCS358.
It was of interest to construct a gacA null mutant of strain WCS358 in order to study the role of this gene in the regulation of quorum sensing and RpoS gene expression. The gacA gene of P. putida WCS358 was identified and consisted of 528 bp encoding a protein with a predicted molecular mass of 19.3 kDa having 81% identity to GacA of P. putida KT2440 (accession number Q88FJ6) and over 70% identity with several other GacA proteins from other Pseudomonas spp. (data not shown). Interestingly, the predicted GacA protein of strain WCS358 is 36 amino acids shorter than the GacA proteins of all other Pseudomonas spp., lacking a stretch of 36 residues from position 25 to position 61 of the protein sequence; the rest of the amino acid sequence is over 95% identical with the sequences of other Pseudomonas spp. GacA proteins. The gacA locus was inactivated in the chromosome of strain WCS358, resulting in construction of a gacA null mutant designated IBE1.
Regulation of the AHL quorum-sensing system of P. putida.
The quorum-sensing system of strain WCS358 was under considerable positive and negative autoregulation. The gene promoters of ppuR, rsaL, and ppuI were cloned in the broad-host-range low-copy-number β-galactosidase promoter probe vector pMP220, yielding pPUR220, pRSA220, and pPUI220, respectively. The activities of the promoters were determined in response to the growth phase in LB medium for strain WCS358 and quorum-sensing, rpoS, and gacA mutant derivatives. The activity of the ppuI promoter in strain WCS358 increased threefold at the onset of the stationary phase, whereas this promoter was silent in P. putida IBE2, indicating that it is under strong positive autoregulation; this was also demonstrated phenotypically since AHLs were not detected in P. putida IBE2 (Fig. 2 and 3A). The activity of the ppuI promoter was dramatically increased in the rsaL mutant IBE3, and the values were 10-fold higher than those in the wild-type strain. This increase took place at the onset of the stationary phase, and the activity quickly decreased after the bacteria reached the stationary phase. This increase in activity also resulted in much higher levels of AHL production in the P. putida IBE3 mutant (Fig. 2A to C). The higher ppuI promoter activity and level of AHL production in IBE3 could both be restored to wild-type levels by introduction of the rsaL gene in trans via plasmid pMQS1 (Fig. 2B and 3A). The activity of the ppuI promoter was considerably decreased in the gacA::Tn5 mutant IBE1 compared to the values observed in strain WCS358; however, AHLs were detected in this mutant (Fig. 2), probably due to the high degree of sensitivity of the AHL detection system, which responds well to small amounts of AHLs. In addition, ppuI promoter activities were higher in P. putida MKO1 (rpoS::Tn5) and P. putida M17 (psrA::Tn5) (Fig. 3A), as indicated by higher AHL levels in these mutants (Fig. 2). Promoter levels could be restored to wild-type levels by addition of pRPO770, which carried the intact rpoS gene, to P. putida MKO1 (data not shown).
The activity of the rsaL promoter was low throughout the growth phase in strain WCS358, mildly increasing at the onset of the stationary phase; however, the values were considerably higher in P. putida MKO1, as well as in the psrA null mutant P. putida M17 (Fig. 3B), and the highest levels were reached in the stationary phase. PsrA is a positive regulator of rpoS (16); thus, the values in mutant M17 were most probably indirect due to the effect of RpoS. Interestingly, in both the rpoS and psrA mutants higher levels of AHLs were also observed (Fig. 2). The promoter levels of pRSA220 in rsaL::Tn5 mutant IBE3 were considerably higher (30-fold higher) (Fig. 3B), indicating that rsaL is under strong negative autoregulation; the promoter levels increased during the growth phase, and the promoter activity and AHL values were reduced dramatically by introduction of pMQS1, which carried an intact rsaL gene (Fig. 3B). The ppuR promoter values were almost constant throughout the growth phase in P. putida WCS358 (Fig. 3C), and the promoter values for all other mutants were comparable to those for parent strain WCS358 (data not shown), indicating that this promoter is not under transcriptional regulation by the global regulators tested here.
Regulation of the rpoS stationary-phase sigma factor RpoS in P. putida.
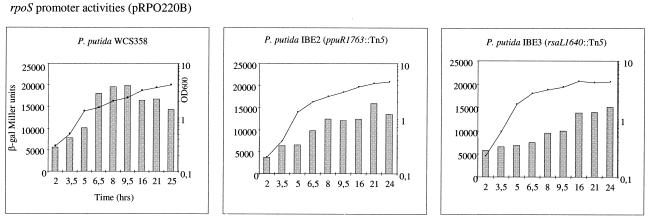
As previously reported, the activity of the promoter of rpoS increased considerably at the onset of the stationary phase in P. putida WCS358 and was regulated at the transcriptional level by the TetR family regulator PsrA (Fig. 4) (18). Interestingly, the rpoS promoter activities, as measured by using plasmid pRPO220B, were reduced by one-half throughout the growth phase in the ppuR and rsaL WCS358 mutants IBE2 and IBE3, implying that quorum sensing has a positive effect on rpoS expression since the absence of both PpuR and RsaL decreased rpoS promoter activity (Fig. 4). The rpoS promoter values in IBE2 with pMQS1 in trans could be restored to wild-type levels; this was not the case for IBE3(pMQS1) (data not shown), and the reason for the latter observation is not known. The response regulator GacA had no effect on rpoS transcription since the levels were comparable to the wild-type levels throughout the growth phase (data not shown).
FIG. 4.
rpoS promoter activities in parent strain P. putida WCS358 and mutant derivatives obtained by using plasmid pRPO220B. All experiments were performed in triplicate, and the means are shown; the standard deviations are not shown and were all within 5% of the values reported. β-Galactosidase activities were determined at various times after a 20-ml LB medium culture was started an inoculum of 1.6 × 108 CFU. The growth curves of all mutants were comparable to the curve obtained for the parent strain, as indicated in each graph. The values for P. putida mutants MKO1 and M17 are shown in the same graph; the darker bars indicate the values for mutant M17. See text for details. β-gal, β-galactosidase; OD600, optical density at 600 nm.
Regulation of the gacA response regulator in P. putida.
The gacA promoter was cloned in the low-copy-number broad-host-range plasmid pMP220, giving rise to pGAC220. The gacA promoter activity increased mildly with the growth phase, reaching the highest levels in the early stationary phase (Fig. 5). The promoter levels were comparable to the values obtained for parent strain WCS358 in all of the genomic mutants tested, indicating that gacA is not under regulation by quorum sensing and RpoS (data not shown).
DISCUSSION
In this paper we describe cloning and regulation of the quorum-sensing system of rhizosphere P. putida WCS358. The quorum-sensing system is identical to the recently reported ppuI-rsaL-ppuR locus system of P. putida IsoF, which has been implicated in regulation of biofilm formation. This locus is most similar to the LasIR-3-oxo-C12-AHL system of P. aeruginosa, with just over 50% identity of the protein components, and responds best to the same AHL even though it results in the synthesis of four 3-oxo-AHL species (32; this study). AHL-dependent quorum sensing is not common in P. putida, and it appears that most strains of this species lack such a system (32). Here we performed studies on the regulation of the system as such studies in general lag behind compared to studies on the regulon that AHL-dependent quorum sensing controls. The novel aspect of our findings is the role of the rsaL repressor located between the ppuIR genes and the linkage of the system in the same species with the global regulators RpoS and GacA.
The quorum-sensing system of P. putida WCS358 is under positive and negative autoregulation. As is often the case with other AHL-dependent systems, the luxI family synthase ppuI is under positive regulation by the cognate luxR family member ppuR, ensuring positive feedback control as ppuI expression is very low and AHLs are not made in the ppuR null mutant (Fig. 2 and 3A). Interestingly, a palindromic sequence with significant identity to the lux box was found in the promoter region of ppuI (data not shown). The system is, however, also under strong negative control by the RsaL repressor since in the rsaL null mutant ppuI expression and AHL concentration increased considerably (Fig. 2 and 3A). Importantly, ppuI expression in the rsaL null mutant is still under growth phase regulation, exhibiting maximal expression in a phase similar to that which occurs in the wild-type parent strain; ppuR expression, on the other hand, is not affected throughout the growth phase in the mutant having constant comparable levels, as occurs in the parent strain. In the rsaL null mutant, the level of ppuI expression is temporal, reaching high values at the onset of the stationary phase, after which it dramatically decreases, bringing into question the role of quorum sensing throughout the stationary phase at high cell densities. A role for RsaL as a negative regulator has also been reported for P. aeruginosa (9), in which it was observed through overexpression studies that RsaL negatively regulates lasI transcription. A model was proposed in which RsaL is in competition with LasR-3-oxo-C12-AHL, ensuring inhibition of lasI at low cell densities, which permits timely expression of quorum sensing in response to the AHL concentration or cell population density. However, this model lacked the evidence of an rsaL null mutant. This is the first report of an rsaL null mutant, and in this mutant we did not observe earlier activation of ppuI and, thus, premature activation of the quorum-sensing system. ppuI expression and AHL increased considerably; however, these things occurred at the same time that they occurred in the parent strain, meaning that some other mechanism ensures a low level of transcription of ppuI at low cell densities. Due to the much higher levels of ppuI transcription when RsaL is lacking, we propose that RsaL and PpuR-3-oxo-C12-AHL are in binding competition at the ppuI promoter at the onset of the stationary phase. PpuR is then able to outcompete RsaL, albeit not very efficiently, increasing the levels of ppuI transcription threefold, which results in activation of the quorum-sensing system. However, we cannot exclude the possibility that RsaL provides a further level of control for the quorum-sensing system and derepresses ppuI expression more efficiently than was observed here under certain environmental conditions which are currently unknown. In addition, RsaL strongly negatively regulates its own transcription (Fig. 3B), thus tightly controlling its expression, and again it is not known whether it does so in response to specific conditions. The results presented here allow us to deduce that most probably it is not necessary to strongly activate ppuI expression, and thus more AHL production is not required, for the quorum-sensing system of P. putida to be activated. In fact, it was observed that the orthologous PpuIR system of P. putida IsoF required only a minimal concentration of AHL (less than 10 nM) for activation (32). It therefore appears that the levels of expression of ppuI are tightly controlled via PpuR-AHL and RsaL at the onset of the stationary phase and that an additional factor(s) represses ppuI expression at low cell densities, avoiding the anticipation of the quorum. Addition to the media of high exogenous levels of AHL (i.e., either 1 μM 3-oxo-C12-AHL or 1 μM 3-oxo-C10-AHL, the AHL molecules that best activate the system) in cultures of the rsaL null mutant did not result in advanced ppuI expression (data not shown), indicating that a critical AHL concentration is still not sufficient to activate transcription in the absence of the RsaL repressor. In P. aeruginosa a repressor called MvaT represses the quorum-sensing system in early phases of growth (10), and a similar repressor or mechanism might be present in P. putida WCS358, which could allow the system to respond not only to AHL concentration but also to the growth phase.
The roles of RpoS and GacA in the regulation of quorum sensing in P. putida were also investigated. Expression of ppuI was decreased in the gacA null mutant, indicating that GacA has a positive effect on ppuI transcription (Fig. 3A). The response regulator GacA was shown to play an important role in P. aeruginosa AHL-dependent quorum-sensing expression by affecting transcription of lasR and rhlR (25). In P. putida WCS358, ppuR and rsaL expression were not affected by GacA, whereas ppuI expression decreased; this decrease was not strong enough to detect a difference in AHL production for cells grown overnight (Fig. 2). It is not known whether this effect of GacA on ppuI transcription is direct or indirect, and GacA is known to regulate the expression of RNA regulators which affect the posttranscriptional activity of target mRNAs (15). Whether the observed effect is direct or through regulatory RNAs which modulate the translation of another regulator(s) needs to be determined. Interestingly, gacA expression itself is growth phase regulated, reaching the highest levels in the stationary phase; the AHL-dependent quorum-sensing system and RpoS were not involved in this growth phase-dependent expression of gacA.
The stationary-phase RpoS sigma factor had a negative effect on ppuI expression. The negative effect of RpoS was also confirmed by ppuI promoter studies with the psrA null mutant; this mutant reduces RpoS levels dramatically since PsrA is a transcriptional activator of rpoS expression (18). In fact, in both psrA and rpoS null mutants AHL production levels increase (Fig. 2), confirming the negative effect of these factors on ppuI expression. Similarly, the promoter of rsaL is also under negative regulation by RpoS and PsrA, having five- to sixfold-higher promoter activities in both null mutants. RpoS does not affect the expression of ppuR and gacA. The molecular mechanism of the negative influence of RpoS on ppuI expression is unknown, and it could be direct or more likely indirect, possibly due to a variation in the metabolic status of the cell resulting from the lack of RpoS. It is evident that RpoS and the AHL-dependent quorum sensing are linked through regulation as each affects expression of the other. Finally, in contrast to what occurs in P. fluorescens (35), GacA does not affect rpoS transcription in P. putida.
This and other studies have recently highlighted the finding that AHL-dependent quorum-sensing systems are under considerable regulation that involves the metabolic status of the cell and other global regulators. Figure 6 summarizes our results in a working model for the regulation of the AHL-dependent quorum-sensing system of P. putida, and future studies need to address how the regulators affect the expression of each other at the molecular level. These controls superimposed on the quorum-sensing system question the role of this system as solely responding to the AHL concentration and cell population density, and understanding this complex regulation should also shed light on the function of AHL quorum sensing in Pseudomonas.
FIG. 6.
Working model for regulation of the AHL-dependent quorum-sensing system of P. putida WCS358 at the onset of the stationary phase when organisms are grown in LB media. ppuI is negatively regulated by the RsaL repressor and positively regulated by PpuR-AHL. RsaL is negatively autoregulated and is negatively regulated by RpoS. GacA has a positive effect either directly or indirectly on ppuI expression, while RpoS has a negative effect. PpuR and RsaL have a positive effect on rpoS expression; it is not known whether this effect is direct. All effects of RpoS observed are also observed in the psrA null mutant, as this is an important positive transcriptional regulator directly regulating rpoS transcription. See text for details.
Acknowledgments
We thank P. Williams and coworkers for providing synthetic AHLs.
This work was supported by the International Centre for Genetic Engineering and Biotechnology, Trieste, Italy, and in part by ISPESL, Rome, Italy, contract B/98-2DIPIA/03. I.B. was supported by a fellowship from the Fondazione per la Ricerca sulla Fibrosi Cistica, Verona, Italy.
REFERENCES
- 1.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, C., A. Friscina, G. Devescovi, M. Kojic, and V. Venturi. 2003. Identification of quorum-sensing-regulated genes of Burkholderia cepacia. J. Bacteriol. 185:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertani, I., M. Kojic, and V. Venturi. 2001. Regulation of the p-hydroxybenzoic acid hydroxylase gene (pobA) in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 147:1611-1620. [DOI] [PubMed] [Google Scholar]
- 4.Better, M., B. Lewis, D. Corbin, G. Ditta, and D. R. Helinski. 1983. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell 35:479-485. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 6.Castaneda, M., J. Sanchez, S. Moreno, C. Nunez, and G. Espin. 2001. The global regulators GacA and σS form part of a cascade that controls alginate production in Azotobacter vinelandii. J. Bacteriol. 183:6787-6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, A., Y. Cui, H. Yang, A. Collmer, J. R. Alfano, and A. K. Chatterjee. 2003. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant-Microbe Interact. 16:1106-1117. [DOI] [PubMed] [Google Scholar]
- 8.Corbin, D., G. Ditta, and D. R. Helinski. 1982. Clustering of nitrogen fixation (nif) genes in Rhizobium meliloti. J. Bacteriol. 149:221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Kievit, T., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Camara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Sayed, A. K., J. Hothersall, and C. M. Thomas. 2001. Quorum-sensing-dependent regulation of biosynthesis of the polyketide antibiotic mupirocin in Pseudomonas fluorescens NCIMB 10586. Microbiology 147:2127-2139. [DOI] [PubMed] [Google Scholar]
- 12.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flavier, A. B., M. A. Schell, and T. P. Denny. 1998. An RpoS (σS) homologue regulates acylhomoserine lactone-dependent autoinduction in Ralstonia solanacearum. Mol. Microbiol. 28:475-486. [DOI] [PubMed] [Google Scholar]
- 14.Geels, F. P., and B. Schippers. 1983. Reduction in yield depression in high frequency potato cropping soil after seed tuber treatments with antagonist fluorescent Pseudomonas spp. Phytopathol. Z. 108:207-221. [Google Scholar]
- 15.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 16.Kojic, M., C. Aguilar, and V. Venturi. 2002. TetR family member psrA directly binds the Pseudomonas rpoS and psrA promoters. J. Bacteriol. 184:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojic, M., G. Degrassi, and V. Venturi. 1999. Cloning and characterisation of the rpoS gene from plant growth-promoting Pseudomonas putida WCS358: RpoS is not involved in siderophore and homoserine lactone production. Biochim. Biophys. Acta 1489:413-420. [DOI] [PubMed] [Google Scholar]
- 18.Kojic, M., and V. Venturi. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 20.Magazin, M. D., J. C. Moores, and J. Leong. 1986. Cloning of the gene coding for the outer membrane receptor protein for ferric pseudobactin, a siderophore from a plant growth-promoting Pseudomonas strain. J. Biol. Chem. 261:795-799. [PubMed] [Google Scholar]
- 21.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 24.Pessi, G., and D. Haas. 2001. Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 200:73-78. [DOI] [PubMed] [Google Scholar]
- 25.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Labpratory, Cold Spring Harbor, N.Y.
- 27.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973-985. [DOI] [PubMed] [Google Scholar]
- 28.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, R. S., and B. H. Iglewski. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 30.Spaink, H. P., R. J. H. Okker, C. A. Wijffelmann, E. Pees, and B. J. J. Lugtemberg. 1987. Promoter in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 31.Stachel, S. E., G. An, C. Flores, and E. W. Nester. 1985. A Tn3 lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 4:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steidle, A., M. Allesen-Holm, K. Riedel, G. Berg, M. Givskov, S. Molin, and L. Eberl. 2002. Identification and characterization of an N-acylhomoserine lactone-dependent quorum-sensing system in Pseudomonas putida strain IsoF. Appl. Environ. Microbiol. 68:6371-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venturi, V. 2003. Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different? Mol. Microbiol. 49:1-9. [DOI] [PubMed] [Google Scholar]
- 34.Venturi, V., F. Zennaro, G. Degrassi, B. C. Okeke, and C. V. Bruschi. 1998. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 144:965-973. [DOI] [PubMed] [Google Scholar]
- 35.Whistler, C. A., N. A. Corbell, A. Sarniguet, W. Ream, and J. E. Loper. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor sigmaS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 38.Yao, F., H. Zhou, and T. G. Lessie. 2002. Characterization of N-acyl homoserine lactone overproducing mutants of Burkholderia multivorans ATCC 17616. FEMS Microbiol. Lett. 206:201-207. [DOI] [PubMed] [Google Scholar]