Abstract
We have developed a novel cell surface display system by employing FadL as an anchoring motif, which is an outer membrane protein involved in long-chain fatty acid transport in Escherichia coli. A thermostable Bacillus sp. strain TG43 lipase (44.5 kDa) could be successfully displayed on the cell surface of E. coli in an active form by C-terminal deletion-fusion of lipase at the ninth external loop of FadL. The localization of the truncated FadL-lipase fusion protein on the cell surface was confirmed by confocal microscopy and Western blot analysis. Lipase activity was mainly detected with whole cells, but not with the culture supernatant, suggesting that cell lysis was not a problem. The activity of cell surface-displayed lipase was examined at different temperatures and pHs and was found to be the highest at 50°C and pH 9 to 10. Cell surface-displayed lipase was quite stable, even at 60 and 70°C, and retained over 90% of the full activity after incubation at 50°C for a week. As a potential application, cell surface-displayed lipase was used as a whole-cell catalyst for kinetic resolution of racemic methyl mandelate. In 36 h of reaction, (S)-mandelic acid could be produced with the enantiomeric excess of 99% and the enantiomeric ratio of 292, which are remarkably higher than values obtained with crude lipase or cross-linked lipase crystal. These results suggest that FadL may be a useful anchoring motif for displaying enzymes on the cell surface of E. coli for whole-cell biocatalysis.
Cell surface display is a technique to display peptides or proteins on the surface of gram-negative and gram-positive bacteria, fungi, or even mammalian cells by appropriately fusing them to surface anchoring motifs (14, 21, 22, 31). The first surface expression system was developed by fusing bacteriophage coat protein with peptides and small proteins (27). This phage display has been widely used in screening of antibodies, epitopes, and high-affinity ligands. However, the size of foreign proteins that can be displayed on the surface of phage is rather limited (4, 7). As an alternative to phage display, microbial cell surface display has been developed. This technique has a wide range of biotechnological and industrial applications, including development of vaccines, peptide and antibody libraries, bioremediation, biocatalysis, and biosensors. Many different proteins, including outer membrane proteins, lipoproteins, autotransporters, subunits of surface appendages, and S-layer proteins, have been successfully employed as anchoring motifs in microbial cell surface display (14, 18, 21). Among these, outer membrane proteins have widely been used as anchoring motifs because they have unique membrane-spanning structures, which provide many potential fusion sites for target proteins. Several membrane proteins, including OmpA, OprF, OmpS, invasin, LamB, PhoE, OmpC, and Lpp-OmpA, have been used as anchoring motifs for displaying relatively small-molecular-weight peptides, antibodies, domains, and receptors (4, 21, 25, 32).
FadL (48.8 kDa) is an outer membrane protein involved in the binding and transportation of long-chain fatty acids and also in the binding of bacteriophage T2 in Escherichia coli (5, 10). It has been reported that FadL is rich in β-structure and spans the outer membrane multiple times to form a long-chain fatty acid-specific channel. FadL consists of 20 antiparallel β-strands which produce a β-barrel structure and are connected by 9 internal loops and 10 external loops (9). These characteristics led us to examine the possibility of employing FadL as a novel anchoring motif for the display of proteins on the E. coli cell surface.
Recently, enzymatic chiral resolution has drawn much attention for obtaining enantiomerically enriched compounds by exploiting the selectivity of enzymes for one form of the enantiomers of a racemic molecule (8, 30). Although many kinds of enzymes can be used for the kinetic resolution of racemic compounds, enzymes including lipase, esterase, and protease have most frequently been used because of their merits such as broad substrate specificity, stability, and no requirement of cofactor (12, 15). Especially, lipase (triacylglycerol hydrolase; EC 3.1.1.3), which generally catalyzes hydrolysis of oils and transesterification of esters, is the most commonly used enzyme for this purpose because of its excellent enantioselectivity, commercial availability, broad substrate specificities to natural and unnatural esters of different structures, and good stability in various media ranging from aqueous to nonaqueous organic solvents (23, 29). Due to these advantages, lipase has been widely applied for the production of enantiomerically pure compounds, which are subsequently used for the synthesis of fine chemicals and drug intermediates. However, the reduced enantioselectivity and product yield and the presence of impurities are the common problems observed. The use of a highly purified enzyme or an immobilized enzyme can partially solve these problems, but the process becomes more expensive and instability problems can arise (20). Therefore, the development of efficient enzyme systems and processes has been an important research objective in this field.
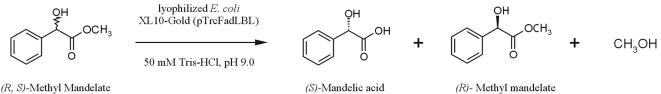
In this paper, we investigated the display of a thermostable Bacillus sp. strain TG43 lipase (44.5 kDa) (28) in an active form on the E. coli cell surface by using FadL as an anchoring motif and its application in enantioselective biocatalysis. As an example, we examined enantioselective resolution of racemic methyl mandelate, as shown in Fig. 1.
FIG. 1.
Reaction scheme for enantioselective resolution of racemic methyl mandelate by using lyophilized cells of recombinant E. coli XL10-Gold displaying the FadLt-lipase fusion protein.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli XL10-Gold (Stratagene cloning system; Stratagene, La Jolla, Calif.) was used as a host strain for general cloning work and gene expression studies. Recombinant cells were cultivated in Luria-Bertani medium (10 g of Bacto Tryptone/liter, 5 g of Bacto yeast extract/liter, and 5 g of NaCl/liter) supplemented with 50 mg of ampicillin/liter at 37°C and 250 rpm. When the optical density at 600 nm (OD600) was 0.4, cells were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for the production of recombinant proteins. After induction, cells were further cultured for 4 h and used for Western blotting and immunofluorescence microscopy.
Construction of plasmids.
PCR was performed with the PCR thermal cycler MP (Takara Shuzo Co., Ltd., Shiga, Japan) using the Expand high-fidelity PCR system (Roche Molecular Biochemicals, Mannheim, Germany). DNA sequencing was carried out using the BigDye terminator cycle sequencing kit (Perkin-Elmer Co., Boston, Mass.), Taq polymerase, and an ABI Prism 377 DNA sequencer (Perkin-Elmer Co.). All DNA manipulations including restriction digestion, ligation, and agarose gel electrophoresis were carried out following standard procedures (24).
PCR primers used in this study are listed in Table 1. Primers for the amplification of the E. coli fadL and Bacillus sp. strain TG43 lipase genes were designed based on the reported E. coli genome sequence (6) and the sequence of Bacillus sp. strain TG43 lipase (28) (GenBank accession no. AF141874), respectively.
TABLE 1.
List of primers used in PCR experiments
| Primer | Sequencea | Gene to be amplified | Template DNA used |
|---|---|---|---|
| Primer 1 | 5′-GGAATTCATGGTCATGAGCCAGAAAACC | Truncated fadL | E. coli W3110 chromosome |
| Primer 2 | 5′-GCTCTAGAACGATTCTGTGCAGGAAC | ||
| Primer 1 | 5′-GGAATTCATGGTCATGAGCCAGAAAACC | Truncated fadL with stop codon | E. coli W3110 chromosome |
| Primer 3b | 5′-GCTCTAGATTAACGATTCTGTGCAGGAAC | ||
| Primer 4 | 5′-GCTCTAGAGCGGCTTCGCGAGCCAAT | Bacillus sp. strain TG43 lipase gene | lipA-pET26b |
| Primer 5 | 5′-CCCAAGCTTTTAAGGCCGCAAACTCGC | ||
| Primer 6 | 5′-CCCAAGCTTTTAATGGTGATGATGGTGAT GAGGCCGCAAACTCGC |
Restriction enzyme sites are shown in bold.
Underlined sequence was added for expression of the truncated fadL gene.
Fractionation of outer membrane proteins.
Culture broth (3 ml) was centrifuged at 3,500 × g for 5 min at 4°C, and the cell pellet was washed with 1 ml of 10 mM Na2HPO4 buffer (pH 7.2), followed by centrifugation at 3,500 × g for 5 min at 4°C. The cell pellet was resuspended in 0.5 ml of 10 mM Na2HPO4 buffer (pH 7.2). Crude extracts of recombinant E. coli cells were prepared by three cycles of sonication (each for 20 s at 15% of maximum output; high-intensity ultrasonic liquid processors; Sonics & Material Inc., Newtown, Conn.). Partially disrupted cells were first removed by centrifugation of sonicated samples at 12,000 × g for 2 min at room temperature. Membrane proteins and lipid layers were isolated by centrifugation at 12,000 × g for 30 min at 4°C, followed by resuspension in 0.5 ml of 10 mM Na2HPO4 buffer (pH 7.2). For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting experiments, 0.5% (wt/vol) sarcosyl was also added. After incubation at 37°C for 30 min, the insoluble pellet containing membrane proteins was obtained by centrifugation at 12,000 × g for 30 min at 4°C. Membrane proteins were obtained by washing the insoluble pellet with 10 mM Na2HPO4 buffer (pH 7.2) followed by resuspending in 50 μl of Tris-EDTA buffer (pH 8.0).
Western blotting.
Since the antibody against the Bacillus sp. strain TG43 lipase was not available, we used anti-His antibody to probe the truncated FadL (FadLt)-lipase-His6 fusion protein. Whole-cell lysates and membrane fractions were analyzed by SDS-12% (wt/vol) PAGE. Western blot analysis was performed following standard protocols (24). For the immunodetection of the fusion protein, rabbit anti-His probe antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) and goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase conjugate (Sigma, St. Louis, Mo.) were used. The light-emitting nonradioactive ECL kit (Amersham Life Sciences, Buckinghamshire, United Kingdom) was used for signal detection.
Immunofluorescence microscopy.
For immunofluorescence microscopy, cells (1 ml) were harvested by centrifugation for 5 min at 3,500 × g and 4°C, washed with phosphate-buffered saline (PBS) solution, and resuspended in PBS solution supplemented with 3% (wt/wt) bovine serum albumin (Sigma). Cells were incubated with the rabbit anti-His probe antibody diluted (1:1,000) in PBS solution containing 3% (wt/wt) bovine serum albumin for 4 h at 4°C. After washing five times with PBS solution, the cell-antibody complex was incubated overnight at 4°C with goat anti-rabbit IgG conjugated with fluorescein isothiocyanate (FITC; Sigma) at a dilution of 1:3,000. Prior to microscopic observation, cells were washed five times with PBS solution to remove unbound goat anti-rabbit IgG conjugated with FITC. Cells were mounted on poly-l-lysine-coated microscopic slides and examined by confocal microscopy (Carl Zeiss, Jena, Germany). Photographs were taken with a Carl Zeiss LSM 410. Samples were excited by a 488-nm argon laser, and images were filtered by a long-pass 505-nm filter.
Measurement of lipase activities.
Cells were cultivated in a 250-ml flask containing 100 ml of Luria-Bertani medium at 37°C and 250 rpm. At an OD600 of 0.4, cells were induced with 0.01, 0.1, or 1 mM IPTG for the production of recombinant proteins. After induction, cells were further cultured for 4 h. Cells were harvested by centrifugation for 5 min at 5,590 × g and 4°C, washed with distilled water, and lyophilized with a freeze dryer (TFD5505; Ilshin Lab., Gyeonggi-do, Korea) for 48 h.
Lipase activity was assayed by a spectrophotometric method using p-nitrophenyl decanoate as a substrate (19). The p-nitrophenyl decanoate was dissolved in acetonitrile at a concentration of 10 mM. Ethanol and 50 mM Tris-HCl (pH 8.0) were subsequently added to make a substrate solution having a volume ratio of 1:4:95 (10 mM p-nitrophenyl decanoate in acetonitrile-ethanol-Tris-HCl). Lyophilized cells (0.15 mg) or culture supernatant (500 μl) was added to 3 ml of substrate solution for the determination of lipase activity. After incubating the reaction mixture at 37°C for 10 min, the reaction was terminated by adding 2 μl of 0.5 M EDTA. The activity was assayed by detecting the product, p-nitrophenol, spectrophotometrically at 405 nm. One unit of lipase activity was defined as the amount of enzyme releasing one micromole of p-nitrophenol per minute (2). The specific activity was defined as the lipase activity per milligram of lyophilized cells. All measurements were carried out in triplicate.
The temperature-dependent lipase activities were examined in the same substrate solution described above at controlled temperatures from 16 to 70°C. The optimal pH was determined at 37°C using substrate solutions having a volume ratio of 1:4:95 (10 mM p-nitrophenyl decanoate in acetonitrile-ethanol-50 mM potassium phosphate or 50 mM Tris-HCl at various pHs ranging from 5 to 10). The effect of substrate chain length was determined by adding a 10 mM solution of p-nitrophenyl caproate or p-nitrophenyl palmitate instead of p-nitrophenyl decanoate. For the examination of thermal stability of cell surface-displayed lipase, 10 mg of lyophilized cells was resuspended in 10 ml of Tris-HCl (pH 8.0) and incubated at 37 or 50°C for a week. The 0.1-ml aliquots were taken, cooled to 37°C, and added to 1 ml of substrate solution for the measurement of residual activity at 37°C for 10 min. For the detection of cell lysis, we measured the OD600 and the enzyme activity in the supernatant during the entire reaction.
Preparation of enantiomerically pure compound.
For the enantioselective hydrolysis, 300 mg of lyophilized cells (prepared by inducing with 0.1 mM IPTG) was resuspended in 30 ml of 50 mM Tris-HCl (pH 9.0), into which 150 mg of racemic methyl mandelate (Aldrich, St. Louis, Mo.) was added. The reaction mixture was incubated at 37°C and 250 rpm. Small aliquots of reaction mixture were removed at 12, 24, and 36 h of the reaction, and the products were analyzed by high-performance liquid chromatography (HPLC; 1100 HPLC system; Agilent, Palo Alto, Calif.).
Analytical methods.
Cell growth was monitored by measuring the OD600 with a spectrophotometer (DU650; Beckman, Fullerton, Calif.). The yield and optical purity of substrate (racemic methyl mandelate) were analyzed by using the HPLC apparatus equipped with a chiral column (Chiralcel OJ-H column; Daicel Chemical Industries, Osaka, Japan). A mixture of hexane and isopropanol having a volume ratio of 90:10 was used as a mobile phase at a flow rate of 1.0 ml/min. For the analysis of product [(S)-mandelic acid], a Chiralcel OD-H column (Daicel) was employed using a mixture of hexane, isopropanol, and trifluoroacetic acid at a volume ratio of 80:20:1 as a mobile phase at a flow rate of 0.5 ml/min. Reaction products and substrates were detected by measuring the absorbance at 230 nm using a diode array detector (1100 HPLC DAD; Agilent).
RESULTS
Construction of cell surface display system.
Based on the predicted structure of FadL (9), two trypsin cleavage sites following Arg93 and Arg384 are exposed at the external face of the outer membrane and are located at the second and ninth loops from the N terminal, respectively. Therefore, these points were considered the potential fusion sites. Between these two fusion sites, the ninth external loop was selected because the ninth loop is the second-to-the-last external loop and, therefore, is not likely to disrupt most of the FadL β-barrel structure.
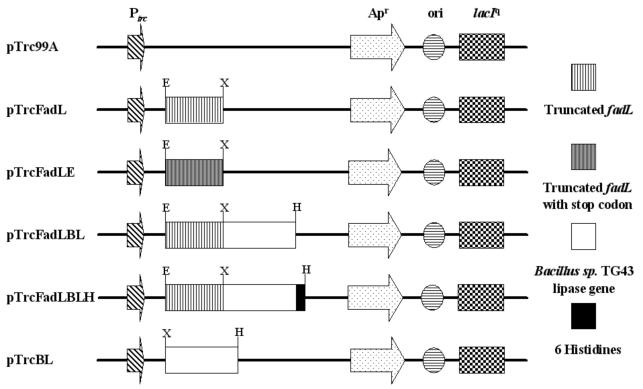
The truncated fadL (fadLt) gene encoding the first 384 amino acids from the N terminus was amplified by PCR using primers 1 and 2 and was cloned into the EcoRI and XbaI sites of pTrc99A to make pTrcFadL (Fig. 2). One arginine was additionally inserted at the C terminus by introducing the XbaI site at the 3′ end of the fadLt gene. The Bacillus sp. strain TG43 lipase gene amplified using primers 4 and 5 was then cloned into the XbaI and HindIII sites of pTrcFadL to make pTrcFadLBL (Fig. 2). For the expression of the fadLt gene without fusion, the fadLt gene containing the stop codon was amplified using primers 1 and 3 and cloned into the EcoRI and XbaI sites of pTrc99A to make pTrcFadLE (Fig. 2). For the immunofluorescence detection of surface-displayed protein, the Bacillus sp. strain TG43 lipase gene fused to a DNA fragment encoding six histidines (His6) at the C terminus was amplified using primers 4 and 6 and was cloned into the XbaI and HindIII sites of pTrcFadL to make pTrcFadLBLH (Fig. 2). The His6 was introduced to serve as an epitope for the rabbit anti-His probe antibody. For the intracellular expression of lipase, the Bacillus sp. strain TG43 lipase gene amplified using primers 4 and 5 was cloned into the XbaI and HindIII sites of pTrc99A to make pTrcBL (Fig. 2).
FIG. 2.
Plasmids used for the display of lipase: pTrc99A (Pharmacia Biotech, Uppsala, Sweden), pTrcFadL, truncated fadL of E. coli; pTrcFadLE, truncated fadL of E. coli containing the stop codon; pTrcFadLBL, truncated fadL-Bacillus sp. strain TG43 lipase gene; pTrcFadLBLH, truncated fadL-Bacillus sp. strain TG43 lipase-His6 fusion gene at the C terminal; pTrcBL, Bacillus sp. strain TG43 lipase. Abbreviations: E, EcoRI; X, XbaI; H, HindIII; Ptrc, trc promoter; Apr, β-lactamase gene.
Confirmation of lipase display on the cell surface.
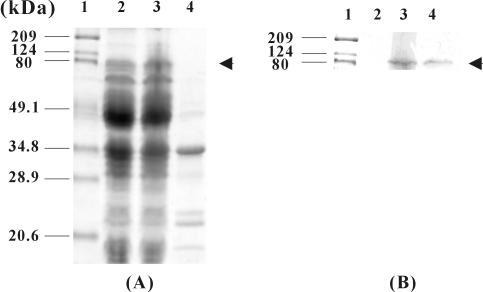
To examine whether lipase was successfully displayed on the cell surface, the whole-cell lysate and outer membrane fraction of recombinant E. coli producing FadLt-lipase fusion protein were analyzed by SDS-PAGE. However, the fusion protein could hardly be detected by Coomassie blue staining, because its expression level was rather low (Fig. 3A). Therefore, Western blot analysis of FadLt-lipase-His6 was carried out using the rabbit anti-His probe antibody, which was subsequently detected with horseradish peroxidase-conjugated goat anti-rabbit IgG (Fig. 3B). The bands corresponding to the 84.2-kDa fusion protein were detected in whole-cell lysates and the outer membrane fraction (Fig. 3B, lanes 2 and 3). No signal was detected in whole-cell lysates of E. coli XL10-Gold (pTrcFadLE) cells producing the FadLt protein (Fig. 3B, lane 1).
FIG. 3.
SDS-PAGE analysis (A) and immunoblotting (B) of recombinant E. coli XL10-Gold cells expressing FadLt and FadLt-lipase-His6 fusion proteins. Lane 1, molecular mass standards; lane 2, whole-cell lysates of E. coli XL10-Gold harboring pTrcFadLE; lane 3, whole-cell lysates of E. coli XL10-Gold harboring pTrcFadLBLH; lane 4, outer membrane fraction of E. coli XL10-Gold harboring pTrcFadLBLH.
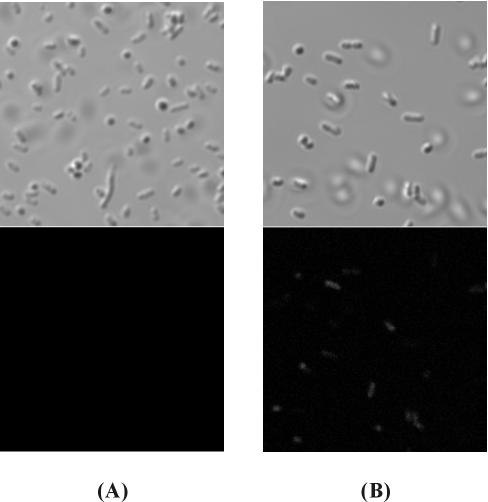
The display of lipase on the cell surface could be more directly confirmed by immunofluorescence microscopy. As shown in Fig. 4, E. coli XL10-Gold cells expressing the FadLt-lipase-His6 fusion protein became fluorescent due to the binding of anti-His probe antibody followed by binding of FITC-conjugated secondary antibody, indicating that lipase was successfully displayed on the cell surface (Fig. 4B). On the other hand, E. coli XL10-Gold cells expressing FadLt were not fluorescent at all (Fig. 4A).
FIG. 4.
Differential interference micrographs (upper) and immunofluorescence micrographs (lower) of XL10-Gold cells harboring pTrcFadLE (A) and pTrcFadLBLH (B). Cells were incubated with rabbit anti-His probe antibody followed by probing with goat anti-rabbit IgG-FITC conjugate.
After confirming that lipase was successfully displayed on the E. coli cell surface, we next examined whether the displayed lipases were active. Whole-cell lipase activities of 71.2 ± 7.9 (mean ± standard deviation), 104.9 ± 9.9, and 66.2 ± 8.1 U were obtained using lyophilized XL10-Gold (pTrcFadLBL) cells prepared by inducing with 0.01, 0.1, and 1 mM IPTG, respectively, while only 3.2 ± 0.6, 3.3 ± 0.92, and 5.4 ± 1.1 U of lipase activity, respectively, was detected in the supernatant. The maximum specific activities of lyophilized XL10-Gold (pTrcBL), XL10-Gold (pTrcFadLBL), and purified Bacillus sp. strain TG43 lipase were 200 U/mg, 2,800 U/mg of lyophilized cells, and 726,700 U/mg of lipase, respectively, which indicated that the expression level of lipase was 0.4% of total cell weight at least (28). To estimate the actual expression level of lipase in each fraction, the activities of whole-cell, soluble, and membrane fractions were compared. The activity of the membrane fraction was 80% of whole-cell activity. The activity of the soluble fraction was below 10% of whole-cell activity. These results suggest that lipases were successfully displayed in an active form by using FadLt as an anchoring motif.
Activity and stability of cell surface-displayed lipase.
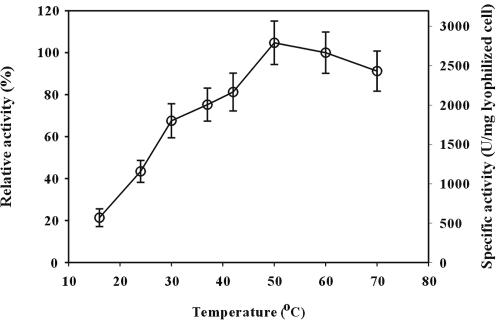
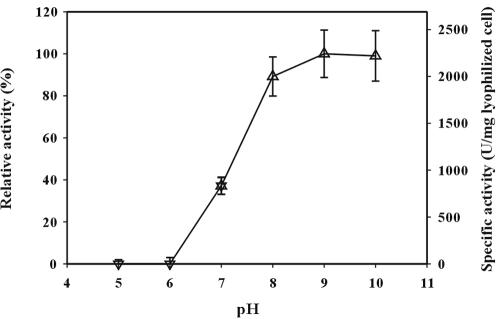
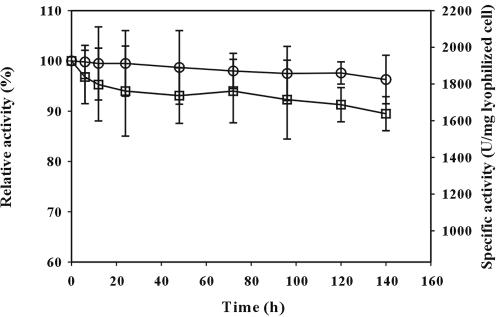
To determine the optimal conditions of cell surface-displayed lipase, reactions were carried out at various temperatures ranging from 16 to 70°C and pHs of 5 to 10. The results are shown in Fig. 5 and 6. Cell surface-displayed lipase showed the maximum activity at 50°C. In the temperature range of 40 to 70°C, the activity was higher than 80% of the maximum activity. The activity of cell surface-displayed lipase was higher under alkaline conditions and was the highest at pH 9. After the optimization of reaction conditions, thermal stability was examined, as it is important for industrial applications. Cell surface-displayed lipase was incubated at 37 and 50°C for 1 week, and the whole-cell activity was measured periodically during the week. As shown in Fig. 7, the cell surface-displayed lipase was quite stable against heat and did not show much loss of activity (less than 10%) at either temperature. When we measured the cell density (OD600) during the reaction, the change of OD600 was negligible during the entire reaction period. Furthermore, the activities in the supernatants were negligible, which indicated that cell lysis did not occur. Finally, substrate specificity was examined using three p-nitrophenyl esters, of caproate, decanoate, and palmitate, having different carbon chain lengths. The lipase used in this study showed the highest activity towards p-nitrophenyl octanoate (C8) when it is used in the free form. The activity towards p-nitrophenyl palmitate (C16) or p-nitrophenyl butyrate (C4) was below 50% of the highest activity (28). As shown in Table 2, the activity obtained with p-nitrophenyl palmitate was below half of that obtained with p-nitrophenyl decanoate or p-nitrophenyl caproate. These results suggest that the enzymatic characteristics of the cell surface-displayed lipase were similar to those of free enzyme (3, 28).
FIG. 5.
Effect of temperature on lipase activity of E. coli XL10-Gold (pTrcFadLBL). The enzyme activity was determined at pH 8.0 by using p-nitrophenyl decanoate as the substrate. Relative activity was calculated by assuming the activity obtained at 60°C was 100%.
FIG. 6.
Effect of pH on lipase activity of E. coli XL10-Gold (pTrcFadLBL). The enzyme activity was determined at 37°C by using p-nitrophenyl decanoate as the substrate. Buffers used were 50 mM potassium phosphate buffer (▿) and 50 mM Tris-HCl (▵). Relative activity was calculated by assuming the activity obtained at pH 9.0 was 100%.
FIG. 7.
Stability of lipase displayed on the cell surface of E. coli XL10-Gold (pTrcFadLBL) cells during prolonged incubation at pH 8.0 and 37°C (○) or 50°C (□). The enzyme activity was determined at 37°C by using p-nitrophenyl decanoate as the substrate. Relative activity was calculated by assuming the initial activity was 100%.
TABLE 2.
Effect of substrate chain length on activity of cell surface-displayed lipase
| Activity parameter | p-Nitrophenyl caproate | p-Nitrophenyl decanoate | p-Nitrophenyl palmitate |
|---|---|---|---|
| Activity (U)a | 96.9 ± 10.2 | 105 ± 10.6 | 43.3 ± 10.6 |
| Specific activity (U/mg of lyophilized cells) | 1,940 ± 200 | 2,000 ± 200 | 870 ± 200 |
| Relative activity (%)b | 92.3 ± 9.72 | 100 ± 10.2 | 41.2 ± 10.1 |
Activity was assayed by adding 0.15 mg of lyophilized cells into the substrate solution consisting of 10 mM substrate in acetonitrile, ethanol, and 50 mM Tris-HCl (pH 8.0) at the volume ratio of 1:4:95.
Relative activity was calculated by assuming the activity obtained with p-nitrophenyl decanoate as 100%.
Enantioselective resolution of racemic methyl mandelate by using cell surface-displayed lipase.
As a potential application, we investigated the possibility of enantioselective resolution of racemic compounds by using cell surface-displayed lipase. Racemic methyl mandelate was used as a model substrate. The scheme for enantioselective resolution with cell surface-displayed lipase is shown in Fig. 1. After 36 h, the enantiomeric excesses of the remaining methyl mandelate and the product, (S)-mandelic acid, reached 33 and 99%, respectively (Table 3). Also, high enantioselectivity (E value of >250) was obtained (Table 3). These results suggest that the lipase displayed on the E. coli cell surface can be used for efficient chiral resolution of racemic compounds.
TABLE 3.
Enantioselective hydrolysis of racemic methyl mandelate by cell surface-displayed lipase
| Time (h) | Enantiomeric excess (%)a
|
Conversion (%)b | Enantiomeric ratioc | |
|---|---|---|---|---|
| Remaining ester | Product | |||
| 12 | 0 | NDd | 0 | |
| 24 | 23.2 ± 1.6 | 99.0 | 19.0 ± 1.0 | 250 |
| 36 | 39.1 ± 2.5 | 99.0 | 28.3 ± 1.3 | 292 |
Enantiomeric excess (ee) = 100 × (A − B)/(A + B), where A and B are enantiomers and A is greater than B.
Percentage of conversion (c) = ees/(ees + eep), where subscripts s and p represent remaining ester and product, respectively.
Enantiomeric ratio (E) = ln[1 − c(1 + eep)]/ln[1 − c(1 − eep)].
ND, not detected.
DISCUSSION
The microbial cell surface display system can be used in a wide range of applications as described earlier. Especially, display of active enzyme has been intensively pursued for its potential to be used as a whole-cell biocatalyst in the fields of pharmaceutical, fine chemical, and agrochemical production. To date, however, only a few enzymes, including levansucrase, organophosphorous hydrolase, lipase, dimeric bovine adrenodoxin, and carboxymethylcellulase, have been displayed on the cell surface by using only a small number of different anchoring motifs (17, 18, 21, 26). With an aim to develop a novel system for the display of enzymes, we examined the E. coli outer membrane protein FadL as a potential anchoring motif. Several strategies have been developed to fuse target proteins to the anchoring motif: N-terminal fusion, sandwich fusion, and C-terminal fusion (11, 16, 26, 32). Among them, we employed a C-terminal deletion-fusion strategy, as it allows display of relatively large proteins of up to 60 kDa (21). Successful display of the 44.5-kDa Bacillus sp. strain TG43 lipase by using FadL as an anchoring motif was confirmed by whole-cell activity measurement, immunofluorescence microscopy, and Western blot analysis.
As shown in Fig. 5, 6, and 7, cell surface-displayed lipase showed good enzymatic characteristics. This performance seems to have been due to the displayed pure lipase being stably anchored at the cellular outer membrane in active form, behaving like an immobilized enzyme system. The most remarkable finding with the FadL surface display system is the heat stability. Lyophilized cells displaying lipase were very stable at high temperature (50°C) and retained 90% of full activity after incubation at 50°C for a week (Fig. 7). Display of enzyme on the cell surface often causes instability of the membrane, which consequently causes cell growth defects, lysis, and/or inactivation of cell surface-displayed enzyme (26). However, the lipase displayed by using FadL as an anchor motif showed high heat stability, with no sign of cell growth defects, lysis, or thermal inactivation.
To apply cell surface-displayed lipase in the production of chiral compounds, we carried out enantioselective hydrolysis of racemic methyl mandelate as an example. During the hydrolysis reaction, no significant cell lysis was observed at 37°C (the change of OD600 and supernatant activity during the reaction were negligible), indicating that hydrolysis of methyl mandelate was carried out by cell surface-displayed lipase, not by the free lipase released. As shown in Table 3 optically pure (S)-mandelic acid could be obtained with an enantiomeric excess of 99% and an E value of 292. These results are remarkably higher than those obtained with crude lipase or cross-linked lipase crystal (1, 20). It has been reported that the substrate structure and the origin of lipase mainly determine the reactivity and selectivity in a lipase-catalyzed reaction (13). This is also true for the cell surface-displayed lipase, because (S)-methyl mandelate is the preferred substrate in this reaction. This further suggests that higher reactivity and selectivity for a desired substrate can be achieved by displaying a different lipase highly active towards that substrate.
In this study, we demonstrated that E. coli FadL can be used as an efficient anchoring motif for the display of a relatively large enzyme (44.5 kDa). Also, displayed lipase could be used for enantioselective biocatalysis with high reactivity, enantioselectivity, and enhanced thermal stability. Moreover, because cell surface-displayed lipase can be simply prepared by cultivation and harvesting of recombinant cells, no additional steps for the purification and immobilization of lipase are required. In conclusion, the cell surface-displayed lipase and, more generally, cell surface-displayed enzyme can be used as a cost-effective system for various biocatalytic reactions in the fields of pharmaceuticals, fine chemicals, agrochemicals, and other demanding industries.
Acknowledgments
We thank Peter L. Bergquist, Macquarie University, New South Wales, Australia, for kindly providing the plasmid lipA-pET26b.
This work was supported by MOCIE grants from the Intelligence Bioinformatics and Application Center (TGW10011093) at the KRIBB, the Center for Ultramicrochemical Process Systems sponsored by KOSEF, an LG Chem Chair Professorship, and by the BK21 project. Further support from IBM through the Shared University Research Program is appreciated.
REFERENCES
- 1.Ahmed, S. N., R. J. Kazlauskas, A. H. Morinville, P. Grochulski, J. D. Schrag, and M. W. Cygler. 1994. Enantioselectivity of Candida rugosa lipase toward carboxylic acids: a predictive rule from substrate mapping and X-ray crystallography. Biocatalysis 9:209-225. [Google Scholar]
- 2.Ahn, J. H., J. G. Pan, and J. S. Rhee. 1999. Identification of the tliDEF ABC transporter specific for lipase in Pseudomonas fluorescens SIK W1. J. Bacteriol. 181:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, P. J. L., H. Nevalainen, H. W. Morgan, and P. L. Bergquist. 1999. Rapid cloning of thermoalkalophilic lipases from Bacillus spp. using PCR. Biotechnol. Lett. 21:1003-1006. [Google Scholar]
- 4.Benhar, I. 2001. Biotechnological applications of phage and cell display. Biotechnol. Adv. 19:1-33. [DOI] [PubMed] [Google Scholar]
- 5.Black, P. N. 1988. The fadL gene product of Escherichia coli is an outer membrane protein required for uptake of long-chain fatty acids and involved in sensitivity to bacteriophage T2. J. Bacteriol. 170:2850-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 7.Boder, E. T., and K. D. Wittrup. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553-557. [DOI] [PubMed] [Google Scholar]
- 8.Burton, S. G., D. A. Cowan, and J. M. Woodley. 2002. The search for the ideal biocatalyst. Nat. Biotechnol. 20:37-45. [DOI] [PubMed] [Google Scholar]
- 9.Cristalli, G., C. C. DiRusso, and P. N. Black. 2000. The amino-terminal region of the long-chain fatty acid transport protein FadL contains an externally exposed domain required for bacteriophage T2 binding. Arch. Biochem. Biophys. 377:324-333. [DOI] [PubMed] [Google Scholar]
- 10.DiRusso, C. C., P. N. Black, and J. D. Weimar. 1999. Molecular inroads into the regulation and metabolism of fatty acids, lessons from bacteria. Prog. Lipid Res. 38:129-197. [DOI] [PubMed] [Google Scholar]
- 11.Dröge, M. J., C. J. Rüggeberg, A. M. van der Sloot, J. Schimmel, D. S. Dijkstra, R. M. D. Verhaert, M. T. Reetz, and W. J. Quax. 2003. Binding of phage displayed Bacillus subtilis lipase A to a phosphonate suicide inhibitor. J. Biotechnol. 101:19-28. [DOI] [PubMed] [Google Scholar]
- 12.Faber, K., and M. C. R. Frassen. 1993. Prospects for the increased application of biocatalysts in organic transformations. Trends Biotechnol. 11:461-470. [DOI] [PubMed] [Google Scholar]
- 13.Gais, H. J., and F. Theil. 2002. Hydrolysis and formation of carboxylic acid esters, p. 335-578. In K. Drauz and H. Waldmann (ed.), Enzyme catalysis in organic synthesis: a comprehensive handbook, 2nd ed. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- 14.Georgiou, G., C. Stathopoulos, P. S. Daugherty, A. R. Nayak, B. L. Iverson, and R. I. Curtiss. 1997. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15:29-34. [DOI] [PubMed] [Google Scholar]
- 15.Jaeger, K. E., B. W. Dijkstra, and M. T. Reetz. 1999. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipase. Annu. Rev. Microbiol. 53:315-351. [DOI] [PubMed] [Google Scholar]
- 16.Jose, J., R. Bernhardt, and F. Hannemann. 2001. Functional display of active bovine adrenodoxin on the surface of E. coli by chemical incorporation of the [2Fe-2S] cluster. ChemBioChem 2:695-701. [DOI] [PubMed] [Google Scholar]
- 17.Jose, J., R. Bernhardt, and F. Hannemann. 2002. Cellular surface display of dimeric Adx and whole cell P450-mediated steroid synthesis on E. coli. J. Biotechnol. 95:257-268. [DOI] [PubMed] [Google Scholar]
- 18.Jung, H. C., J. M. Lebeault, and J. G. Pan. 1998. Surface display of Zymomonas mobilis levansucrase by using the ice-nucleation protein of Pseudomonas syringae. Nat. Biotechnol. 16:576-580. [DOI] [PubMed] [Google Scholar]
- 19.Kouker, G., and K. E. Jaeger. 1987. Specific and sensitive plate assay for bacterial lipases. Appl. Environ. Microbiol. 53:211-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lalonde, J. J., M. A. Navia, and A. L. Margolin. 1997. Cross-linked enzyme crystals of lipases as catalysts for kinetic resolution of acids and alcohols. Methods Enzymol. 286:443-464. [Google Scholar]
- 21.Lee, S. Y., J. H. Choi, and Z. Xu. 2003. Microbial cell surface display. Trends Biotechnol. 21:45-52. [DOI] [PubMed] [Google Scholar]
- 22.Li, M. 2000. Applications of display technology in protein analysis. Nat. Biotechnol. 18:1251-1256. [DOI] [PubMed] [Google Scholar]
- 23.Reetz, M. T. 2002. Lipases as practical biocatalysts. Curr. Opin. Chem. Biol. 6:145-150. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Samuelson, P., E. Gunneriusson, P. A. Nygren, and S. Ståhl. 2002. Display of proteins on bacteria. J. Biotechnol. 96:129-154. [DOI] [PubMed] [Google Scholar]
- 26.Shimazu, M., A. Mulchandani, and W. Chen. 2001. Cell surface display of organophosphorus hydrolase using ice nucleation protein. Biotechnol. Prog. 17:76-80. [DOI] [PubMed] [Google Scholar]
- 27.Smith, G. P. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315-1317. [DOI] [PubMed] [Google Scholar]
- 28.Sunna, A., L. Hunter, C. A. Hutton, and P. L. Bergquist. 2002. Biochemical characterization of a recombinant thermoalkalophilic lipase and assessment of its substrate enantioselectivity. Enzyme Microbiol. Technol. 31:472-476. [Google Scholar]
- 29.Svendsen, A. 2000. Lipase protein engineering. Biochim. Biophys. Acta 1543:223-238. [DOI] [PubMed] [Google Scholar]
- 30.Thomas, S. M., R. DiCosimo, and V. Nagarajan. 2002. Biocatalysis: applications and potentials for the chemical industry. Trends Biotechnol. 20:238-242. [DOI] [PubMed] [Google Scholar]
- 31.Wittrup, K. D. 2001. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 12:395-399. [DOI] [PubMed] [Google Scholar]
- 32.Xu, Z., and S. Y. Lee. 1999. Display of polyhistidine peptides on the Escherichia coli cell surface by using outer membrane protein C as an anchoring motif. Appl. Environ. Microbiol. 65:5142-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]