Abstract
A d-erythorbic acid-forming soluble flavoprotein, gluconolactone oxidase (GLO), was purified from Penicillium cyaneo-fulvum strain ATCC 10431 and partially sequenced. Peptide sequences were used to isolate a cDNA clone encoding the enzyme. The cloned gene (accession no. AY576053) exhibits high levels of similarity with the genes encoding other known eukaryotic lactone oxidases and also with the genes encoding some putative prokaryotic lactone oxidases. Analysis of the coding sequence of the GLO gene indicated the presence of a typical secretion signal sequence at the N terminus of GLO. No other targeting or anchoring signals were found, suggesting that GLO is the first known lactone oxidase that is secreted rather than targeted to the membranes of the endoplasmic reticulum or mitochondria. Experimental evidence, including the N-terminal sequence of mature GLO and data on glycosylation and localization of the enzyme in native and recombinant hosts, supports this analysis. The GLO gene was expressed in Pichia pastoris, and recombinant GLO was produced by using the strong methanol-induced AOX1 promoter. In order to evaluate the suitability of purified GLO for production of d-erythorbic acid, we immobilized it on N-hydroxysuccinimide-activated Sepharose and found that the immobilized GLO retained full activity during immobilization but was rather unstable under reaction conditions. Our results show that both soluble and immobilized forms of GLO can, in principle, be used for production of d-erythorbic acid from d-glucono-δ-lactone or (in combination with glucose oxidase and catalase) from glucose. We also demonstrated the feasibility of glucose-d-erythorbic acid fermentation with recombinant strains coexpressing GLO and glucose oxidase genes, and we analyzed problems associated with construction of efficient d-erythorbic acid-producing hosts.
d-Erythorbic acid (d-EA) (d-araboascorbic acid, isovitamin C) is a C-5 epimer of l-ascorbic acid. It has chemical properties very similar to those of ascorbic acid but very low vitamin C activity. d-EA and erythorbates have been used as food antioxidants for many years, particularly in processed meat and soft drinks. Currently, d-EA is manufactured by a two-step process; glucose is first converted into d-2-ketogluconic acid by fermentation, and this is followed by chemical lactonization similar to the lactonization of l-2-ketogulonic acid in the Reichstein process. In the early 1960s, direct conversion of glucose into d-EA by Penicillium fungi was discovered (48). This discovery was followed by an extensive mutagenesis and selection program that resulted in strains capable of converting glucose into d-EA with a yield of approximately 40% over a week-long fermentation (44, 56). These process parameters are still not sufficient to make the direct fermentation of glucose into d-EA economically feasible. In the last decade, much research effort has been spent on development of biotechnology-based processes for l-ascorbic acid manufacture (12, 40). Most of the studies aimed at a two-step process which included fermentation of glucose into l-2-ketogulonic acid and then a chemical lactonization step; production of ascorbic acid directly from glucose by fermentation remains elusive. Since conversion of glucose into d-EA can be done in a single step, this process has the potential of being the lowest-cost technology for production of food antioxidants. To realize this potential, significant improvements in both product yield and process productivity are required. Towards this end, we applied biotechnological techniques to the development of methods for enzymatic and fermentative production of d-EA.
The metabolic pathway from glucose to d-EA in Penicillium is known (30, 47). It comprises two steps; glucose is first oxidized into d-glucono-1,5-lactone by glucose oxidase (GOD) (E.C. 1.1.3.4), and this is followed by further oxidation of d-gluconolactone into d-EA by d-gluconolactone oxidase (GLO) (E.C. 1.1.3.X). While GOD is a well-known enzyme which is widely distributed among fungi, GLO appears to be produced by only a few species of Penicillium (56). GLO has been purified from cell extracts of Penicillium cyaneo-fulvum and characterized biochemically (49). Like other known lactone oxidases, GLO contains covalently bound flavin adenine dinucleotide (FAD) as the prosthetic group (13). A unique property of GLO is that it can use as substrates both glucono-γ-lactone (d-glucono-1,4-lactone) and glucono-δ-lactone (d-glucono-1,5-lactone) even though d-EA itself is a γ-lactone. GLO reacts with glucono-γ-lactone to form d-EA having a γ ring and with glucono-δ-lactone to form an unknown analog having a δ-lactone ring that is nonenzymatically converted to d-EA (30). All other known oxidases acting on aldonolactones are specific only towards γ-lactones. No information on amino acid sequences or the gene structure of GLO is available.
In this study we purified the GLO of P. cyaneo-fulvum, cloned and sequenced the corresponding gene, and characterized the recombinant enzyme. We also confirmed the feasibility of producing d-EA by using recombinant GLO in two ways: by converting glucose to d-EA enzymatically with GOD and recombinant GLO and by direct fermentation with recombinant Pichia pastoris coexpressing both GLO and Aspergillus niger GOD. The results presented below suggest that unlike the other known lactone oxidases, GLO is synthesized in the form of a preprotein having a 20-amino-acid N-terminal secretion signal. This signal sequence can also act as an efficient secretory signal in the yeast Pichia pastoris.
MATERIALS AND METHODS
Materials and microbial strains.
The microbial strains used in this study were P. cyaneo-fulvum ATCC 10431, Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.), and P. pastoris GS115 (Invitrogen, Carlsbad, Calif.). The following cloning vectors were used: pCR2.1-TOPO, pPIC3.5K, pGAPZA, and pGAPZB (all obtained from Invitrogen), as well as pBluescript SK(+/-) (Stratagene). Restriction enzymes were obtained from Boehringer (Mannheim, Germany). The chemicals used were analytical grade.
General biochemical and genetic techniques.
Protein concentrations were determined with the protein assay reagent (Bio-Rad, Richmond, Calif.) by using bovine serum albumin as the standard. Analytical sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed with 12% Tris-glycine Ready gels (Bio-Rad). The gels were stained with Coomassie blue R-250 (41). Endoglycosidase H treatment was performed by incubating 7 μg of purified GLO with 50 mU of endoglycosidase H (Sigma, St. Louis, Mo.) in 10 mM sodium phosphate buffer (pH 6.5) with 0.1 mM EDTA in the presence of 0.05% SDS and 25 mM β-mercaptoethanol at 37°C overnight.
A PCR with degenerate primers was performed by using the following reaction conditions: each primer at concentration of 7.5 μM, 1× PCR buffer with Mg2+ (Boehringer), each deoxynucleoside triphosphate at a concentration of 0.2 mM, 50 mU of Taq polymerase (Boehringer), and 25 ng of P. cyaneo-fulvum genomic DNA. The amplification was performed by using an annealing temperature of 50°C for the first 10 cycles and 53°C for the subsequent 30 cycles. Other PCRs were carried out under similar reaction conditions by using an annealing temperature of 60°C and a total of 30 cycles.
The PCR products were cloned by using a TOPO TA cloning kit (Invitrogen). A DNA sequencing service was purchased from MedProbe S/A (Oslo, Norway).
Yeast transformation was done by electroporation (1). P. pastoris was manipulated according to the instructions in the Multi-Copy Pichia Expression Kit manual (Invitrogen).
Enzyme assays.
GLO activity was measured spectrophotometrically by monitoring the reduction of 2,6-dichlorophenolindophenol (DCIP) by d-EA at 600 nm and 30°C. The reaction mixture (freshly prepared for each experiment) consisted of 50 mM potassium biphthalate buffer (pH 6.2) containing 2 mM hydroxyquinoline, 12 μM DCIP, and 70 mM d-glucono-δ-lactone (added in a 10% stock solution in dimethylformamide). A calibration curve was obtained by adding known amounts of d-EA to the reaction mixture. One unit of activity was defined as 1 μmol of d-EA produced per min under the enzyme assay conditions used.
GOD activity was determined by measuring the formation of H2O2 by the method described by Witteveen et al. (54), and 1 U of GOD activity was defined as 1 μmol of H2O2 oxidized per min under the reaction conditions used.
Purification of GLO.
The native enzyme was purified by using a modification of a procedure described previously (49). Briefly, P. cyaneo-fulvum ATCC 10431 was cultivated for 60 h in a 15-liter fermentor (Biostat E; Medical Braun, Melsungen, Germany) at 30°C with agitation at 300 rpm; the aeration rate was 5 liters/min, and the working volume of a mineral medium described by Yagi et al. (56) was 10 liters. The mycelium was collected by filtration, washed with buffer A (10 mM sodium phosphate buffer [pH 6.5] with 0.1 mM EDTA), and homogenized with a bead beater (Biospec Products, Bartlesville, Okla.) or with a flowthrough bead beater (Netzsch; Feinmahltechnik, Selb, Germany) in the presence of buffer A containing 1 mM phenylmethylsulfonyl fluoride. The homogenate was centrifuged at 19,000 × g and 4°C for 30 min. Eighty milliliters of DEAE-Sepharose FF (Pharmacia Biotech, Uppsala, Sweden) was added per liter of supernatant (conductivity, 2.5 mS/cm) and stirred at 4°C overnight. The resin was removed by filtration, and the enzyme was precipitated from the filtrate by adding ammonium sulfate to 60% saturation. After dialysis against buffer A, the crude GLO preparation (conductivity, 1.2 mS/cm) was fractionated on DEAE-Sepharose FF by using a 5- by 50-cm column. Elution was performed with a linear gradient of 0 to 100 mM sodium chloride in buffer A. The active fractions were pooled, concentrated by ultrafiltration, and applied to a Sephacryl S-300 HR column (1.6 by 200 cm; Pharmacia Biotech) equilibrated and eluted with buffer A. A peak containing the GLO activity was collected and applied to a Bio-Gel HT hydroxyapatite column (1.6 by 20 cm; Bio-Rad) equilibrated with buffer A and eluted with a linear gradient of 0 to 7.5% ammonium sulfate in buffer A. The fractions with the highest specific activity contained essentially homogeneous GLO.
For preparative purification of recombinant GLO, the recombinant P. pastoris strain was cultivated in 15-liter fermentor by using a fed-batch mode essentially as described by Sreekrishna et al. (46). The culture medium containing GLO was concentrated 5- to 10-fold by ultrafiltration, dialyzed extensively (conductivity, 1.2 mS/cm) against buffer A, and stirred overnight with 50 ml of DEAE-Sepharose FF per liter. The resin was collected by filtration, transferred to a column (5 by 50 cm), and eluted with a linear gradient of 0 to 100 mM sodium chloride in buffer A. Active fractions were pooled and subjected to hydroxyapatite chromatography as described above.
Partial sequencing of GLO.
The N-terminal sequence of GLO was obtained by automated Edman degradation of the purified GLO. The single protein band of purified GLO was cut out from the SDS-PAGE gel, and the protein in the gel was alkylated and digested with protease Lys-C. The peptides resulting from protease cleavage were extracted, and their quality was determined by matrix-assisted laser desorption ionization—time of flight mass spectrometry. Partial sequences of GLO were then determined by liquid chromatography-electrospray tandem mass spectrometry. The details of these procedures have been described elsewhere (37).
Isolation of total RNA and Northern blotting.
Three grams of washed mycelium of P. cyaneo-fulvum was ground in a mortar under liquid nitrogen. The mycelium was suspended in 10 ml of ice-cold RNA extraction buffer (4 M guanidine thiocyanate, 0.5% sodium lauroylsarcosine, 25 mM sodium citrate [pH 7], 0.1 M β-mercaptoethanol) and centrifuged for 6 min at 12,000 × g and 4°C. Cesium chloride was added to a final concentration of 0.4 g/ml to the resulting supernatant. The solution was layered carefully on top of 1.2 ml of a 5.7 M cesium chloride-0.1 M EDTA (pH 7) solution and centrifuged at 130,000 × g and 15°C for about 20 h. The pellet was washed quickly with a small amount of water and dissolved in 100 μl of water. For Northern blotting, the DNA probe was radioactively labeled with [α-32P]dCTP (Amersham Life Sciences, Little Chalfont, United Kingdom) by using a random primed DNA labeling kit (Boehringer). Blotting and hybridization were carried out as described by Sambrook et al. (41).
Construction and screening of the cDNA library.
mRNA was isolated from the total RNA of P. cyaneo-fulvum with an Oligotex mRNA midi kit (QIAGEN, Hilden, Germany). A cDNA library of P. cyaneo-fulvum was constructed with a ZAP-cDNA synthesis kit and a ZAP-cDNA Gigapack III Gold cloning kit (both kits were obtained from Stratagene). The phage library was plated on 20 15-cm petri dishes at a density of about 50,000 PFU/plate and transferred onto duplicate nitrocellulose membranes (Protran BA 85; Schleicher & Schuell, Keene, N.H.). Hybridization was performed according to the protocol of Sambrook et al. (41) by using 0.2× SSC-0.2% SDS for 30 min at 65°C as the final washing step (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membranes were exposed to Kodak Biomax MR film (Amersham Life Sciences) with an enhancer screen at −20°C. The positive phage clones were purified and converted to the phagemid form by following the manufacturer's instructions.
Construction of recombinant yeast strains for GLO production.
The DNA fragment corresponding to the coding region of the GLO gene was amplified by PCR by using the oligonucleotide primers oGLOCD51 (5′-CCAACAATTGATGCTGAGCCCTAAGCCGGCTTTCCTGC-3′) and oGLO3 (5′-CAAAGCTTCTAGAGCCTCAGACCACTCATATCACATC-3′). The cloned cDNA plasmid obtained from the cDNA library screening procedure was used as the template. The resulting PCR product was digested with restriction endonucleases XbaI and MfeI and ligated with plasmid pPIC3.5K (Multi-Copy Pichia Expression Kit; Invitrogen) digested with AvrII and EcoRI. The pPIC3.5K(GLO) expression vector contained the complete coding region of the GLO gene under control of the P. pastoris AOX1 promoter.
Construction of recombinant yeast strains for d-EA production.
pPIC3.5K(GLO) was digested with restriction endonucleases SnaBI and NotI, and a 1,481-bp DNA fragment carrying the GLO gene was isolated by agarose electrophoresis. This DNA fragment was ligated with pGAPZA digested with PmlI and NotI, resulting in the expression vector pGAPZA(GLO). To change the selection marker to the HIS4 gene of pPICK3.5K, pGAPZA(GLO) was digested with BglII and NotI. A 1,985-bp DNA fragment was isolated and ligated with the 3,568-bp XhoI-BglII fragment from pPIC3.5K and with the 6,403-bp NotI-XhoI fragment from the same plasmid. The resulting expression vector, pGIC(GLO), contained the GLO gene under control of the GAP promoter instead of the AOX1 promoter of pPIC3.5K. The coding region for the A. niger GOD gene was amplified by PCR by using the oligonucleotide primers oGOE5 (5′-TTGAATTCATGCAGACTCTCCTTGTGAGCTCGCTTG-3′) and oGO3 (5′-TTGCGGCCGCTCACTGCATGGAAGCATAATCTTCC-3′) and A. niger genomic DNA as the template. The resulting PCR product was digested with NotI and partially digested with EcoRI. The purified 1,826-bp fragment was ligated with plasmid pGAPZB digested with NotI and EcoRI. The resulting expression vector, pGAPZB(GOD), contained the complete coding region of the GOD gene under control of the P. pastoris GAP promoter. P. pastoris strain GS115 was transformed with pGIC(GLO), and about 130 transformants were screened for clones expressing the highest levels of GLO activity. One such clone was transformed further with pGAPZB(GOD), and both the GOD and GLO activities were assayed in a random set of about 10 transformants. One clone producing both GOD and GLO activities was used to study the fermentation of glucose into d-EA.
Cultivation of recombinant strains.
Recombinant strains of P. pastoris expressing GLO under control of the AOX1 promoter were grown in a rotary shaker at 30°C and 200 rpm in BMGY medium (10 g of yeast extract per liter, 20 g of peptone per liter, 100 mM potassium phosphate [pH 6], 13.4 g of yeast nitrogen base [Difco, Detroit, Mich.] per liter, 10 g of glycerol per liter, 4 × 10−4 g of biotin per liter). After the cells reached the early stationary phase, they were pelleted and resuspended in BMMY medium (similar to BMGY medium, except that the glycerol was replaced with 5 ml of methanol per liter). Cultivation was continued for 96 h, during which time additional methanol (5 ml/liter) was added to the culture and aliquots were withdrawn for testing the GLO activity after every 24 h of fermentation.
GLO expression level under control of the GAP promoter in P. pastoris/pGIC(GLO) was tested on three different media. The media used were YPD medium, YPD medium buffered with 20 g of CaCO3 per liter, and BMGY medium. The fermentation was carried out in 250-ml Erlenmeyer flasks containing 50 ml of medium at 30°C and 200 rpm. Cultivation was continued for 10 days, during which time aliquots were withdrawn to test the GLO activity after every 24 h of fermentation.
Enzymatic properties of recombinant GLO.
The experiments described below were performed by using purified recombinant GLO. The pH activity profile of GLO was determined by using the following buffers at a concentration of 100 mM in an otherwise standard GLO assay: sodium acetate (pH 5 to 6.5), potassium biphthalate (pH 5 to 6.5), and sodium phosphate (pH 6.5 to 7.5). The pH stability profile was determined by incubating the enzyme overnight at room temperature in the following buffers: 100 mM glycine (pH 2.5 to 4.5), 100 mM sodium acetate (pH 4.5 to 6.5), 100 mM potassium phosphate (pH 6.5 to 8.5), and 100 mM Tris-HCl (pH 8.5 to 9.5). After incubation the residual activity was measured. Thermostability and temperature activity profiles were determined in 50 mM potassium biphthalate buffer (pH 6.2) containing 2 mM hydroxyquinoline. To study thermostability, the enzyme (100-μl aliquots containing approximately 10 U/ml) was incubated for 30 min at temperatures between 25 and 95°C, chilled in an ice-water bath, and assayed immediately. Also, the effects of 2 and 5 mM CaCl2 on the thermostability of GLO were tested by adding an appropriate amount of CaCl2 to the assay buffer. The temperature activity profile was determined by performing the standard GLO assay at a defined temperature for 5 min. The Km for recombinant GLO was calculated by using a Lineweaver-Burk plot (5).
Enzymatic conversion of glucose to d-EA.
A reaction mixture containing 72 U of GOD per ml, 1.2 U of catalase per ml, and 1% glucose in 100 mM potassium phosphate buffer (pH 6.0) was incubated for 1 h at 35°C. Next, an equal volume of a solution containing 0.8 U of GLO per ml in 50 mM potassium biphthalate buffer (pH 6.2) containing 2 mM hydroxyquinoline was added, and the reaction was allowed to proceed for an additional 1 h. The reaction was terminated by freezing the reaction mixture, and d-EA was analyzed spectrophotometrically by measuring the reduction of DCIP or by thin-layer chromatography (TLC) (see below).
TLC.
TLC was performed with Silica Gel 60 plates (Merck, Rahway, N.J.). Before use, the TLC plates were first impregnated with 0.3 M NaH2PO4 and then dried. The mobile phase consisted of acetone, 1-butanol, and H2O (8:1:1) (9). d-EA was detected by spraying the plates with a 0.08% DCIP solution.
Immobilization of the recombinant GLO.
One milliliter of N-hydroxysuccinimide-activated Sepharose 4 FF (Pharmacia, Uppsala, Sweden) was incubated with 0.5 ml of a 0.35-mg/ml solution of purified recombinant GLO in buffer A and adjusted to pH 8. The coupling reaction and the subsequent treatment were carried out according to the instructions of the manufacturer of the resin, and the activity of the immobilized GLO was assayed by the standard enzyme assay. The GLO-Sepharose stability was assayed by a method based on the standard enzyme assay. One milliliter of the GLO-Sepharose resin was added to 9 ml of the standard enzyme assay solution, and the reaction mixture was incubated at 30°C and 200 rpm. Samples were withdrawn at 30-min intervals for 120 min, the GLO-Sepharose was washed twice with buffer A, and the residual GLO activity was measured by the standard GLO assay.
Production of d-EA from glucose by fermentation.
Direct conversion of glucose to d-EA by a P. pastoris strain coexpressing GLO and GOD was studied on the following media: YPD medium containing 2 and 5% glucose and BMDY medium (similar to BMGY medium, except that the glycerol was replaced with 2% glucose). The fermentation was performed in 250-ml Erlenmeyer flasks containing 50 ml of medium at 30°C and 200 rpm. Cultivation was continued for 7 days, during which time aliquots were withdrawn and the d-EA content and activities of both enzymes were analyzed daily.
Nucleotide sequence accession number.
The nucleotide sequence of the open reading frame determined in this study has been deposited in the GenBank database under accession number AY576053.
RESULTS AND DISCUSSION
Cloning of the GLO gene.
GLO was purified to apparent homogeneity from extracts of P. cyaneo-fulvum mycelia by a variation of the procedure described by Takahashi et al. (49). Purified enzyme appeared on an SDS-PAGE gel as a diffuse band at 66 to 90 kDa. This pattern changed to a single sharp band at 56 kDa after purified GLO was treated with endoglycosidase H. By using gel filtration, the molecular mass of native GLO was estimated to be 160 kDa, which is similar to the previous estimate of 150 kDa (49). A comparison of the molecular mass of native GLO with the SDS-PAGE data clearly suggested that GLO is a homodimer. This is an unexpected property for a lactone oxidase since all other known enzymes of this class are monomers (18, 19, 35). The purified enzyme had a specific activity towards d-glucono-1,5-lactone of 6.1 U/mg, which is similar to the previously reported value of 5.6 U/mg (49). The N-terminal amino acid sequence (YRWFNWQFEVTXQSDAYIAPHNEH) of purified GLO was determined, and in addition, three partial or complete sequences of internal peptides were determined (Pep1 [EHDRMTVCGPHFDYNAK], Pep2 [EYICYDEVTDAASCSPQGVV], Pep3 [CQFVNEFLVEQLGITR]) as described in Materials and Methods. Degenerate oligonucleotides were designed by reverse translation of these sequences and used to prime PCR with chromosomal DNA of P. cyaneo-fulvum as the template. The largest PCR product obtained with primers TAYCGITGGTTYAAYTGGCA (sense) and CCIARYTGYTCIACIARRAAYTCRTTIACRAAYTGRCA (antisense) was approximately 1.2 kb long. The fragment was cloned into pCR2.1TOPO and partially sequenced. The partial sequences revealed that this fragment was derived from a gene encoding a protein homologous to FAD-dependent dehydrogenases. By using this DNA fragment as a probe, a cDNA library of P. cyaneo-fulvum was screened by hybridization, and a number of hybridization-positive clones were isolated, purified, and analyzed by restriction mapping. The size of the largest cloned cDNA (1.8 kb) was in accordance with the size of GLO mRNA estimated by Northern blot analysis. This DNA fragment was sequenced and was found to contain a 1,443-bp open reading frame encoding a polypeptide consisting of 480 amino acid residues. The calculated molecular mass of this polypeptide (54.3 kDa) matches the molecular mass of deglycosylated GLO estimated experimentally by SDS-PAGE (56 kDa). The open reading frame is preceded by a 5′ noncoding region consisting of 79 nucleotides and is followed by a 3′ noncoding region consisting of 267 nucleotides. Analysis of the sequence features of the cloned cDNA indicated that it contains a full-length coding sequence of the GLO gene. The deduced amino acid sequence contains the sequenced internal peptides and the sequenced NH2 terminus of the mature GLO. The open reading frame is followed by a 3′ noncoding region ending in a poly(A) tail and includes a putative yeast polyadenylation signal, TATATA (14), approximately at the expected position 43 nucleotides before the poly(A) tail.
Cellular localization of GLO.
The experimentally determined N-terminal sequence of the mature enzyme is within the sequence of GLO starting at position 21. This finding correlates with the analysis of the deduced amino acid sequence of pre-GLO performed with the SignalP-HMM model (http://www.cbs.dtu.dk/services/SignalP) (32). This model predicts the presence of a signal peptide with a probability of 0.999 and places the most probable location of the cleavage site between residues 20 and 21. Further in silico analysis of the GLO amino acid sequence by using two prediction algorithms, PSORTII (http://psort.ims.u-tokyo.ac.jp) (31) and TargetP (http://www.cbs.dtu.dk/services/TargetP) (7), did not reveal any additional sorting or targeting signals. A Kyte-Doolittle hydropathy plot (23) constructed by using a window size of 17 to 21 amino acids did not reveal any transmembrane regions in the mature GLO protein, as it did for amino acid sequences of the known transmembrane proteins l-arabinonolactone oxidase and l-gulonolactone oxidase. Also, no transmembrane regions were found by TMpred (a prediction algorithm for membrane-spanning regions based on the statistical analysis of TMbase, a database of naturally occurring transmembrane proteins [http://www.ch.embnet.org/software/TMPRED_form.html]) (16). Thus, the sequence analysis strongly suggested that GLO is a secreted enzyme. This conclusion was also supported (see below) by recombinant expression of GLO in P. pastoris, in which the native signal sequence was sufficient for complete secretion of GLO. These findings are somewhat surprising since both Takahashi et al. (49) and our group isolated GLO as a soluble enzyme from cell extracts of P. cyaneo-fulvum. Moreover, all other known aldonic acid lactone oxidases show completely different targeting patterns. Animal l-gulonolactone oxidases are integral membrane proteins of the endoplasmic reticulum (6, 20, 39), plant l-galactonolactone oxidases are localized in the inner mitochondrial membrane (25, 34, 45), and yeast l-arabinonolactone oxidases are also membrane-bound mitochondrial enzymes (18). We obtained no evidence of membrane association for GLO. Supplementation of the homogenization buffers with 1% Triton X-100 did not influence the yield of GLO activity. When the insoluble fraction of P. cyaneo-fulvum homogenate was extensively washed with homogenization buffer and subsequently extracted with the same buffer containing Triton X-100, no GLO activity was found in the extract. In the purification of the other lactone oxidases and dehydrogenases detergents have been used to solubilize the enzyme, but both our group and Takahashi et al. (49) were able to purify GLO without detergents.
In fungi, secreted enzymes tend to be glycosylated to a significant extent. This is also true of GLO; as mentioned above, GLO contains 20 to 30% carbohydrate. However, we were able to measure only low GLO activity (0.01 to 0.03 mU/ml) in concentrated culture broth of P. cyaneo-fulvum. According to our crude estimate, about 1/10 of the total GLO activity was secreted under the shake flask fermentation conditions. This is not a very rare property of a secreted enzyme. For a long time another fungal flavoprotein, GOD from A. niger, was believed to be an intracellular (36) or peroxisomal (51) enzyme, while a similar enzyme from Penicillium is secreted (22). However, in later studies workers concluded that a variable fraction (about 20 to 70%) of A. niger GOD is also secreted and the remaining enzyme is distributed between cell wall and intracellular locations (28). A similar variable distribution of enzymes between cell wall and culture medium is common in S. cerevisiae (43). Finally, enzyme kinetics and substrate chemistry suggest that the whole d-EA pathway in Penicillium is located extracellularly. Unlike gulono-γ-lactone and galactono-γ-lactone, glucono-δ-lactone is quite unstable at a neutral or slightly alkaline intracellular pH (29). Gluconolactone is formed outside the fungal cells by GOD (28), and it would be difficult to imagine a mechanism by which it can be transported into the cells without being irreversibly hydrolyzed spontaneously or by the action of gluconolactonase (EC 3.1.1.17) (55). In conclusion, we believe that most probable location of the bulk of GLO in P. cyaneo-fulvum is within the cell wall, and any prospective recombinant host for fermentative production of d-EA should produce GLO in the secreted form.
Homology comparisons and some features of the GLO sequence.
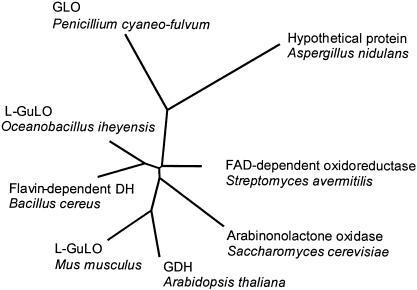
Comparison of the predicted amino acid sequence of GLO to the sequences of other aldonic acid lactone oxidases suggested that GLO is only a distant relative of l-gulono-, l-galactono-, and l-arabinonolactone oxidases (Fig. 1). Notably, there is no indication of a fungal lactone oxidase family. The sequences of the two known fungal lactone oxidases (l-arabinonolactone oxidases from Candida albicans and S. cerevisiae) are no more homologous to the GLO sequence than to the sequence of the rat l-gulonolactone oxidase. The phylogenetic tree in Fig. 1 also includes the sequences of one presumptive fungal (Aspergillus nidulans) and three presumptive prokaryotic (Oceanobacillus iheyensis, Bacillus cereus, and Streptomyces avermitilis) lactone oxidases. These sequences have been identified by a BLAST search of the GenBank sequences as the closest homologues of GLO. There is hardly any information in the literature about the production of ascorbic acid analogues by prokaryotes, and the nonenzymatic defense system against free radicals is believed not to be essential for prokaryotic survival (10, 17). It would be very interesting to investigate whether the oxidases of O. iheyensis, S. avermitilis, and B. cereus are indeed aldonic acid lactone specific and do produce ascorbic acid analogues.
FIG. 1.
Phylogenetic relationships among amino acid sequences of several aldonic acid lactone oxidases and putative FAD-dependent oxidases. L-GuLO, l-gulonolactone oxidase; GDH, l-galactono-1,4-lactone dehydrogenase. The National Center for Biotechnology Information accession numbers for the sequences used for phylogenetic comparison are as follows: Mus musculus l-gulonolactone oxidase, AAR15891; B. cereus flavin-dependent dehydrogenase, NP_830486; S. avermitilis putative FAD-dependent oxioreductases, NP_823585; O. iheyensis l-gulonolactone oxidase, NP_692632; A. nidulans hypothetical protein, EAA64236; S. cerevisiae l-galactono-γ-lactone oxidase, BAA23804; and Arabidopsis thaliana putative l-galactone-1,4-lactone dehydrogenase, BAC42562.
Previous studies indicated that FAD is covalently attached to GLO (13). According to a National Center for Biotechnology Information conserved domain database search (26), the deduced amino acid sequence of GLO has a conserved putative binding site for covalently bound FAD of oxygen-dependent oxioreductases (PROSITE pattern PS00862). This domain corresponds to amino acid residues 34 to 196 in GLO and can also be found in d-arabinono-1,4-lactone oxidase and l-gulonolactone oxidase. In the domains of d-arabinono-1,4-lactone oxidase and l-gulonolactone oxidase histidine residues 56 and 54, respectively, are thought to be the sites of cofactor attachment (18, 19). This histidine (His-67) is also conserved in the conserved domain of GLO.
Based on the experimentally observed amount of N-linked oligosaccharides in GLO (20 to 30%), the predicted amino acid sequence of GLO was expected to contain multiple consensus sequences for N-glycosylation sites (Asn-X-Ser/Thr). Eight such sites are in the GLO deduced amino acid sequence.
Overexpression of GLO in P. pastoris.
Taxonomically related filamentous fungi with established gene expression systems (such as A. niger) seem to be an obvious choice as hosts for recombinant production of GLO and construction of metabolically engineered strains to produce d-EA by fermentation. However, an analysis of the previously published data indicated that A. niger grown aerobically on glucose-containing media produces extremely high levels of d-gluconolactonase, about 40 U/mg (dry weight) (55). This is about 30 times higher than the level of GOD activity produced by the same host and about 104 times higher than the GLO expression level in wild-type P. cyaneo-fulvum. We tried to solve this problem by searching for strains of filamentous fungi which have significant GOD activity but little or no d-gluconolactonase activity. Unfortunately, both literature searches and a small-scale laboratory screening (data not shown) failed to identify suitable candidates. Gluconolactonase activity is also found in yeast (2), but the enzyme is located only in the yeast cytosol and is present at a level that is remarkably lower than the levels in filamentous fungi. We tested the culture medium of P. pastoris for gluconolactonase activity and were not able to detect any activity. Therefore, we decided to use P. pastoris as a model host for GLO production and to study the feasibility of direct fermentation of glucose into d-EA. The results of the studies of expression of both the GLO and A. niger GOD genes under control of different promoters in P. pastoris are summarized in the Table 1. The level of GLO expression under control of the AOX1 promoter in a shake flask culture was 72-fold higher than the level of GLO expression in a fermentor culture of wild-type P. cyaneo-fulvum, about 25 mg/liter. Both extra- and intracellular GLO activities were determined, and the activity was found only in the culture medium. No GLO activity was found in the fraction of lysed cells containing both intracellular protein and protein from the cell wall.
TABLE 1.
Levels of GLO expression in wild-type P. cyaneo-fulvum and in recombinant P. pastoris and level of expression of GOD in recombinant P. pastorisa
| Organism | Construct | Promoter | Enzyme | Activity (U/liter) |
|---|---|---|---|---|
| P. cyaneo-fulvum | Wild type | GLO | GLO | 5.0 ± 2.1 |
| P. pastoris | pPICK3.5K(GLO) | AOX1 | GLO | 360 ± 21 |
| P. pastoris | pGIC(GLO) | GAP | GLO | 65 ± 6.4 |
| P. pastoris | pGAPZB(GOD) | GAP | GOD | 12,200 ± 720 |
The data were derived from at least three separate expression experiments.
Characterization of recombinant GLO produced in P. pastoris.
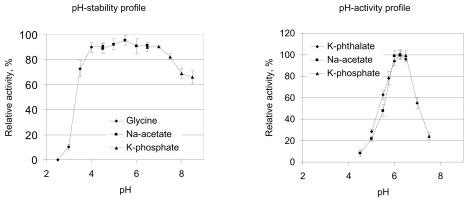
Recombinant GLO produced by P. pastoris/pPIC3.5K(GLO) was purified to homogeneity by a combination of DEAE-Sepharose and hydroxyapatite chromatography as described in Materials and Methods. Several properties of the recombinant enzyme were studied. Recombinant heterologous (particularly fungal) enzymes expressed in yeast cells are commonly known to be overglycosylated. This was also expected to be the case for GLO, which is extensively glycosylated in its native host and has in its sequence eight consensus sites for attachment of N-linked oligosaccharides. However, the glycosylation level of recombinant GLO produced by P. pastoris was similar to that of native GLO. The molecular mass of the recombinant GLO as determined by SDS-PAGE was found to be 66 to 80 kDa, and after endoglycosidase H treatment the molecular mass was approximately 54 kDa, which corresponds to the molecular mass of the mature 52.2-kDa GLO predicted on the basis of DNA translation. The pH stability profile of recombinant GLO (Fig. 2) was found to be essentially similar to that of the native GLO (49). The optimum pH of both recombinant and native GLO was found to be 6.25 (Fig. 2), which differs slightly from the optimum pH reported by Takahashi et al. (49) (pH 5.6 to 6). The temperature stability of recombinant GLO was measured and was found to be rather typical for an enzyme derived from a mesophilic microorganism, and the optimum temperature was 42°C. At temperatures above 45°C the activity was quickly lost. Addition of 2 or 5 mM Ca2+ did not have any effect on the enzyme's temperature stability. The recombinant enzyme exhibited rather good storage stability, like the native GLO. Both forms of the enzyme remained fully active after eight cycles of freezing and thawing. No loss of activity was detected after several months of storage at −20°C.
FIG. 2.
(Left panel) pH stability profile of recombinant GLO. (Right panel) pH activity profile of recombinant GLO. The different buffers used are indicated (see Materials and Methods for details). Each data point is the mean for three experiments, and the error bars indicate standard deviations.
Somewhat surprisingly, the specific activity of recombinant GLO (approximately 14.4 U/mg of protein) was found to be more than two times higher than that of the native enzyme. The Km values for d-gluconolactone were very similar for the two forms of the enzyme (2.2 ± 0.2 mM for the recombinant enzyme and 1.7 ± 0.2 mM for the native enzyme, as determined by Takahashi et al. [49]). One possible explanation for the higher specific activity of the recombinant GLO is that the native enzyme is partially inactivated by hydrogen peroxide that is produced during P. cyaneo-fulvum fermentation by GOD.
In native yeast d-arabinono-1,4-lactone oxidase, FAD is covalently attached, while the same enzyme produced in E. coli contains noncovalently bound FAD (24). We compared the attachment modes of FAD in native and recombinant GLO using a fluorescence-based method (33) and found no difference between the two forms of enzyme, which suggests that both forms contained only covalently bound FAD. This conclusion was confirmed by the fact that no activation of GLO was observed when additional free FAD (0.1 to 1 mM) was added to the assay mixture for GLO.
The effects of d-glucose and d-gluconic acid on GLO activity are important because these compounds are expected to be present in the reaction mixtures of any prospective d-EA manufacturing process involving GLO. We tested several concentrations of glucose (up to 25%) and gluconic acid (up to 5%) and found no inhibition of GLO.
Production of d-EA by recombinant GLO.
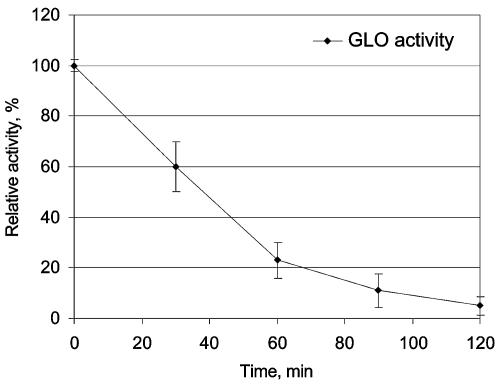
Enzymatic conversion of glucose to d-EA was demonstrated by using purified recombinant GLO in a reaction mixture containing commercial GOD and catalase. The reaction was performed without pH control. Under the conditions described in Materials and Methods 33% conversion (yield, 0.1 g/liter) of glucose and 70% conversion of gluconolactone to d-EA were obtained, which is similar to the results that Takahashi et al. obtained with native GLO (49). One of the most likely limiting factors for the reaction yield was the decrease in pH to values below 4, which was mainly caused by the hydrolysis of gluconolactone (we were able to detect lactonase activity in commercial preparations of glucose oxidase and catalase) to gluconic acid and also by the formation of d-EA from glucose. The possibility of immobilizing both GOD and catalase and using them in various applications has been studied extensively (27, 38, 50, 53). Using immobilized GLO together with GOD and catalase in an enzymatic process in order to catalyze the conversion of glucose to d-EA is one possible method for d-EA production. We found that GLO retains nearly 100% of its enzymatic activity when it is coupled to N-hydroxysuccinimide-activated Sepharose. However, the inactivation of the immobilized GLO turned out to be considerable during the conversion reaction (Fig. 3). This inactivation could have been due to the H2O2 formed during the enzymatic reaction. H2O2 has previously been shown to cause inactivation of immobilized GOD, and the extent of this inactivation was decreased by removal of H2O2 from the microenvironment of GOD by coimmobilized catalase (11, 38).
FIG. 3.
Reaction stability of immobilized GLO as determined by studying conversion of gluconolactone to d-EA for 120 min. Samples were withdrawn at 30-min intervals, immobilized GLO was washed, and the residual GLO activity was measured. Each data point is the mean for three experiments, and the error bars indicate standard deviations.
Coexpression of GLO and GOD.
Because the AOX1 promoter is repressed by glucose, it cannot be used in hosts designed for efficient conversion of glucose into d-EA by fermentation. The glyceraldehyde-3-phosphate dehydrogenase (GAP) promoter has been reported not to be repressed by glucose and to be comparable in strength to the AOX1 promoter in P. pastoris (42, 52). In our study, the level of expression of GLO under control of the GAP promoter was five to six times lower than the level of expression of GLO under control of the AOX1 promoter (Table 1), and the level of expression obtained with the GAP promoter when glycerol was used as a carbon source was approximately 10 times higher than the level of expression obtained when glucose was used as a carbon source in an otherwise similar medium. Previously published findings suggest that the GOD gene can be expressed in yeast quite efficiently (4, 8, 15), and this suggestion was supported by our study. We observed almost 10-fold-higher levels of GOD (30 to 40 mg/liter) than of GLO (4 to 5 mg/liter) when the two genes were expressed under control of the same promoter (GAP) and under the same cultivation conditions. The imbalance in the levels of activity of the two enzymes was even greater due to the higher specific activity of purified GOD (approximately 360 U/mg) than of purified GLO (14.4 U/mg for the yeast-derived GLO). The GOD activity gained (12,200 U/liter) was over 185-fold greater than the activity of GLO (65 U/liter). The highest GOD and GLO activities under control of the GAP promoter were observed in BMGY medium after 7 and 8 days of growth, respectively.
Production of d-EA by fermentation of glucose with recombinant P. pastoris expressing the GLO and GOD genes.
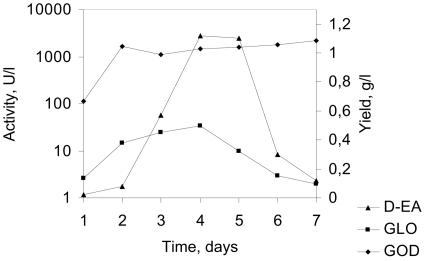
Production of d-EA from glucose by direct fermentation of P. pastoris strains coexpressing both the GLO and GOD genes was carried out in shake flasks by using several different glucose-containing media (YPD medium with 2 or 5% glucose and BMDY medium). The highest conversion yield observed in these experiments was 12% (yield, 5.9 g/liter) after 144 h of cultivation in YPD medium containing 5% glucose. The productivity obtained with recombinant P. pastoris was 0.04 g liter−1 h−1. The most likely reason for the rather inefficient productivity is the imbalance in the activity levels of GLO and GOD (Fig. 4) and, as a result of this, accumulation of gluconolactone and the practically irreversible spontaneous hydrolysis of the lactone ring. The difference in the activity levels of GLO and GOD in a typical conversion experiment was nearly 40-fold in favor of GOD. Also, the absence of a sufficient amount of catalase activity may have had an effect on the half-lives of both enzymes. Especially GLO was found to be rather unstable in the reaction conditions used (Fig. 3). In any case, this result proves our concept, which is the fermentation of d-EA from glucose in yeast, although strain engineering work on the coexpression of GLO, GOD, and catalase and work to investigate high-cell-density fermentation optimization are needed to obtain P. pastoris strains that produce sufficient amounts of d-EA. In the ideal strain for production of d-EA endogenous GOD and catalase would be efficiently expressed at high glucose concentrations, but the strain would be devoid of gluconolactonase activity. Penicillium and Aspergillus are known to produce endogenous glucose oxidase and catalase but also gluconolactonase. In A. niger the gluconolactonase is known to exist in the form of isoenzymes (3). This finding is supported by the sequencing data for two other filamentous fungi, A. nidulans and Neurospora crassa. According to the GenBank database, these two organisms both possess two separate previously uncharacterized hypothetical proteins whose sequences are homologous to Fusarium oxysporum lactonohydrolase amino acid sequences (21). This information provides a good basis for pathway engineering of second-generation d-EA-producing strains, possibly including a strain of filamentous fungi in which GLO is strongly secreted and gluconolactonase activity is abolished.
FIG. 4.
Typical scheme for the imbalance between the expression levels of GLO and GOD during d-EA production in YPD medium (containing 5% glucose) by recombinant P. pastoris. Cultivation was performed in shake flasks containing 50 ml of the medium at 30°C and 200 rpm. The data are the data from one experiment in a series of cultivations as described in Materials and Methods. The imbalance between the two recombinant enzymes was observed to be essentially the same in these experiments.
Acknowledgments
We thank M. Karhunen and S. Jaakola for help with experiments and I. Ylikangas for assistance with laboratory work. We also thank H. Ojamo for stimulating discussions, C. Bridges and A. Morgan for support, and A. Nyyssölä and V. Kumar for critically reading the manuscript.
REFERENCES
- 1.Becker, D., and L. Guarente. 1991. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 194:182-187. [DOI] [PubMed] [Google Scholar]
- 2.Brodie, A. F., and F. Lipman. 1955. Identification of a gluconolactonase. J. Biol. Chem. 212:677-685. [PubMed] [Google Scholar]
- 3.Bruchmana, E. E., H. Schach, and H. Graf. 1987. Role and properties of lactonase in a cellulase system. Biotechnol. Appl. Biochem. 9:146-159. [Google Scholar]
- 4.De Baetselier, A., A. Vasavada, P. Dohet, V. Ha-Thi, M. De Beukelaer, T. Erpicum, L. De Clerck, J. Hanotier, and S. Rosenberg. 1991. Fermentation of a yeast producing A. niger glucose oxidase: scale-up, purification and characterization of the recombinant enzyme. Biotechnology 9:559-561. [DOI] [PubMed] [Google Scholar]
- 5.Dixon, M., and E. C. Webb. 1979. Enzymes, 3rd ed. Longman, London, United Kingdom.
- 6.Eliceiri, G. L., E. K. Lai, and P. B. McCay. 1969. Gulonolactone oxidase. J. Biol. Chem. 244:2641-2645. [PubMed] [Google Scholar]
- 7.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 8.Frederick, K., J. Tung, R. Emerick, F. Masiarz, S. Chamberlain, A. Vasavada, and S. Rosenberg. 1990. Glucose oxidase from Aspergillus niger cloning, gene sequence, secretion from Saccharomyces cerevisiae and kinetic analysis of a yeast-derived enzyme. J. Biol. Chem. 265:3793-3802. [PubMed] [Google Scholar]
- 9.Ghebregzabher, M., S. Rufini, B. Monaldi, and M. Lato. 1976. Thin-layer chromatography of carbohydrates. J. Chromatogr. 127:133-162. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg, J. T., and B. Demple. 1986. Glutathione in Escherichia coli is dispensable for resistance to H2O2 and gamma radiation. J. Bacteriol. 168:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenfield, P. F., J. R. Kittrell, and R. L. Laurence. 1975. Inactivation of immobilized glucose oxidase by hydrogen peroxide. Anal. Biochem. 65:109-124. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. D., and R. Viola. 2002. Biotechnological approaches for l-ascorbic acid production. Trends Biotechnol. 20:299-305. [DOI] [PubMed] [Google Scholar]
- 13.Harada, Y., M. Shimizu, S. Murakawa, and T. Takahashi. 1979. Identification of FAD of d-gluconolactone dehydrogenase: d-erythorbic acid producing enzyme of Penicillium cyaneo-fulvum. Agric. Biol. Chem. 43:2635-2636. [Google Scholar]
- 14.Helden, J., M. Olmo, and J. Perez-Ortin. 2000. Statistical analysis of putative yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 28:1000-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodgins, M., D. Mead, D. J. Balance, A. Goodey, and P. Sudbery. 1993. Expression of the glucose oxidase gene from Aspergillus niger in Hansenula polymorpha and its use as a reporter gene to isolate regulatory mutations. Yeast 9:625-635. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, K., and W. Stoffel. 1993. TMBASE—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347:166-170. [Google Scholar]
- 17.Huh, W., B. Lee, S. Kim, Y. Kim, G. Rhie, Y. Baek, C. Hwang, J. Lee, and S. Kang. 1998. d-Erythroascorbic acid is an important molecule in Saccharomyces cerevisiae. Mol. Microbiol. 30:895-903. [DOI] [PubMed] [Google Scholar]
- 18.Huh, W. H., S. T. Kim, K. S. Yang, Y. J. Seok, Y. C. Hah, and S. O. Kang. 1994. Characterisation of d-arabinono-1,4-lactone oxidase from Candida albicans ATCC 10231. Eur. J. Biochem. 225:1073-1079. [DOI] [PubMed] [Google Scholar]
- 19.Imai, T., S. Karita, G. Shiratori, M. Hattori, T. Nunome, K. Oba, and M. Hirai. 1998. l-Galactone-γ-lactone dehydrogenase from sweet potato: purification and cDNA sequence analysis. Plant Cell Physiol. 39:1350-1358. [DOI] [PubMed] [Google Scholar]
- 20.Kiuchi, K., M. Nishikimi, and K. Yagi. 1982. Purification and characterisation of l-gulonolactone oxidase from chicken liver microsomes. Biochemistry 21:5076-5082. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, M., M. Shinohara, C. Sakoh, M. Kataoka, and S. Shimizu. 1998. Lactone-ring-cleaving enzyme: genetic analysis, novel RNA editing and evolutionary implications. Proc. Natl. Acad. Sci. USA 95:12787-12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusai, K., I. Sekuzu, B. Hagihara, K. Okunuki, S. Yamaguchi, and M. Nakai. 1960. Crystallisation of glucose oxidase from Penicillium amagasakiense. Biochim. Biophys. Acta 40:555-557. [DOI] [PubMed] [Google Scholar]
- 23.Kyte, J., and R. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 24.Lee, B., W. Huh, S. Kim, J. Lee, and S. Kang. 1999. Bacterial production of d-erythroascorbic acid and l-ascorbic acid through functional expression of Saccharomyces cerevisiae d-arabinono-1,4-lactone oxidase in Escherichia coli. Appl. Environ. Microbiol. 65:4685-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mapson, L. W., and E. Breslow. 1958. Biological synthesis of l-ascorbic acid: l-galactono γ-lactone dehydrogenase. Biochem. J. 68:359-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, R. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messing, R. A. 1974. Simultaneously immobilized glucose oxidase and catalase in controlled-pore titania. Biotechnol. Bioeng. 16:897-908. [DOI] [PubMed] [Google Scholar]
- 28.Mischak, H., C. P. Kubichek, and M. Röhr. 1985. Formation and location of glucose oxidase in citric acid producing mycelia of Aspergillus niger. Appl. Microbiol. Biotechnol. 21:27-31. [Google Scholar]
- 29.Mitchell, R. E., and F. R. Duke. 1970. Kinetics and equilibrium constants of the gluconic acid-gluconolactone equilibrium. Ann. N. Y. Acad. Sci. 172:129-138. [Google Scholar]
- 30.Murakawa, S., and T. Takahashi. 1977. Biosynthesis of a new ascorbic acid analogue by d-gluconolactone dehydrogenase of Penicillium cyaneo-fulvum. Agric. Biol. Chem. 41:2103-2104. [Google Scholar]
- 31.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-35. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen, H., S. Brunak, and G. von Heijne. 1999. Machine learning approaches to the prediction of signal peptides and other protein sorting signals. Protein Eng. 12:3-9. [DOI] [PubMed] [Google Scholar]
- 33.Nishikimi, M., K. Kiuchi, and K. Yagi. 1977. Detection of l-gulono-γ-lactone oxidase on SDS-polyacrylamide gels by the fluorescence of its covalently bound flavin. FEBS Lett. 81:323-325. [DOI] [PubMed] [Google Scholar]
- 34.Oba, K., S. Ishikawa, M. Nishikawa, H. Mizuno, and T. Yamomoto. 1995. Purification and properties of l-galactono-γ-lactone dehydrogenase, a key enzyme for ascorbic acid biosynthesis, from sweet potato roots. J. Biochem. 117:120-124. [DOI] [PubMed] [Google Scholar]
- 35.Ostergaard, J., G. Persiau, M. Davey, G. Bauw, and M. Montagu. 1997. Isolation of a cDNA coding for l-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. J. Biol. Chem. 272:30009-30016. [DOI] [PubMed] [Google Scholar]
- 36.Pazur, J. H. 1966. Glucose oxidase from Aspergillus niger. Methods Enzymol. 9:82-86. [Google Scholar]
- 37.Poutanen, M., L. Salusjärvi, L. Ruohonen, M. Penttilä, and N. Kalkkinen. 2001. Use of matrix assisted laser desorption/ionization time-of-flight mass mapping and nanospray liquid chromatography/electrospray ionization tandem mass spectrometry sequence tag analysis for high sensitivity identification of yeast proteins separated by two-dimensional gel electrophoresis. Rapid Commun. Mass Spectrom. 15:1685-1692. [DOI] [PubMed] [Google Scholar]
- 38.Prenosil, J. E. 1979. Immobilized glucose oxidase-catalase and their deactivation in a differential-bed loop reactor. Biotechnol. Bioeng. 2:89-109. [DOI] [PubMed] [Google Scholar]
- 39.Puskas, F., L. Braun, M. Csala, T. Kardon, P. Marcolongo, A. Bendetti, J. Mandl, and G. Banhegyi. 1998. Gulonolactone oxidase activity-dependent intravesicular glutathione oxidation in rat liver microsomes. FEBS Lett. 430:293-296. [DOI] [PubMed] [Google Scholar]
- 40.Saito, Y., Y. Ishii, H. Hayashi, K. Yoshikawa, Y. Noguchi, S. Yoshida, S. Soeda, and M. Yoshida. 1998. Direct fermentation of 2-keto-l-gulonic acid in recombinant Gluconobacter oxydans. Biotechnol. Bioeng. 58:309-315. [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sears, I., J. O'Connor, O. Rossanese, and B. Glick. 1998. A versatile set of vectors for constitutive and regulated gene expression in Pichia pastoris. Yeast 14:783-790. [DOI] [PubMed] [Google Scholar]
- 43.Sheckman, R., and P. Novick. 1982. The secretory process and yeast cell-surface assembly, p. 361-398. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Shimizu, K., K. Nishiyama, T. Inoue, N. Takano, M. Mikata, Y. Masataka, T. Azuma, and S. Osawa. 1967. Studies on erythorbic acid production by fermentation. Part II. Erythorbic acid production by jar fermentor. Agric. Biol. Chem. 31:346-352. [Google Scholar]
- 45.Siendones, E., J. A. González-Reyes, C. Santos-Oscaña, P. Navas, and F. Córdoba. 1999. Biosynthesis of ascorbic acid in kidney bean. l-Galactonolactone dehydrogenase is an intrinsic protein localised in mitochondrial inner membrane. Plant Physiol. 120:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sreekrishna, K., L. Nelles, R. Potenz, J. Cruze, P. Mazzaferro, W. Fish, M. Fuke, K. Holden, D. Phelps, P. Wood, and K. Parker. 1989. High-level expression, purification and characterization of recombinant human tumor necrosis factor synthesized in the methylotrophic yeast Pichia pastoris. Biochemistry 28:4117-4125. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi, T. 1969. Erythorbic acid fermentation. Biotechnol. Bioeng. 11:1157-1171. [Google Scholar]
- 48.Takahashi, T., M. Mitsumoto, and H. Kayamori. 1960. Production of d-araboascorbic acid by Penicillium. Nature 188:411-412. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi, T., H. Yamashita, E. Kato, M. Mitsumoto, and S. Murakawa. 1976. Purification and some properties of d-glucono-γ-lactone dehydrogenase d-erythorbic acid producing enzyme of Penicillium cyaneo-fulvum. Agric. Biol. Chem. 40:121-129. [Google Scholar]
- 50.Tzanov, T., S. A. Costa, G. M. Gubitz, and A. Cavaco-Paulo. 2002. Hydrogen peroxide generation with immobilized glucose oxidase for textile bleaching. J. Biotechnol. 93:87-94. [DOI] [PubMed] [Google Scholar]
- 51.Van Dijken, J. P., and M. Veenhuis. 1980. Cytochemical localisation of glucose oxidase in peroxisomes of Aspergillus niger. Eur. J. Appl. Microbiol. Biotechnol. 9:275-283. [Google Scholar]
- 52.Waterham, H., M. Digan, P. Koutz, S. Lair, and J. Cregg. 1997. Isolation of the Pichia pastoris glyceraldehydes-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 186:37-44. [DOI] [PubMed] [Google Scholar]
- 53.Weibel, M. K., and H. J. Bright. 1971. Insolubilized enzymes. Kinetic behaviour of glucose oxidase bound to porous glass particles. Biochem. J. 124:801-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Witteveen, C. F. B., P. van de Vondervoort, K. Swart, and J. Visser. 1990. Glucose oxidase overproducing and negative mutants of Aspergillus niger. Appl. Microbiol. Biotechnol. 33:683-686. [Google Scholar]
- 55.Witteveen, F. B., P. J. van de Vondervoort, H. C. van den Broeck, A. C. van Engelenburg, L. H. de Graaff, M. H. Hillebrand, P. J. Schaap, and J. Visser. 1993. Induction of glucose oxidase, catalase, and lactonase in Aspergillus niger. Curr. Genet. 24:408-416. [DOI] [PubMed] [Google Scholar]
- 56.Yagi J., T. Yamashita, K. Kati, Y. Takagi, and H. Sakai. 1967. Studies on erythorbic acid production by fermentation. Part I. Erythorbic acid producing strain and culture conditions. Agric. Biol. Chem. 31:340-345. [Google Scholar]