Abstract
Oxalyl coenzyme A (CoA) decarboxylase (Oxc) is a key enzyme in the catabolism of the highly toxic compound oxalate, catalyzing the decarboxylation of oxalyl-CoA to formyl-CoA. The gene encoding a novel oxalyl-CoA decarboxylase from Bifidobacterium lactis DSM 10140 (oxc) was identified and characterized. This strain, isolated from yogurt, showed the highest oxalate-degrading activity in a preliminary screening with 12 strains belonging to Bifidobacterium, an anaerobic intestinal bacterial group largely used in probiotic products. The oxc gene was isolated by probing a B. lactis genomic library with a probe obtained by amplification of the oxalyl-CoA decarboxylase gene from Oxalobacter formigenes, an anaerobic bacterium of the human intestinal microflora. The oxc DNA sequence analysis revealed an open reading frame of 1,773 bp encoding a deduced 590-amino-acid protein with a molecular mass of about 63 kDa. Analysis of amino acid sequence showed a significant homology (47%) with oxalyl-CoA decarboxylase of O. formigenes and a typical thiamine pyrophosphate-binding site that has been reported for several decarboxylase enzymes. Primer extension experiments with oxc performed by using RNA isolated from B. lactis identified the transcriptional start site 28 bp upstream of the ATG start codon, immediately adjacent to a presumed promoter region. The protein overexpressed in Escherichia coli cross-reacted with an anti-O. formigenes oxalyl-CoA decarboxylase antibody. Enzymatic activity, when evaluated by capillary electrophoresis analysis, demonstrated that the consumption substrate oxalyl-CoA was regulated by a product inhibition of the enzyme. These findings suggest a potential role for Bifidobacterium in the intestinal degradation of oxalate.
Oxalate is ubiquitous in the plant kingdom and is consumed in normal human diets as a component of fruits, vegetables, grains, and nuts (19). Moreover, oxalic acid is one of the most highly oxidized organic compounds and acts as a strong chelator of cations, especially Ca2+. These properties result in limited possibilities for its catabolism and energy production but also make oxalate toxic for most forms of life, especially mammals. In humans, an accumulation of oxalic acid can result in a number of pathological conditions, including hyperoxaluria, calcium oxalate nephrolithiasis, cardiomyopathy, and cardiac conductance disorders (24, 39, 51). In addition, several pathological conditions, including Crohn's disease, steatorrhea, and cystic fibrosis, or medical procedures such as jejuno-ilean bypass surgery are associated with enteric hyperoxaluria due to enhanced oxalic acid absorption in the colon (12, 22, 32). The importance of the colonic segment in regulating oxalic acid homeostasis has focused attention on the possible role oxalate-degrading colonic anaerobic bacteria may have in oxalate-related diseases.
Oxalobacter formigenes is a common inhabitant of the gastrointestinal tract of vertebrates, including humans (1); this bacterium is unique among oxalate-degrading organisms, having evolved a total dependence on oxalate metabolism for energy (13). In O. formigenes, oxalic acid catabolism requires two enzymes: formyl coenzyme A (CoA) transferase, which activates an oxalate molecule to oxalyl-CoA (5, 44), and oxalyl-CoA decarboxylase (Oxc), which decarboxylates the oxalyl-CoA molecule to formyl-CoA (4, 30). Persistence of Oxalobacter in the gut leads to the degradation of intestinal oxalate, thereby limiting oxalate absorption and reducing the oxalate concentration in plasma and urine (14). The role of O. formigenes for scavenging dietary oxalate has been confirmed by studies of the urinary oxalate excretion in both rats fed oxalate and hyperoxaluric rats administered O. formigenes (45, 46). Furthermore, Campieri et al. (9) reported that variation of the intestinal microbiota composition, due to the oral administration of the probiotic bacteria Bifidobacterium and Lactobacillus, reduced the urinary oxalate excretion in patients with idiopathic calcium-oxalate urolithiasis and mild hyperoxaluria.
Bifidobacteria are important members of the gastrointestinal tract flora of humans and animals. Evidence exists that these bacteria exert a health-promoting activity, because they play an important role in the control of the intestinal microflora and in the maintenance of its normal state (37). In particular, their presence has been associated with important metabolic, trophic, and protective functions, e.g., fermentation of nondigestible dietary residues, production of short-chain fatty acids, control of epithelial cell proliferation, and enhancement of the intestinal barrier effect, as well as development and homeostasis of the mucosal immune system (7, 11, 16, 25, 27).
Because of their beneficial properties, Bifidobacterium strains are commonly used in dairy and pharmaceutical probiotic preparations. Bifidobacterium lactis, due to its elevated oxygen tolerance, is one of the Bifidobacterium species most widely used in industrial processes, including fermented foods, yogurt, cheese, beverages, sausages, cereals, infant formula, and pharmaceutical products (34). Despite their widespread use, understanding the roles of this group of bacteria in the gut continues to be a significant challenge. For this reason, genetic characterization of intestinal bifidobacteria is essential to determine the traits considered important for their functional roles as probiotics. The recent description of the genome sequence of Bifidobacterium longum NCC2705 (43) represents a major step forward in bifidobacterial biology, providing crucial information on genes that direct important functional properties of these probiotic bacteria.
In the present study, Bifidobacterium lactis DSM 10140, demonstrated in our screening to have oxalate-degrading activity, has been used to isolate the gene(s) involved in its oxalate catabolism. We report that the gene responsible for its ability to degrade oxalate encodes an oxalyl-CoA decarboxylase (oxc) family enzyme. Enzyme activity is shown for the purified overexpressed protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bifidobacterial strains used in this study are listed below in Table 1. Bifidobacteria strains were grown anaerobically in MRS medium (Difco Laboratories, Detroit, Mich.) supplemented with 0.05% (wt/vol) l-cysteine at 37°C. O. formigenes DSM 4420 was cultured anaerobically in Oxalobacter medium (medium 419; Deutsche Sammlung von Mikroorganismen und Zellkulturen) at 37°C. The anaerobic conditions were achieved in anaerobic jars supplemented with a pad of Anaerocult C (Merck, Milan, Italy). Escherichia coli strains were cultured at 37°C in Luria-Bertani medium with shaking. Kanamycin (50 μg/ml) and ampicillin (100 μg/ml) were added as selective agents when appropriate.
TABLE 1.
Oxalate-degrading activity of Bifidobacterium strains
| Straina | Oxalate degraded (mM) | % Oxalate degraded |
|---|---|---|
| B. lactis DSM 10140 | 4.029 | 60.6 |
| B. animalis ATCC 27536 | 2.454 | 49 |
| B. breve MB 283 | 1.892 | 37.8 |
| B. breve MB 151 | 0.058 | 1 |
| B. longum MB 282 | 1.760 | 35.2 |
| B. longum ATCC 15707 | 1.780 | 35 |
| B. longum MB 58 | 1.387 | 27.7 |
| B. longum MB 229 | 1.207 | 24 |
| B. longum MB 112 | 0.083 | 2 |
| B. infantis MB 57 | 1.335 | 26.7 |
| B. infantis MB 111 | 0.225 | 4 |
| B. adolescentis MB 238 | 2.850 | 57 |
| O. formigenes DSM 4420 | 4,992 | 100 |
| E. coli ATCC 11105 | 0.089 | 1.8 |
ATCC, American Type Culture Collection (Rockville, Md.); DSM, Deutsche Sammlung von Mikroorganismen and Zellkulturen (Braunschweig, Germany); MB, collection of our laboratory (University of Bologna, Bologna, Italy).
Evaluation of oxalate degradation activity in B. lactis DSM 10140.
To evaluate the oxalate-degrading property, Bifidobacterium strains were cultured for 5 days with 5 mM sodium oxalate. In order to remove possible interfering substances, the bacterial cultures were subjected to the following inactivation and purification steps: (i) addition of 2.7 mM EDTA to bind ions, (ii) addition of 10 mM trichloroacetic acid to precipitate proteins, (iii) sterilization and centrifugation steps to remove viable cells, and (iv) addition of activated charcoal to eliminate phenolic derivatives. The oxalate concentration in the supernatants was measured in triplicate by using an enzymatic kit (diagnostic oxalate; Sigma, St. Louis, Mo.), based on the oxidation of oxalate by oxalate oxidase. Oxalate recovery assays were carried out to validate the experimental procedure. An O. formigenes culture was used as a positive control, whereas an E. coli ATCC 11105 culture was used as the negative control.
Isolation and sequencing of the oxc gene from B. lactis DSM 10140.
Chromosomal DNA from B. lactis DSM 10140 was isolated according to the procedure described by Rossi et al. (40). Chromosomal DNA from E. coli and O. formigenes was isolated using the QIAGEN DNeasy tissue kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions.
PCR was carried out in a Biometra thermal cycler T gradient (Biometra, Göttingen, Germany); Dynazyme II (Celbio, Milan, Italy) was used as thermostable polymerase, as per the manufacturer's procedures. A first primer set, Oxadec/L (5′-CCATGTATGGTGTTGTCGGCAT-3′) and Oxadec/R2n (5′-TTCAGGAAGCCAGGGGCGGA-3′), designed on the oxalyl-CoA decarboxylase gene (oxc) from O. formigenes, was used to amplify B. lactis genomic DNA. The nucleotide sequence of the amplicon obtained (150 bp) was used for the development of the new forward primer, oxL (5′-GGTCACTGATTTCGCACGTAT-3′). This last primer was used with the reverse primer oxR3 (5′-GACATCGGCAGGAAGGGAAT-3′), designed on the oxc sequence of O. formigenes, for a new amplification reaction of B. lactis genomic DNA. The sequence analysis of the resulting amplicon (640 bp) allowed us to design the right primer, boxR (5′-TCGGTCTTCTCGACGAATTCA-3′). oxL and boxR primers, both derived from the bifidobacterial chromosomal DNA sequence, were used to amplify the genome of B. lactis to obtain a PCR product (622 bp) which served as probe in colony hybridization experiments.
To construct the genomic library, 30 μg of B. lactis DSM 10140 chromosomal DNA was partially digested with Bsp143I. After electrophoresis of the chromosomal digest in ultrapure agarose (Bio-Rad Laboratories, Hercules, Calif.), fragments of 4 to 6 kb in size were isolated from the gel and ligated into the vector pDG7 (33) that was BamHI linearized. Chemically competent E. coli DH5α cells were transformed with the resulting constructs according to standard procedure (41). The screening of the B. lactis genome library was performed by using the 622-bp amplicon, digoxigenin-dUTP labeled (Roche Diagnostics, Mannheim, Germany) following the supplier's instructions. The colony hybridization was carried out according to standard procedure (41). The primers LpDG7 (5′-CAGTCCTGCTCGCTTCGCTA-3′) and RpDG7 (5′-CGATCTTCCCCATCGGTGAT-3′), based on the pDG7 cloning site flanking regions, were employed for amplifying the B. lactis genome fragments from positive clones. Nucleotide sequencing of both DNA strands from the clone of the B. lactis genomic library, presenting the inserted fragment with highest molecular weight, was performed by the dideoxy chain termination method using BigDye terminators (ABI Perkin-Elmer, Foster City, Calif.) and a 377 sequencer (ABI) for analysis. The primary DNA sequence data were analyzed and assembled using GCG version 10 (Wisconsin package; Genetics Computer Group, Madison, Wis.). BLAST (3) and ClustalW (48) network services were used to search for homologous DNA and protein sequences.
Overexpression and purification of the T7-Oxc fusion protein.
Amplification of the entire oxc gene from the B. lactis DSM 10140 genome was performed with the primers OxcbL (5′-GGATGTTTGCAATGGTTGAT-3′) and BamoxcbR (5′-AAAGGATCCAACGCCATGATGACGAT-3′). The BamoxcbR primer was designed in order to introduce a BamHI restriction site at the 3′ end of the amplicon (the BamHI site is underlined). The PCR product (1,773 bp) was cloned into the pCR2.1 vector (Invitrogen, Carlsbad, Calif.), following the protocol supplied by the manufacturer. The oxc gene, recovered from the recombinant pCR2.1 construct by BamHI restriction, was sequenced and successively ligated into the pET9a vector (Novagen, Madison, Wis.) BamHI linearized in frame with the T7-tag codons. E. coli JM109 was used as the transformation host. The correct construct, identified by digestion with SalI, EcoRI, and SphI, was transferred into E. coli BL21(DE3) for expression. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM to cultures with an optical density at 600 nm of 0.6 to induce the expression of the recombinant protein. Bacteria were grown at 37°C until IPTG addition and then transferred at 28°C during expression time (3 h) to avoid formation of inclusion bodies. Cells, harvested by centrifugation, were sonicated (Brandson sonifier W-250; Heinemann, Schwäbisch Gmünd, Germany), and the soluble fraction containing the recombinant protein was collected by centrifugation. The T7-tagged fusion protein was purified using the T7 · Tag affinity purification kit (Novagen) according to the supplier's protocol. The yield of fusion protein was determined by the Bradford method using the Bio-Rad protein assay kit (Bio-Rad). Bovine serum albumin was used as the standard. Approximately 50 μg of total proteins and 2 μg of the purified protein were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as described by Laemmli (28), using a 12% polyacrylamide running gel. The gels were stained with Coomassie brilliant blue R (Sigma).
Evaluation of oxalyl-CoA decarboxylase activity.
The activity of the oxalyl-CoA decarboxylase was measured by monitoring the consumption of oxalyl-CoA and the production of formyl-CoA by capillary electrophoresis. The reaction mixture contained phosphate buffer (0.1 M; pH 6.8), thiamine pyrophosphate (85 μM), MgCl2 (8.5 mM), oxalyl-CoA (∼0.2 mM), and NAD (0.95 mM). Oxalyl-CoA was synthesized as described by Quayle et al. (38). The reaction was started by adding the purified B. lactis oxalyl-CoA decarboxylase enzyme, recovered from the recombinant E. coli clone at the final concentration of 15 ng/μl. Each mixture was incubated at 37°C for a period ranging from 5 to 40 min.
Electrophoresis experiments were performed using the HPCE 3D system from Agilent Technologies (Waldbronn, Germany). The data were collected on a personal computer equipped with HPCE version A 09 software from Agilent Technologies. The separation was obtained using conventional fused silica capillaries of 50 μm internal diameter with a total length of 38.5 cm. The applied voltage was maintained at 15 kV at a controlled temperature of 15°C. Samples were injected with a hydrodynamic system employing a pressure of 50 mbar for 5 s. The detection wavelength was 200 nm. In order to have high reproducibility of the migration times, the capillary was flushed, alternating water (3 min) and background electrolyte (3 min).
The activity of the cloned oxalyl-CoA decarboxylase enzyme was further evaluated by capillary electrophoresis, measuring the consumption of the substrate (oxalyl-CoA). In this case, the reaction mixture described above was enriched with succinate (10 mM) and cytoplasmic extracts (0.35 μg/μl) from a recombinant E. coli clone overexpressing formyl-CoA transferase of O. formigenes (44).
Oxalyl-CoA and formyl-CoA, the latter synthesized by ester interchange (38) between CoA and thiocresyl formate (8), were employed as standards in capillary electrophoresis measurements. Cytoplasmic extracts from a recombinant E. coli clone which overexpressed the oxc gene of O. formigenes (30) were used as a positive control.
Western blotting.
Proteins of the soluble fraction from E. coli cells overexpressing the oxalyl-CoA decarboxylase of B. lactis DSM 10140 were separated on SDS-PAGE using a 10% polyacrylamide running gel and then transferred to a nitrocellulose membrane (Bio-Rad) by mini-blot system (Bio-Rad). Membranes were blocked by incubation in 1% gelatin in 10 mM Tris-HCl [pH 8], 150 mM NaCl, and 0.1% Tween 20. The recombinant oxalyl-CoA decarboxylase was detected by using pooled anti-oxalyl-CoA decarboxylase monoclonal antibodies (29) and visualized with rabbit anti-mouse immunoglobulin M alkaline phosphatase secondary antibody. As a positive control, lysates of E. coli cells overexpressing oxalyl-CoA decarboxylase of O. formigenes were used.
Primer extension analysis.
Total RNA was isolated from a culture of B. lactis DSM 10140 at late exponential phase by using a Nucleospin RNA II kit (Macherey-Nagel, Germany) as per the manufacturer's instructions. The primer extension product of the oxc transcript was obtained by using oligonucleotide Pex2 (5′-GCGAGGTAGTGGGGAGAATC-3′), corresponding to complementary nucleotide positions 49 to 68 starting from the ATG codon (Fig. 1). The oligonucleotide was labeled with [γ-32P]ATP and T4 polynucleotide kinase, and the reverse transcriptase reaction was performed by annealing 6 and 8 pmol of labeled primer to 60 and 80 μg of total RNA, respectively (primer extension system, AMV reverse transcriptase; Promega, Madison, Wis.). The extension product was separated by electrophoresis on a 7% polyacrylamide-urea sequencing gel, and it was visualized by autoradiography. As a reference, sequencing reactions were performed by the dideoxy chain termination method (42), using the same primer as in the primer extension experiment.
FIG. 1.
Nucleotide sequence and deduced amino acid sequence of the oxc gene encoding oxalyl-CoA decarboxylase in B. lactis (EMBL accession number AB163432). The TPP binding site is indicated by a double underline, and its site-specific amino acids are in bold.
Nucleotide sequence accession number.
The complete sequence of the oxc gene from B. lactis DSM 10140 has been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession number AB163432.
RESULTS
Oxalate-degrading activity of Bifidobacterium strains.
The oxalate-degrading activities for 12 Bifidobacterium strains belonging to human species B. longum, B. breve, B. infantis, and B. adolescentis and to the closely related species B. lactis and B. animalis, widely used as probiotics in dairy and pharmaceutical products, were determined. As reported in Table 1, a high variability in the oxalate-degrading capacity, ranging from 1 to 60%, was evident. B. lactis DSM 10140 was the most active strain, degrading 60% of the oxalate added to the culture medium after 5 days of incubation. It is noteworthy that the enzymatic method employed for evaluating the oxalate concentration presents a low precision and accuracy in all culture media analyzed, as demonstrated by the recovery tests in which 93 to 107% of added oxalate was recovered. However, this error did not appear to compromise the validity of the screening, as the relative oxalate-degrading capacity of the various Bifidobacterium strains was observed, while E. coli, expected to show no oxalate catabolism, exhibited a degrading activity of only 1.8%. O. formigenes, a well-known oxalate-degrading bacterium, consumed the oxalate completely.
Identification and characterization of oxc from B. lactis DSM 10140.
The genomic library of B. lactis DSM 10140 was screened for the oxc gene by hybridization with a B. lactis digoxinenin-labeled probe of 622 bp obtained by three subsequent chromosomal amplifications employing different primer sets, the first of which was based on the oxc gene sequence of O. formigenes (see Materials and Methods). Of approximately 2,000 colonies tested, 10 resulted in positive hybridizations. By amplification of these positive clones with primers designed on the pDG7 sequences flanking the insertion site, a bifidobacterial fragment of 5,000 bp was selected for sequencing. The high molecular weight of this insert supported the probability of finding the entire oxc coding sequence. Genetic analysis of the first 3,060 bp revealed the presence of a 1,773-bp open reading frame (ORF) (Fig. 1) encoding a hypothetical 590-amino-acid protein, with a deduced molecular mass of 63.360 kDa and a calculated isoelectric point of pH 4.65. This DNA segment possessed a G+C content of 62.5%, which was in good agreement with the previous estimated values of 55 to 64% reported for Bifidobacterium DNA (36) and with the value of 60% determined by the complete genome sequence of B. longum NCC2705 (43). The oxalyl-CoA decarboxylase function was attributed to the product of the ORF based on amino acid similarity with proteins of known function. The oxalyl-CoA decarboxylase of O. formigenes (accession no. M77128) presented the highest nucleotide homology (56%) and amino acid similarity (identities, 47%; positives, 64%). Furthermore, most of the decarboxylase enzymes described to date, including the oxalyl-CoA decarboxylase of O. formigenes, present a conserved thiamine pyrophosphate (TPP)-binding region (18). A similar site was present at positions 454 to 483 of the deduced protein product of the B. lactis ORF (Fig. 1). Finally, the hydrophobicity plot of this hypothetical protein did not reveal any region involved in membrane sorting or anchoring, suggesting its cytoplasmic localization.
Primer extension analysis was attempted to elucidate the transcriptional start site of the B. lactis DSM 10140 oxc transcript. The transcriptional start site was identified as a guanine base, situated 28 bases upstream of the assumed ATG start codon of oxc (Fig. 2). Examination of the sequence downstream of the transcriptional start point revealed the canonical gram-positive ribosome-binding site AGGAGG (50) at positions −15 to −9 relative to the ATG. Screening for promoter consensus sequences upstream of the transcriptional start site did not reveal any typical −10 and −35 sequences, but probable consensus regions could be identified at −17 bp (AAAAGT) and −34 bp (TTCTGC), respectively. No terminator region was found downstream of the stop codon TGA, and no stem-loop structure or poly(T) sequence that could reveal the presence of a rho-independent terminator was identified.
FIG. 2.
Primer extensions analysis of the transcriptional start site for oxc mRNA from B. lactis. The arrow indicates the position of the extension products obtained by using oligonucleotide PEX2 with 80 μg (lane 1) and 60 μg (lanes 2 and 3 [two different primer extension experiments]) of total RNA from B. lactis. The assumed ribosome-binding site (RBS) and the start codon (ATG) are indicated in bold. The transcriptional start site (TS) is indicated by a triangle. Proposed upstream −10 and −35 motifs are underlined.
Characterization of the T7-Oxc fusion protein expressed in E. coli.
Isolation of the oxc gene from B. lactis DSM 10140 was achieved by amplification of chromosomal DNA with the OxcbL and BamoxcbR primers. The 1,940-bp amplicon was subcloned into pCR2.1-TOPO, recovered by digestion with BamHI, and ligated into BamHI-linearized pET9a. The recombinant pETOxcc and pETOxco vectors, presenting the correct and opposite orientation of the oxc insert, respectively, were transferred in E. coli BL21(DE3) for the expression of a fusion protein with an N-terminal T7 tag. SDS-PAGE analysis of the crude extracts of these recombinant clones revealed the presence of a 67-kDa protein in the induced cultures of E. coli harboring pETOxcc (Fig. 3). This protein was not detected in the uninduced E. coli/pETOxcc cultures, nor in the uninduced and induced recombinant E. coli/pETOxco cultures. The molecular weight of the fusion protein determined by SDS-PAGE was in good agreement with the calculated molecular mass of the putative protein encoded by the B. lactis ORF (63 kDa) with the fusion tag (1.8 kDa) and a short stretch of amino acids (1.7 kDa) derived from the subcloning step.
FIG. 3.
SDS-PAGE analysis of total protein of cell lysates from E. coli transformed with pETOxcc, pETOxco, or purified recombinant B. lactis oxalyl-CoA decarboxylase. Lane 1, cell lysate of uninduced E. coli/pETOxco; lane 2, cell lysate of induced E. coli/pETOxco; lane 3, cell lysate of uninduced E. coli/pETOxcc; lane 4, cell lysate of induced E. coli/pETOxcc; lanes 5 and 6, two different elution fractions of purified T7-tagged oxalyl-CoA decarboxylase; lane M, molecular mass marker (Bio-Rad).
Cell extracts of E. coli clones transformed with pETOxco and pETOxcc were subjected to Western immunoblotting. Immunoscreening with a pooled anti-oxalyl-CoA decarboxylase monoclonal antibody revealed that the 67-kDa protein expressed by E. coli harboring pETOxcc reacted with the antibody preparation (Fig. 4). The 67-kDa protein, purified from induced E. coli/pETOxcc cultures via its T7 tag using T7 tag affinity immunochromatography, was used in enzyme activity tests.
FIG. 4.
Immunoblot analysis of cloned B. lactis oxalyl-CoA decarboxylase. Cell lysates of E. coli overexpressing the oxc from O. formigenes (lane 1) and E. coli transformed with either pETOxcc (lane 2) or pETOxco (lane 3) were size fractionated by SDS-PAGE and transferred by Western blotting to a nitrocellulose membrane. The oxalyl-CoA decarboxylase enzymes were detected by using pooled anti-oxalyl-CoA decarboxylase monoclonal antibodies and visualized with alkaline phosphatase-conjugated rabbit anti-mouse immunoglobulin. Molecular mass markers (Bio-Rad) were run in lane M.
Evaluation of the oxalyl-CoA decarboxylase activity by capillary electrophoresis.
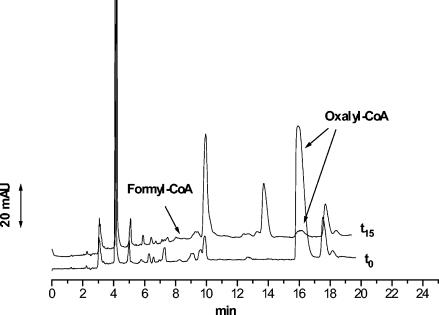
A capillary electrophoresis technique was used for the determination of oxalyl-CoA decarboxylase activity of the 67-kDa purified protein. This analytical approach allows one to evaluate enzymatic mixture variations in concentrations of both substrate oxalyl-CoA and product formyl-CoA. The migration times and UV spectra of both compounds were determined by analyzing chemically synthesized oxalyl-CoA and formyl-CoA standards. The quantification of the analytes was carried out by the determination of corrected peak area A′, which is defined as the ratio between the peak area and the correspondent migration time. The electropherograms obtained by analyzing the reaction mixture before and after incubation for 15 min showed a small decrease of the corrected peak area of oxalyl-CoA and a small increase of the peak related to formyl-CoA (data not shown). Variation of the reaction mixture pH, in the range of 5.5 to 7.5, and of incubation time, prolonged until 40 min, did not imply a further decrease of the substrate, suggesting a product inhibition of the enzyme. In order to confirm this enzymatic activity, it was necessary to show that formyl-CoA produced in the decarboxylation reaction was consumed. To this end, the previous reaction mixture, containing the purified oxalyl-CoA decarboxylase, was enriched with crude extracts of recombinant E. coli overexpressing formyl-CoA transferase of O. formigenes (44) and succinate as the CoA acceptor. The capillary electrophoresis analysis of this last enzymatic mixture was focused on the detection of oxalyl-CoA and formyl-CoA peaks, whereas the succinyl-CoA peak could not be identified lacking the standard. Electropherograms, shown in Fig. 5, obtained by analyzing the reaction mixture before (t0) and after incubation for 15 min (t15) exhibited a marked decrease of the oxalyl-CoA peak area and a complete lack of the formyl-CoA peak. In light of these results, the time course of oxalyl-CoA substrate consumption in the presence of O. formigenes formyl-CoA transferase and succinate was determined (Fig. 6). A decrease of 72 and 92% in the oxalyl-CoA concentration occurred in 1 and 7 min, respectively.
FIG. 5.
Electropherograms of enzymatic reaction mixture at t0 and after 15 min of incubation (t15) at 37°C. The arrows indicate the peaks of oxalyl-CoA and the position at which the peaks of formyl-CoA occur.
FIG. 6.
Degradation kinetics of oxalyl-CoA by B. lactis oxalyl-CoA decarboxylase. The peak areas of oxalyl-CoA detected by capillary electrophoresis are plotted over 30 min of the enzymatic reaction.
Reproducibility of the electrophoretic system was determined by six replicate injections (n = 6) of a reaction mixture after 7 min of incubation. The relative standard deviation percentage of migration time and corrected peak area of oxalyl-CoA were found to be 1.79% (tm = 16.76) and 2.31% (A′ = 5.40), respectively.
DISCUSSION
This report describes, for the first time, an in-depth genetic and functional characterization of the oxalyl-CoA decarboxylase present in B. lactis DSM 10140. To our knowledge, this is also the first enzyme involved in the oxalate degradation to be described in the genus Bifidobacterium.
As bifidobacteria are common inhabitants of the intestinal tract, the finding that these bacteria possess an oxalate-degrading potential could represent an important alternative to O. formigenes in regulating oxalate homeostasis. The colon is the major site of absorption of oxalate (6), a potentially toxic compound widely distributed in food (21). An increase in intestinal absorption of oxalate is known to lead to hyperoxaluria, with a significantly enhanced risk of urinary stone formation (10, 15, 47). Several studies have already demonstrated the presence of oxalate-degrading bacteria in the human intestine (2, 20, 23) and their ability to control oxalate levels by influencing intestinal absorption of dietary oxalate. Identification of intestinal bacteria with oxalate-degrading activity can offer unique opportunities to provide this capacity to individuals suffering from an increased body burden of oxalate and oxalate-associated disorders. Bifidobacterium strains, used both in pharmaceutical and dairy products due to their wide probiotic activity (7, 27), are therefore a potentially important source of oxalate-degrading microorganisms.
Preliminary screening carried out in this study concerning the bifidobacterial oxalate-degrading capacity allowed us to identify a number of strains of Bifidobacterium consuming more than 50% of the oxalate added to the culture medium. B. lactis DSM 10140, a strain employed in several probiotic dairy products (C. Bonaparte and G. Reuter, Proc. Symp. Probiotics Man Anim., p. 33, 1996), proved to be the most active strain in oxalate degradation.
It can be hypothesized that Bifidobacterium has a mechanism for oxalate catabolism similar to that in other intestinal anaerobic bacteria, such as Clostridium and Oxalobacter (13). In O. formigenes, whose intestinal absence has been associated with enteric hyperoxaluria (17) and recurrent oxalate urolithiasis (26), oxalic acid is catabolized by an activation-decarboxylation reaction which yields formate and CO2. The key enzyme is the oxalyl-CoA decarboxylase, which decarboxylates oxalyl-CoA to formyl-CoA (30). Oxalate must be activated to oxalyl-CoA before the decarboxylation takes place, and the formyl-CoA transferase catalyzes the transfer of the CoA moiety from formyl-CoA to oxalate (44).
Based on the nucleotide sequence of the O. formigenes oxc gene, we have been able to identify a genetic element of B. lactis DSM 10140 which presented a significant nucleotide sequence similarity (56%) with oxc of O. formigenes. Interestingly, the sequence of this putative B. lactis oxc gene is not found in the recently published genome sequence of B. longum NCC2705 (43). However, the lack of an oxc gene in the B. longum NCC2705 genome is not unexpected, since among 12 bifidobacterial strains tested in this study, 3 strains were demonstrated to lack the oxalate-degrading capacity and 1 of them was a B. longum species.
Up to now, there have been only a few reports of the primer extension technique being used in Bifidobacterium for the determination of a transcriptional start site, as well as examination of the DNA sequence immediately upstream of the transcriptional start site to identify a potential promoter region (31, 40, 49). No promoter consensus sequences similar to those of other bacteria (i.e., −10 TATAAT and −35 TTGACA) were revealed in the sequence upstream of the transcriptional start site identified in the B. lactis oxc. This result is in accordance with previously reported studies concerning the screening of promoter consensus sequences in Bifidobacterium spp. which failed to identify canonical −10 and −35 regions and suggested no conserved putative consensus sequences (31, 35, 40, 49).
At this time, we would speculate that the bifidobacterial RNA polymerase recognition sites either tolerate a significant amount of degeneracy or that the recognition sites of the vegetative RNA polymerase in Bifidobacterium spp. are dissimilar to those reported for a variety of bacterial species, as suggested by MacConaill et al. (31). Further experimentation will be required to identify which alternative is correct.
The amino acid sequence deduced from the B. lactis oxc gene sequence showed a 47% identity with O. formigenes Oxc. The homology between the two proteins was confirmed by the comparative analysis of their molecular masses and isoelectric points, which revealed very similar values (63.36 kDa versus 65 kDa and pI 4.65 versus 4.9, respectively, for B. lactis and O. formigenes). Furthermore, based on analysis of the amino acid sequences, both enzymes possess a TPP-binding motif (18). This region is conserved in most of the decarboxylases, as TPP is an essential enzymatic cofactor for the cleavage of carbon-carbon bonds adjacent to an Oxo function. As expected, the TPP motif of B. lactis Oxc spans 29 amino acid residues, ends with a conserved NN sequence, includes conserved residues E and P residing at positions 13 and 21, respectively, and presents five hydrophobic amino acids immediately preceding the NN residues. The starting GDG sequence, highly conserved in TPP regions of most bacterial decarboxylases, in the B. lactis TPP motif is replaced with GDS residues, the same starting sequence found in the TPP motif of the O. formigenes Oxc. Furthermore, this TPP-binding motif is located approximately 100 amino acid residues upstream of the C-terminal end in both the B. lactis and O. formigenes decarboxylase.
Evidence that the protein encoded by the putative B. lactis oxc is an oxalyl-CoA decarboxylase came from immunoblotting analysis, which showed that anti-O. formigenes oxalyl-CoA decarboxylase antibody reacted with the recombinant B. lactis Oxc.
In order to verify that this B. lactis Oxc protein possessed oxalate-degrading enzymatic activity, we developed a rapid and sensitive method based on capillary electrophoresis for evaluation of oxalyl-CoA consumption. This analytical approach suggested that B. lactis Oxc is negatively controlled by the reaction product formyl-CoA and that the oxalyl-CoA degradation level is relevant (95%).
In summary, this is the first report suggesting a potential role of Bifidobacterium in the intestinal degradation of oxalate. Unlike the “specialist” O. formigenes, whose growth depends upon oxalate, B. lactis DSM 10140 can be considered a “generalist” oxalate-degrading bacterium, since it is able to ferment other substrates as well as oxalate. Identification and genetic characterization of oxalyl-CoA decarboxylase in B. lactis should increase our knowledge of mechanisms and molecular pathways involved in its health-promoting activities, which apparently include the regulation of oxalic acid.
REFERENCES
- 1.Allison, M. J., K. A. Dawsons, W. R. Mayberry, and J. G. Foss. 1985. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch. Microbiol. 141:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Allison, M. J., C. M. Hook, D. B. Milne, S. Gallagher, and R. V. Clayman. 1986. Oxalate degradation by gastrointestinal bacteria from humans. J. Nutr. 116:455-460. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baetz, A. L., and M. J. Allison. 1989. Purification and characterization of oxalyl-CoA decarboxylase from Oxalobacter formigenes. J. Bacteriol. 171:2605-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baetz, A. L., and M. J. Allison. 1990. Purification and characterization of formyl-coenzyme A transferase from Oxalobacter formigenes. J. Bacteriol. 172:3537-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaji, K. C., and M. Menson. 1997. Mechanism of stone formation. Urol. Clin. North Am. 24:1-11. [DOI] [PubMed] [Google Scholar]
- 7.Ballongue, J. 1998. Bifidobacteria and probiotic action, p. 519-587 In S. Salminen and A. Von Wright (ed.), Lactic acid bacteria: microbiology and functional aspects. Marcel Decker, Inc., New York, N.Y.
- 8.Bax, P. C., and W. Stevens. 1970. Mixed carboxylic anhydrides. VIII. Synthesis of aryl thioformates. Recl. Trav. Chim. Pays-Bas Belg. 89:265-269. [Google Scholar]
- 9.Campieri, C., M. Campieri, V. Bertuzzi, E. Swennen, D. Matteuzzi, S. Stefoni, F. Pirovano, C. Centi, S. Ulisse, G. Famularo, and C. De Simone. 2001. Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int. 60:1097-1105. [DOI] [PubMed] [Google Scholar]
- 10.Chadwick, V. S., K. Modhs, and R. H. Dowling. 1973. Mechanism for hyperoxaluria in patients with ileal dysfunction. N. Engl. J. Med. 289:172-176. [DOI] [PubMed] [Google Scholar]
- 11.Chiang, B. L., Y. H. Sheih, L. H. Wang, C. K. Liao, and H. S. Gill. 2000. Enhancing immunity by dietary consumption of probiotic lactic acid bacterium (Bifidobacterium lactis HN019): optimisation and definition of cellular immune responses. Eur. J. Clin. Nutr. 54:849-855. [DOI] [PubMed] [Google Scholar]
- 12.Clark, J. H., J. F. Fitzgerald, and J. M. Bergstein. 1985. Nephrolithiasis in childhood inflammatory bowel disease. J. Pediatr. Gastroenterol. Nutr. 4:829-834. [DOI] [PubMed] [Google Scholar]
- 13.Dawson, K. A., M. J. Allison, and P. A. Hartman. 1980. Isolation and some characteristic of anaerobic oxalate-degrading bacteria from rumen. Appl. Environ. Microbiol. 40:833-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doane, L. T., M. Liebman, and D. R. Caldwell. 1989. Microbial oxalate degradation: effects on oxalate and calcium balance in humans. Nutr. Res. 9:957-964. [Google Scholar]
- 15.Earnest, D. L. 1979. Enteric hyperoxaluria. Adv. Intern. Med. 25:407-427. [PubMed] [Google Scholar]
- 16.Fukushima, Y. S., S. T. Li, H. Hara, A. Terada, and T. Mitsuaoka. 1997. Effect of follow-up formula containing bifidobacteria (NAN BF) on fecal flora and fecal metabolites in healthy children. Biosci. Microflora 16:65-72. [Google Scholar]
- 17.Goldkin, L., D. R. Cave, B. Jaffin, W. Robinson, and C. M. Bliss. 1985. A new factor in enteric hyperoxaluria: Oxalobacter formigenes. Am. J. Gastroenterol. 80:860. [Google Scholar]
- 18.Hawkins, C. F., A. Borger, and R. N. Perham. 1989. A common structural motif in thiamine pyrophosphate-binding enzymes. FEBS Lett. 255:77-82. [DOI] [PubMed] [Google Scholar]
- 19.Hodgkinson, A. 1977. Oxalic acid in biology and medicine. Academic Press, Inc., New York, N.Y.
- 20.Hokama, S., Y. Honma, C. Toma, and Y. Ogawa. 2000. Oxalate-degrading Enterococcus faecalis. Microbiol. Immunol. 44:235-240. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, R. P., H. O. Goodman, and D. G. Assimos. 2001. Contribution of dietary oxalate to urinary oxalate excretion. Kidney Int. 59:270-276. [DOI] [PubMed] [Google Scholar]
- 22.Hylander, E., S. S. Jarnun, H. J. Jensen, and M. Thale. 1978. Enteric hyperoxaluria: dependence on small intestinal resection, colectomy and steatorrhea in chronic inflammatory bowel disease. Scand. J. Gastroenterol. 13:577-588. [DOI] [PubMed] [Google Scholar]
- 23.Ito, H., T. Kotake, and M. Masai. 1996. In vitro degradation of oxalic acid by human feces. Int. J. Urol. 3:207-211. [DOI] [PubMed] [Google Scholar]
- 24.James, L. F. 1972. Oxalate toxicosis. Clin. Toxicol. 5:231-243. [DOI] [PubMed] [Google Scholar]
- 25.Klaver, F. A., and R. van der Meer. 1993. The assumed assimilation of cholesterol by lactobacilli and Bifidobacterium bifidum is due to their bile salt-deconjugating activity. Appl. Environ. Microbiol. 59:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleinschmidt, K., A. Mahlmann, and R. Hautmann. 1994. Microbial degradation of dietary oxalate in the human gut and urinary oxalate concentrations in patients with calcium oxalate urolithiasis and control persons. Investig. Urol. (Berlin). 5:222-224. [PubMed] [Google Scholar]
- 27.Kurmann, J. A., and J. L. Rasic. 1991. The health potential of products containing bifidobacteria, p. 117-157. In R. Robinson (ed.), Therapeutic properties of fermented milks. Elsevier Applied Science, London, United Kindom.
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lung, H. Y., J. G. Cornelius, and A. B. Peck. 1991. Cloning and expression of the oxalyl-CoA decarboxylase gene from the bacterium, Oxalobacter formigenes: prospects for gene therapy to control Ca-oxalate kidney stone formation. Am. J. Kidney Dis. 17:381-385. [DOI] [PubMed] [Google Scholar]
- 30.Lung, H. Y., A. L. Baetz, and A. B. Peck. 1994. Molecular cloning DNA sequence, and gene expression of the oxalyl-CoA decarboxylase gene, oxc, from the bacterium Oxalobacter formigenes. J. Bacteriol. 176:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacConaill, L. E., D. Butler, M. O'Connell-Motherway, G. F. Fitzgerald, and D. Van Sinderen. 2003. Identification of two-component regulatory systems in Bifidobacterium infantis by functional complementation and degenerate PCR approaches. Appl. Environ. Microbiol. 69:4219-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews, L. A., C. F. Doershuk, R. C. Stern, and M. I. Resnick. 1996. Urolithiasis in cystic fibrosis. J. Urol. 155:1563-1564. [PubMed] [Google Scholar]
- 33.Matteuzzi, D., P. Brigidi, M. Rossi, and D. Di Gioia. 1990. Characterization and molecular cloning of Bifidobacterium longum cryptic plasmid pMB1. Lett. Appl. Microbiol. 11:220-223. [DOI] [PubMed] [Google Scholar]
- 34.Meile, L., W. Ludwig, U. Rueger, C. Gut, P. Kaufmann, G. Dasen, S. Wenger, and M. Teuber. 1997. Bifidobacterium lactis sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 20:57-64. [Google Scholar]
- 35.Minowa, T., S. Ieata, H. Sakai, H. Masaki, and T. Otha. 1989. Sequence and characteristics of the Bifidobacterium longum gene encoding l-lactate dehydrogenase and the primary structure of the enzyme: a new feature of the allosteric site. Gene 85:161-168. [DOI] [PubMed] [Google Scholar]
- 36.O'Riordan, K., and G. F. Fitzgerald. 1997. Determination of genetic diversity within the genus Bifidobacterium and estimation of chromosomal size. FEMS Lett. 156:259-264. [DOI] [PubMed] [Google Scholar]
- 37.Orrhage, K., and C. E. Nord. 2000. Bifidobacteria and lactobacilli in human health. Drugs Exp. Clin. Res. 26:95-111. [PubMed] [Google Scholar]
- 38.Quayle, J. R. 1962. Chemical synthesis of oxalyl-CoA and its reduction to glyoxilate. Biochim. Biophys. Acta 57:398-400. [DOI] [PubMed] [Google Scholar]
- 39.Rodby, R. A., T. S. Tyazka, and J. W. Williams. 1991. Reversal of cardiac dysfunction secondary to type I primary hyperoxaluria after combined liver-kidney transplantation. Am. J. Med. 90:498-504. [PubMed] [Google Scholar]
- 40.Rossi, M., L. Altomare, A. Gonzàles Vara, P. Brigidi, and D. Matteuzzi. 2000. Nucleotide sequence, expression and transcriptional analysis of the Bifidobacterium longum MB 219 lacZ gene. Arch. Microbiol. 174:74-80. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning and laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arrigoni. 2002. The genome sequence of Bifidobaterium longum reflects its adaptation to the human gastrointestinal tracts. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sidhu, H., S. D. Ogden, H. Y. Lung, B. G. Luttge, and A. B. Peck. 1997. DNA sequencing and expression of the formyl-coenzyme A transferase gene, frc, from Oxalobacter formigenes. J. Bacteriol. 179:3378-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidhu, H., M. E. Schmidt, J. G. Cornelius, M. E. Thamilselvan, S. R. Khan, A. Hesse, and A. B. Peck. 1999. Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract-dwelling bacterium Oxalobacter formigenes: possible prevention by gut recolonization or enzyme replacement therapy. J. Am. Soc. Nephrol. 10:S334-S340. [PubMed] [Google Scholar]
- 46.Sidhu, H., M. J. Allison, J. M. Chow, A. Clark, and A. B. Peck. 2001. Rapid reversal of hyperoxaluria in a rat model after probiotic administration of Oxalobacter formigenes. J. Urol. 166:1487-1491. [PubMed] [Google Scholar]
- 47.Smith, L. H., H. Fromm, and A. Hoffmann. 1972. Acquired hyperoxaluria, nephrolithiasis and intestinal disease: description of a syndrome. N. Engl. J. Med. 286:1371-1375. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgens, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trindade, M. I., V. R. Abratt, and S. Reid. 2003. Induction of sucrose utilization genes from Bifidobacterium lactis by sucrose and rafinose. Appl. Environ. Microbiol. 69:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vellanoweth, R. L. 1993. Translation and its regulation, p. 699-711. In A. Sonenshein, J. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 51.Williams, H. E., and L. H. Smith. 1968. Disorders of oxalate metabolism. Am. J. Med. 45:715-735. [DOI] [PubMed] [Google Scholar]