Abstract
Natural attenuation of the environmental contaminant perchlorate is a cost-effective alternative to current removal methods. The success of natural perchlorate remediation is dependent on the presence and activity of dissimilatory (per)chlorate-reducing bacteria (DPRB) within a target site. To detect DPRB in the environment, two degenerate primer sets targeting the chlorite dismutase (cld) gene were developed and optimized. A nested PCR approach was used in conjunction with these primer sets to increase the sensitivity of the molecular detection method. Screening of environmental samples indicated that all products amplified by this method were cld gene sequences. These sequences were obtained from pristine sites as well as contaminated sites from which DPRB were isolated. More than one cld phylotype was also identified from some samples, indicating the presence of more than one DPRB strain at those sites. The use of these primer sets represents a direct and sensitive molecular method for the qualitative detection of (per)chlorate-reducing bacteria in the environment, thus offering another tool for monitoring natural attenuation. Sequences of cld genes isolated in the course of this project were also generated from various DPRB and provided the first opportunity for a phylogenetic treatment of this metabolic gene. Comparisons of the cld and 16S ribosomal DNA (rDNA) gene trees indicated that the cld gene does not track 16S rDNA phylogeny, further implicating the possible role of horizontal transfer in the evolution of (per)chlorate respiration.
Widespread perchlorate contamination in the United States, especially throughout the southwest, is a direct effect of unregulated ammonium perchlorate disposal practices from 1950 to 1997 (22). While the debate continues over the final maximum concentration limit to be set for perchlorate, the Environmental Protection Agency (EPA), the Department of Defense, the Department of Energy, and the National Aeronautics and Space Industry have asked the National Science Association to review the EPA 2002 Draft Health Assessment on Perchlorate (29). Regardless of this debate, the neurodevelopmental toxicity of perchlorate has been established (29), and methods of perchlorate removal are actively being pursued, with bioremediation technologies emerging as a cost-effective and less-invasive alternative to physical or chemical practices (28).
Within the last 7 years, more than 40 different strains of dissimilatory (per)chlorate-reducing bacteria (DPRB) have been isolated from a diverse range of environments (5, 6, 13, 16, 18, 23, 31). Because of the metabolic capability and ubiquity of DPRB (6), natural attenuation of perchlorate is garnering more and more interest. While studies by various groups have shown the ability of microbes to remediate perchlorate under environmental conditions (10, 13, 27), a quick, reliable method for detecting DPRB is needed to determine the natural attenuation candidacy of a contaminated site as well as for monitoring active degradation.
Traditionally, contaminant site evaluation for the presence of DPRB is done by enumeration and isolation. However, it is well known that cultivation techniques are time consuming and often prove unsuccessful in isolating the target bacteria due to both media selectivity and organism culturability (7, 11). To alleviate the limitations of cultivation-based methods, molecular techniques using the 16S rRNA gene have been employed to examine bacterial diversity in the environment (2, 20), and numerous primer sets have been developed for the 16S rRNA gene that target specific groups of bacteria. However, based on the phylogenetic diversity of DPRB and their close phylogenetic relationships to non-(per)chlorate-reducing relatives, detection of DPRB using 16S ribosomal DNA (rDNA) primers is problematic (1). A more inclusive approach would be to target a gene essential to the metabolic pathway. A primer set for a metabolic gene not only would be of value for the detection of DPRB in the environment but could be used in RNA-based studies to assess functional activity in situ.
Metabolic primer sets have been applied to a variety of bioremediative studies for the detection of specific bacteria. For example, since many denitrifiers are able to degrade toluene and xylene, Braker and colleagues developed primer sets targeting two nitrate reductase genes that allowed for the qualitative detection of denitrifiers in the environment (4). And while primers for the catechol 2,3-dioxygenase were used to detect bacteria capable of aerobically degrading benzene, toluene, and xylene, they were also used in quantitative PCR to show an increase in gene copy number after soil samples were amended with petroleum (17).
An ideal target for the environmental detection of DPRB is the chlorite dismutase gene, cld. This is based on previous studies indicating that chlorite dismutation is essential to the (per)chlorate reduction pathway (5, 6, 30). To date, no other enzyme has been isolated that is capable of converting chlorite to oxygen and chloride ions. Hybridization analysis using a chlorite dismutase immunoprobe indicated that all DPRB tested possess the chlorite dismutase enzyme, and the chlorite dismutase antibody did not bind to close non-(per)chlorate-reducing relatives (19). Similarly, a DNA probe targeting the cld gene only hybridized to genomic DNA (gDNA) from DPRB and the non-(per)chlorate reducer Magnetospirillum magnetotacticum. The probe did not hybridize to any other close phylogenetic relatives incapable of (per)chlorate reduction (3). [The M. magnetotacticum anomaly was explained by subsequent genome analysis which indicated the presence of the cld gene in the M. magnetotacticum genome (3) but the absence of the (per)chlorate reductase genes, thus rendering this organism incapable of reducing (per)chlorate.] These studies suggest that the chlorite dismutase gene is unique to and required by all DPRB. Thus, a metabolic primer set targeting this gene would be useful for molecular detection of DPRB in the environment. However, the efficacy of this metabolic primer set is dependent upon regions of sequence conservation within the cld gene, information which is currently unavailable due to the paucity of cld gene sequences in the database. Sequence analysis of the cld gene would also allow phylogenetic inferences to be drawn, providing data to support or refute the hypothesis that (per)chlorate metabolism is horizontally transferred. Here we report the development of cld primer sets for the environmental detection of (per)chlorate-reducing bacteria as well as the first phylogenetic analysis of the cld gene.
MATERIALS AND METHODS
Bacterial strains, environmental samples, and DNA extraction.
The bacterial strains and environmental samples used in this study are listed in Table 1. Genomic DNA from pure cultures was extracted by using the PUREGENE DNA isolation kit (Gentra Systems Inc., Minneapolis, Minn.). DNA was extracted from environmental samples by using the Fast DNA Spin kit for soil (Qbiogene, Carlsbad, Calif.). DNA for PCR from the Los Alamos well most-probable-number samples was obtained by harvesting the cell pellet from 1.5 ml of culture, adding 40 μl of sterile H20 and 5 μl of chloroform, and lysing the cells by heating them at 95°C for 10 min.
TABLE 1.
Bacterial strains and environmental samples used in this study
| Origin of DNAs tested | Description | Reference or source |
|---|---|---|
| Strains | ||
| Dechloromonas agitata | (Per)chlorate reducer | 6 |
| “Dechloromonas aromatica” | (Per)chlorate reducer | 6 |
| Dechloromonas sp. strain LT1 | (Per)chlorate reducer | J. Coates |
| Dechlorosoma suillum | (Per)chlorate reducer | 6 |
| “Dechloromarinus chlorophilus” strain NSS | Chlorate reducer | 6 |
| “Dechlorospirillum anomalous” strain WD | (Per)chlorate reducer | 6 |
| Pseudomonas sp. strain PK | Chlorate reducer | 6 |
| Strain CR | (Per)chlorate reducer | J. Coates |
| Dechlorospirillum sp. strain DB | (Per)chlorate reducer | J. Coates |
| Ideonella dechloratans | Chlorate reducer | ATCC 51718 |
| Pseudomonas stutzeri | Non-perchlorate reducer | ATCC 14405 |
| Magnetospirillum magnetotacticum | Non-perchlorate reducer | D. Bazylinski |
| Rhodocyclus tenuis | Non-perchlorate reducer | M. Madigan |
| Escherichia coli | Non-perchlorate reducer | ATCC 55151 |
| Environmental samples | ||
| Vida | Antarctica, diesel-contaminated site | G. Gilbert |
| Lake Fryxell sediment | Antarctica, pristine site | 12 |
| Lake Fryxell water column | Antarctica, pristine site | 12 |
| Lake Hoare mat | Antarctica, pristine site | L. Karr |
| Lake Hoare water column | Antarctica, pristine site | L. Karr |
| campus lake | Southern Illinois University | W. Fugate |
| library pond | Southern Illinois University | W. Fugate |
| library soil | Southern Illinois University | W. Fugate |
| Los Alamos wells | MPNs from perchlorate-contaminated site in Los Alamos, N.M.a | J. Pollock |
MPNs, most-probable-number samples.
PCR primers and reaction conditions.
Primer sets targeting the chlorite dismutase gene were designed based on areas of amino acid and nucleotide sequence conservation. These areas of conservation were visualized by manual sequence alignment using the Se-Al (21) program. The primers DCD-F [5′-GA(A/G)CGCAA(A/G)(A/G)GNGCNGCNG(A/C)NGA(A/G)GT-3′] and DCD-R [5′-TC(A/G)AA(A/G)TANGT(A/T/G)AT(A/G)AA(A/G)TC-3′] were developed based on the amino acid conservation of the cld sequences from Dechloromonas agitata, “Dechloromonas aromatica,” Ideonella dechloratans, and M. magnetotacticum. The primers UCD-238F [5′-T(C/T)GA(A/C/G)AA(A/G)CA(C/T)AAGGA(A/T/C)AA(A/C/G)GT-3′] and UCD-646R [5′-GAGTGGTA(A/C/G)A(A/G)(C/T)TT(A/C/G)CG(C/T)TT-3′] were developed from an expanded alignment that also included the cld gene sequences from Pseudomonas sp. strain PK, Dechloromonas sp. strain LT1, Dechlorosoma suillum, “Dechlorospirillum anomalous ” strain WD, and “Dechloromarinus chlorophilus” sp. strain NSS. Primers were synthesized by Integrated DNA Technologies, Coralville, Iowa.
To optimize PCR conditions, annealing temperatures ranging from 42 to 55°C, MgCl2 concentrations ranging from 1.0 to 3.0 mM, primer amounts ranging from 15 to 60 pmol, and PCR additives, such as 0.25 mg of bovine serum albumin (BSA)/ml, 5% (vol/vol) dimethyl sulfoxide, and 1 M betaine, were tested. PCRs were performed in a Perkin-Elmer 2400 thermocycler (Applied Biosystems, Foster City, Calif.). All reaction mixtures consisted of 1× Mg-free buffer, 1.0 to 3.0 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates, 2.5 U of Taq polymerase (Sigma, St. Louis, Mo.), 1 μl of gDNA or environmental DNA, and nuclease-free double-distilled H20 to a final volume of 50 μl. All components were purchased from Promega (Madison, Wis.) except for the polymerase. The following PCR conditions produced amplicons of the desired size using the DCD-F/DCD-R primer set: 44°C annealing temperature, 60 pmol (each) primer, 1.5 mM MgCl2, and 0.25 mg of BSA/ml. Optimal PCR conditions for the UCD-238F/UCD-646R primer set were 50°C annealing temperature, 40 pmol (each) primer, 1.5 mM MgCl2, and 0.25 mg of BSA/ml. Normal cycling parameters were as follows: reactions were initially heated to 94°C for 2 min, followed by 30 cycles of 94°C (1 min), annealing temperature (1 min), 72°C (1 min), with a final 10-min 72°C extension period. For touchdown cycling, the parameters consisted of a denaturation step at 94°C for 1 min, a primer-annealing step for 1 min, and an extension step at 72°C for 1 min. After 38 cycles, a final 10-min incubation was performed at 72°C. During the first 18 cycles, the annealing temperature was decreased by 1.0°C every two cycles, starting at 59°C, until reaching a touchdown temperature of 50°C. To verify the integrity of the amplification, both positive and negative (no template DNA) reactions were included. PCR results were checked using agarose gel electrophoresis on a 2% agarose gel containing 1× Tris-acetate-EDTA buffer.
Cloning and sequencing of PCR products.
PCR products of the appropriate size were gel extracted and subjected to the Geneclean Spin kit (Qbiogene) for subsequent analysis. Products from gel purification were directly cloned into the pCR 2.1 TOPO vector (Invitrogen, Carlsbad, Calif.). The inserts were sequenced with vector primers, using a ThermoSequenase cycle sequencing kit (U.S. Biochemicals, Cleveland, Ohio) and [α-35S]dATP as the label. Sequencing reactions were analyzed by electrophoresis through a 6% polyacrylamide-bisacrylamide gel.
Chlorite dismutase nucleotide sequences were manually entered using the MacVector 7.0 computer program (Oxford Molecular Group) and then transferred to Se-Al (21) for alignment. For 16S rDNA analysis, gene sequences were obtained from GenBank and 1,424 bases were aligned in the Seq-App computer program (8). Phylogenetic trees based on these alignments were constructed by using the PAUP* software package (beta version 4.0) (26). Unrooted trees for the chlorite dismutase gene and the 16S rDNA gene were constructed by using the absolute-number-of-differences parameter within the distance criterion. This parameter was chosen based on the closely related protein coding sequence of the cld gene (25). However, separate analyses using the Kimura 2 parameter to correct for evolutionary distances as well protein alignments were also performed for comparison. Gaps were removed from the 16S rDNA data set. Trees were drawn using neighbor joining, and 100 replicates were performed in bootstrap analysis.
GenBank sequence accession numbers.
Chlorite dismutase sequences generated from this study have been deposited in the GenBank database under the accession numbers AY540957 to AY540971. Chlorite dismutase gene sequences from the following strains were used in primer development: D. agitata (accession number AY124796), I. dechloratans (AJ296007), and M. magnetotacticum (ZP_00053098). 16S rDNA sequences from the following strains were used for phylogenetic tree construction: D. agitata (AF047462), “D. aromatica” (AY032610), D. suillum (AF170348), “D. anomalous” strain WD (AF170352), “D. chlorophilus” sp. strain NSS (AF170359), Pseudomonas sp. strain PK (AF170358), Dechloromonas sp. strain LT1 (AY124797), strain CR (AY530552), Dechlorospirillum sp. strain DB (AY530551), I. dechloratans (X72724), and M. magnetotacticum (Y10110).
RESULTS AND DISCUSSION
Primer design.
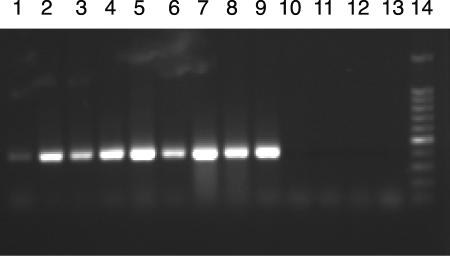
To develop universal primers targeting the cld gene, complete gene sequences were aligned from D. agitata, “D. aromatica” (identified from the complete genome sequence provided by the Department of Energy Joint Genome Institute), I. dechloratans, and M. magnetotacticum (identified by cld hybridization and analysis of the complete genome) (3). Visual alignment indicated sequence divergence at the 5′ end, while the 3′ end of the cld gene appeared more conserved (Fig. 1). This observation was expected based on previous hybridization analysis of several DPRB gDNAs using the D. agitata cld gene probe (3). From the four aligned sequences, two areas of amino acid conservation were chosen, and PCR primers targeting all corresponding codons were developed (Fig. 1). Due to the limited alignment file, primer DCD-F contained 9 degenerate sites out of 27 nucleotide positions, while DCD-R contained 6 degenerate sites out of 20 nucleotide positions. This primer set was tested on five other DPRB for accuracy and ability to amplify the cld gene. While a band at 484 bp resulted with all DPRB tested, an abundance of spurious by-products were also observed (Fig. 2). When the gDNA template was diluted, no increase in specificity occurred with the DCD-F/DCD-R primer set (data not shown); thus, the spurious by-products were most likely caused by the extreme degeneracy of the DCD-F/DCD-R primer set. Based on this lack of specificity of the DCD-F/DCD-R primer set, no negative control strains were tested using this primer set.
FIG. 1.
Amino acid alignment of cld gene products from D. agitata (D.agit), I. dechloratans (I.dech), M. magnetotacticum (M.magn), “D. aromatica” (D.arom), Dechloromonas sp. strain LT1 (DcmLT1), Pseudomonas sp. strain PK (Ps. PK), “D. chlorophilus” (D.chlo), D. suillum (D.suil), “D. anomalous” (D.anom), Dechlorospirillum sp. strain DB (st. DB), and strain CR (st. CR). Numbers correspond to residues from the D. agitata mature protein. “∼” denotes alignment gaps; “-” denotes unknown sequence data; “.” denotes identical residues. The DCD-F/DCD-R primer set targeted the lightly shaded regions, while the UCD-238F/UCD-646 primer set targeted the darkly shaded regions.
FIG. 2.
Amplification of a 484-bp internal region of the cld gene using the DCD-F/DCD-R primer set. Lane 1, “D. aromatica”; lane 2, M. magnetotacticum; lane 3, I. dechloratans; lane 4, Pseudomonas sp. strain PK; lane 5, D. suillum; lane 6, “D. chlorophilus”; lane 7, “D. anomalous”; lane 8, D. strain LT1; lane 9, negative control (no DNA); lane 10, 1-kb ladder.
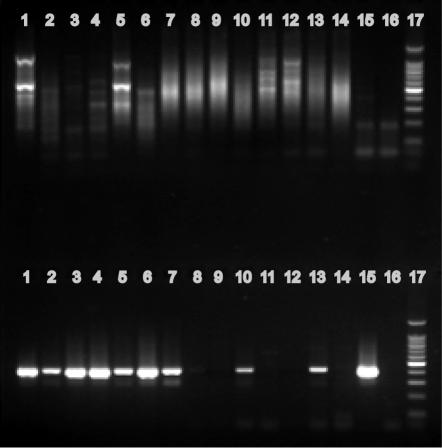
To increase the number of cld sequences represented in the alignment file and potentially reduce primer degeneracy, the 484-bp amplification product was excised from the gel, purified, cloned, and sequenced from DPRB Pseudomonas sp. strain PK, D. suillum, “D. chlorophilus,” “D. anomalous,” and Dechloromonas sp. strain LT1. These cld sequences, excluding priming sites, were added to the alignment file, and two areas of nucleotide conservation were targeted for primer design (Fig. 1). Primer UCD-238F contained 6 degeneracies out of 22 bases, while primer UCD-646R contained 5 degeneracies out of 20 bases. A 408-bp product was visible in all DPRB gDNAs tested with little background amplification (Fig. 3). No amplification occurred in non-(per)chlorate-reducing strains, including Rhodocyclus tenuis and Pseudomonas stutzeri, both close phylogenetic relatives of DPRB strains but unable to reduce (per)chlorate. However, this primer set was unsuccessful in amplifying cld gene sequences from environmental samples known to contain DPRB (data not shown). This may be explained by a lower concentration of target DNA in the environmental sample versus gDNA from pure cultures, as well as interference by nontarget DNA likely present in the environmental samples. Since specific products were obtained via PCR amplification using 16S rDNA primers on the environmental DNAs (data not shown), it is unlikely that PCR inhibitors affected the detection process.
FIG. 3.
Amplification of a 408-bp internal region of the cld gene using the UCD-238F/UCD-646R primer set. Lane 1, D. agitata; lane 2, “D. aromatica”; lane 3, M. magnetotacticum; lane 4, I. dechloratans; lane 5, Pseudomonas sp. strain PK; lane 6, D. suillum; lane 7, “D. chlorophilus”; lane 8, “D. anomalous”; lane 9, Dechloromonas species strain LT-1; lane 10, R. tenuis; lane 11, P. stutzeri; lane 12, Escherichia coli; lane 13, negative control (no DNA); lane 14, 100-bp ladder.
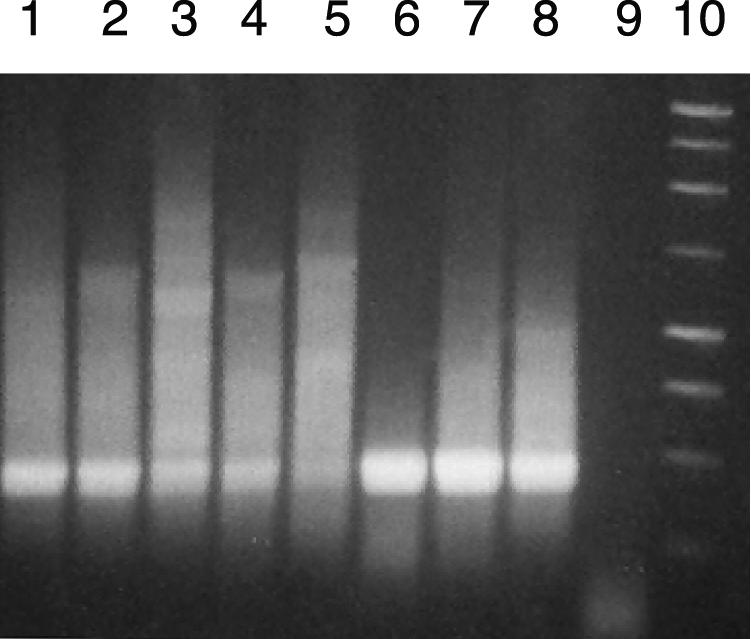
This problem was addressed by employing a nested PCR technique using the DCD-F/DCD-R primer set in an initial PCR, followed by a second amplification using the internal UCD-238F/UCD-646R primer set. Touchdown PCR cycling parameters were used to reduce the number of nontarget amplicons in the first PCR. Reaction products from the first amplification were diluted 1:10 and used as templates for the second round of amplification with the UCD-238F/UCD-646R primer set. Results from the nested procedure indicated that this technique was successful in amplifying cld sequences from Pseudomonas sp. strain PK and Los Alamos well samples as positive controls and from certain experimental environmental samples (Fig. 4). While spurious reaction products were observed from most samples in the first round of amplification, a second-round product of 408 bp was clearly visible in several of the environmental samples, including the Southern Illinois University campus library pond, the pristine Lake Fryxell sediment and Lake Hoare 12-m water column, the diesel-contaminated Vida, and all five samples obtained from a perchlorate-contaminated site in Los Alamos, N.M. (Fig. 4) known to contain DPRB. While no products of the appropriate size were evident in the first-round reaction for the Los Alamos-well 3 and Vida samples, an intense signal was present following the nested reaction. This result indicates that a low concentration of product was present in the first-round reaction, likely caused by a lower concentration of target DNA in these two samples. Thus, the nested procedure increases the sensitivity of this detection method.
FIG. 4.
Testing of the universal cld gene primer sets on environmental DNAs. Top of gel, touchdown PCR using the DCD-F/DCD-R primer set, corresponding to a 484-bp internal region of the cld gene. Bottom of gel, nested PCR on the above reactions, using the UCD-238F/UCD-646R primer set, corresponding to a 408-bp internal region of the cld gene. Lane 1, Pseudomonas sp. strain PK (positive control); lane 2, Los Alamos well 2; lane 3, Los Alamos well 3; lane 4, Los Alamos well 4; lane 5, Los Alamos well 5; lane 6, Los Alamos well 7; lane 7, campus library pond; lane 8, campus library soil; lane 9, campus lake; lane 10, Lake Fryxell sediment; lane 11, Lake Fryxell 7-m water column; lane 12, Lake Fryxell 12-m water column; lane 13, Lake Hoare 12-m water column; lane 14, Lake Hoare mat; lane 15, Vida; lane 16, negative control (no DNA); lane 17, 100-bp ladder.
Sequence analysis of the products.
Sequence analysis of the nested amplification products from Los Alamos-well 3, Los Alamos-well 4, Lake Fryxell sediment, Lake Hoare 12-m water column, and Vida samples indicated that all of the products were indeed cld gene sequences. While the Los Alamos well 3 clone was identical to the “D. aromatica” cld sequence, the Los Alamos well 4, Lake Fryxell sediment, Lake Hoare 12-m water column, and Vida clones were all most similar (amino acid similarity, 98.4 to 81.3%) to sequences from “D. anomalous” and strain DB (Table 2). The presence of cld sequences in the Antarctic samples was expected due to previously obtained DPRB isolates from these sites (data not shown). Sequence analysis indicated that more than one phylotype was present in samples collected from Vida, the Lake Hoare 12-m water column, and the Lake Fryxell sediment. Although these differences were only one or two nucleotides, the predicted protein products reflected these changes (Table 2). The observation of different cld gene sequences from the same environmental sample indicates the presence of more than one DPRB strain, and as such, denaturing gradient gel electrophoresis may be a useful tool in determining the number of and prevalent phylotypes in a given sample (12). Since denaturing gradient gel electrophoresis could also be used to address the effect of ecological changes on the diversity of cld sequences present, the nested cld primer sets could be used to analyze and monitor DPRB populations in the environment.
TABLE 2.
Amino acid identities of partial chlorite dismutase protein sequences
| Source or organism and ID no.a | % Amino acid identity with sample no:
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
| 1. D. agitata | 74.8 | 76.4 | 64.2 | 78.9 | 65.0 | 63.4 | 64.2 | 62.6 | 69.1 | 68.3 | 64.2 | 74.0 | 74.8 | 73.2 | 72.4 | 73.2 | 69.1 | 69.9 | |
| 2. M. magneto | 74.8 | 80.5 | 65.9 | 77.2 | 66.7 | 65.0 | 65.9 | 64.2 | 81.3 | 82.1 | 65.9 | 82.1 | 81.3 | 79.7 | 78.9 | 79.7 | 82.9 | 82.9 | |
| 3. I. dechloratans | 76.4 | 80.5 | 64.2 | 78.9 | 65.0 | 63.4 | 63.4 | 63.4 | 72.4 | 72.4 | 64.2 | 71.5 | 72.4 | 70.7 | 69.9 | 70.7 | 72.4 | 73.2 | |
| 4. “D. aromatica” | 64.2 | 65.9 | 64.2 | 64.2 | 98.4 | 98.4 | 97.6 | 95.9 | 66.7 | 65.9 | 100 | 65.9 | 66.7 | 65.9 | 64.2 | 65.0 | 66.7 | 67.5 | |
| 5. Dcm. strain LT1 | 78.9 | 77.2 | 78.9 | 64.2 | 65.0 | 63.4 | 64.2 | 62.6 | 73.2 | 73.2 | 64.2 | 77.2 | 78.0 | 76.4 | 76.4 | 77.2 | 73.2 | 74.0 | |
| 6. Pseud. strain PK | 65.0 | 66.7 | 65.0 | 98.4 | 65.0 | 98.4 | 95.9 | 95.1 | 68.3 | 66.7 | 98.4 | 66.7 | 67.5 | 65.9 | 65.0 | 65.9 | 68.3 | 69.1 | |
| 7. “D. chlorophilus” | 63.4 | 65.0 | 63.4 | 98.4 | 63.4 | 98.4 | 95.9 | 95.1 | 66.7 | 65.0 | 98.4 | 65.0 | 65.9 | 65.0 | 63.4 | 64.2 | 66.7 | 67.5 | |
| 8. D. suillum | 64.2 | 65.9 | 63.4 | 97.6 | 64.2 | 95.9 | 95.9 | 95.1 | 66.7 | 65.9 | 97.6 | 65.9 | 66.7 | 65.9 | 64.2 | 65.0 | 66.7 | 67.5 | |
| 9. Strain CR | 62.6 | 64.2 | 63.4 | 95.9 | 62.6 | 95.1 | 95.1 | 95.1 | 65.0 | 64.2 | 95.9 | 64.2 | 65.0 | 64.2 | 62.6 | 63.4 | 65.0 | 65.9 | |
| 10. “D. anomalous” | 69.1 | 81.3 | 72.4 | 66.7 | 73.2 | 68.3 | 66.7 | 66.7 | 65.0 | 95.9 | 66.7 | 82.9 | 83.7 | 82.1 | 81.3 | 82.1 | 97.6 | 98.4 | |
| 11. Dsp. strain DB | 68.3 | 82.1 | 72.4 | 65.9 | 73.2 | 66.7 | 65.0 | 65.9 | 64.2 | 95.9 | 65.9 | 82.9 | 83.7 | 82.1 | 81.3 | 82.1 | 96.7 | 97.6 | |
| 12. Los Alamos well 3 | 64.2 | 65.9 | 64.2 | 100 | 64.2 | 98.4 | 98.4 | 97.6 | 95.9 | 66.7 | 65.9 | 65.9 | 66.7 | 65.9 | 64.2 | 65.0 | 66.7 | 67.5 | |
| 13. Los Alamos well 4 | 74.0 | 82.1 | 71.5 | 65.9 | 77.2 | 66.7 | 65.0 | 65.9 | 64.2 | 82.9 | 82.9 | 65.9 | 99.2 | 97.6 | 96.7 | 96.7 | 83.7 | 83.7 | |
| 14. Lake Hoare 12m-A | 74.8 | 81.3 | 72.4 | 66.7 | 78.0 | 67.5 | 65.9 | 66.7 | 65.0 | 83.7 | 83.7 | 66.7 | 99.2 | 98.4 | 97.6 | 97.6 | 83.7 | 84.6 | |
| 15. Lake Hoare 12m-B | 73.2 | 79.7 | 70.7 | 65.9 | 76.4 | 65.9 | 65.0 | 65.9 | 64.2 | 82.1 | 82.1 | 65.9 | 97.6 | 98.4 | 95.9 | 95.9 | 82.1 | 82.9 | |
| 16. Lake Fryxell sed-A | 72.4 | 78.9 | 69.9 | 64.2 | 76.4 | 65.0 | 63.4 | 64.2 | 62.6 | 81.3 | 81.3 | 64.2 | 96.7 | 97.6 | 95.9 | 98.4 | 81.3 | 82.1 | |
| 17. Lake Fryxell sed-B | 73.2 | 79.7 | 70.7 | 65.0 | 77.2 | 65.9 | 64.2 | 65.0 | 63.4 | 82.1 | 82.1 | 65.0 | 96.7 | 97.6 | 95.9 | 98.4 | 82.1 | 82.9 | |
| 18. Vida-A | 69.1 | 82.9 | 72.4 | 66.7 | 73.2 | 68.3 | 66.7 | 66.7 | 65.0 | 97.6 | 96.7 | 66.7 | 83.7 | 83.7 | 82.1 | 81.3 | 82.1 | 99.2 | |
| 19. Vida-B | 69.9 | 82.9 | 73.2 | 67.5 | 74.0 | 69.1 | 67.5 | 67.5 | 65.9 | 98.4 | 97.6 | 67.5 | 83.7 | 84.6 | 82.9 | 82.1 | 82.9 | 99.2 | |
ID no., identification no. for sample (numbers correspond to numbers in column heads); Dcm. strain LT1, Dechloromonas sp. strain LT1; Pseud. strain PK, Pseudomonas sp. strain PK; Dsp. strain DB, Dechlorospirillum sp. strain DB; Lake Hoare 12m-A and 12m-B, clones A and B from 12-m water column of Lake Hoare; Lake Fryxell sed-A and sed-B, clones A and B from Lake Fryxell sediment; Vida-A and Vida-B, clones A and B from Vida.
Aside from the biases of PCR, this detection method is more inclusive than 16S rDNA primer sets, which can detect only a few genera of DPRB. However, a limitation of these primer sets is that only cld genes with sequences similar to those of the priming sites will be detected. This detection method would overlook extremely diverse sequences due to primer development from what is believed to be a minimum sampling of cld genes. Because the primer sets can detect cld genes in a DNA sample, the nested PCR approach does not require that the cells be actively reducing (per)chlorate and, as such, is useful for assessing the (per)chlorate-reducing potential of an environment. Although DNA:DNA hybridization studies have also be used to detect the cld gene (3), this approach requires more target DNA than a PCR-based approach, and hybridization signals could be affected by interference from environmental constituents.
Because this detection method targets a single gene in the metabolic pathway, it is possible to obtain false positives, as evidenced by M. magnetotacticum, an organism that harbors the cld gene but lacks other genes, such as those for (per)chlorate reductase, required for perchlorate reduction. However, subsequent analyses of cld-positive environmental samples, using (per)chlorate reductase probes (K. S. Bender and L. A. Achenbach, unpublished data), should eliminate these false positives from further consideration. While the nested PCR approach is efficient at detecting cld genes in the environment, traditional PCR cannot be used to determine the relative abundance or activity of DPRB in a given site. Based on the lack of perchlorate in most environments and the ability of DPRB to use alternate metabolisms, there is some question that the organisms detected using these primers are actively reducing perchlorate. For these analyses, the cld primer sets could be used in quantitative and reverse transcription-PCR. These strategies could also be used to monitor the sustainability of natural attenuation over long periods of time. Smets and colleagues observed that biodegradation of chlorinated solvents decreased over a 2-month period due to physiological changes of the bacteria in response to the environment (24). Thus, the cld primer sets could be used in an RNA approach to assess the long-term attenuation potential of a bacterial community. Quantitative PCR using this primer set could also determine if an increase in catabolic gene copy number occurs after a growth amendment is exogenously supplied. An increase in gene copy number would imply that the perchlorate-reducing potential of the site had been enhanced and that stimulation of these bacteria may lead to the natural attenuation of perchlorate. Thus, quantitative PCR using a metabolic primer set could aid in monitoring the effectiveness of a bioremediative strategy.
Phylogeny of the cld gene.
From the development of the degenerate cld primer set, the first library of cld gene sequences was generated. Included in this library were cld sequences from strains DB and CR, two perchlorate-reducing strains isolated during the cld primer development. Both strains originated from perchlorate-contaminated sites in Los Alamos, N.M. (J. Coates, unpublished data). From the 16S rDNA sequence, strain DB was designated a Dechlorospirillum species within the Rhodospirillaceae of the α-Proteobacteria, and strain CR was designated a member of the Rhodocyclus assemblage within the β-Proteobacteria.
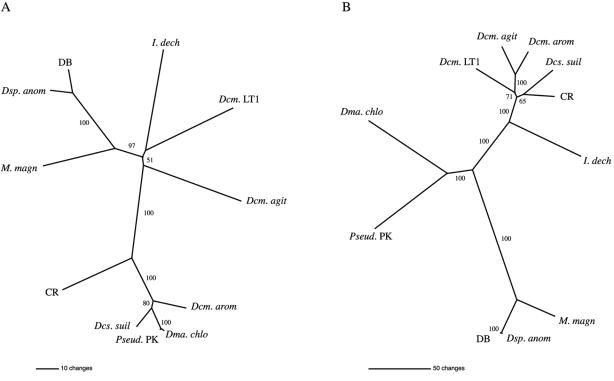
To determine if the cld gene phylogeny tracked that of the 16S rDNA gene and to possibly gain some insight into the evolution of (per)chlorate reduction, unrooted phylogenetic trees were compared. Comparison of the cld and 16S rDNA gene trees resulted in incongruent topologies (Fig. 5). While M. magnetotacticum, “D. anomalous,” and Dechlorospirillum strain DB, all members of the α-Proteobacteria, form a distinct cluster on both trees, the cld gene sequences from “D. aromatica,” D. suillum, and strain CR (all members of the β-Proteobacteria) cluster with those from the γ-Proteobacteria Pseudomonas sp. strain PK and “D. chlorophilus.” This aberration indicates that although D. agitata, “D. aromatica,” and Dechloromonas sp. strain LT1 are all members of the same genus, their respective cld gene sequences are not monophyletic. In addition, extremely short branch lengths on the cld tree among D. suillum, “D. aromatica,” Pseudomonas sp. strain PK, and “D. chlorophilus” reflect the high sequence similarity of these genes and indicate possible transfer of the cld gene among these members of the β- and γ-Proteobacteria (Fig. 5). These incongruent tree topologies suggest a role for horizontal gene transfer in the evolution of the (per)chlorate reduction pathway. This conclusion is based on previous studies regarding incongruent tree topologies and the occurrence of gene conservation among diverse hosts as evidence of horizontal transfer (9, 14, 15). Preliminary G+C content analysis of “D. aromatica” and M. magnetotacticum genomes also implicates the involvement of horizontal transfer with the spread of the cld gene. The G+C content of the “D. aromatica” genome is 59.2% (http://genome.ornl.gov/microbial/daro/), while the G+C content of the cld gene is 49.7%. Similarly, the G+C content of the M. magnetotacticum genome is 64.0% (http://genome.ornl.gov/microbial/mmag/), while the G+C content of the cld gene is 52.4%.
FIG. 5.
cld and 16S rDNA phylogenetic trees. (A) cld phylogenetic tree generated from an alignment of 369 bp. (B) 16S rDNA phylogenetic tree generated from an alignment of 1,424 bp. The numbers correspond to bootstrap values from 100 replicates. D. agitata (Dcm.agit), “D. aromatica” (Dcm.arom), D. strain LT1 (Dcm.LT1), D. suillum (Dcs. suil), “D. anomalous” (Dsp.anom), “D. chlorophilus” (Dma.chlo), I. dechloratans (I.dech), M. magnetotacticum (M.magn), Pseudomonas sp. strain PK (Pseud.PK), Dechlorospirillum sp. strain DB (DB), and strain CR (CR) were analyzed.
Due to the conserved nature of chlorite dismutase and the unambiguous nucleotide sequence alignment (Fig. 1), it is doubtful that the tree topology is incorrect. Trees constructed utilizing the Kimura 2 parameter and those constructed from amino acid alignments for the cld gene product resulted in similar topologies (data not shown). The incongruent tree topologies could alternatively be explained by a series of gene duplication and deletion events. However, in this case, the resulting cld gene sequences would still be expected to be similar to those of close phylogenetic relatives. Thus, both the cld gene sequence diversity and metabolic diversity of DPRB may be a direct result of horizontal transfer. Since DPRB can grow by alternate metabolisms, the cld gene may not be subject to intense selective pressure. As such, mutation may occur until the gene sequence becomes functional with respect to the codon usage and regulation of the host. However, more extensive data are needed on the codon biases and G+C content of housekeeping genes in other DPRB isolates before further conclusions can be drawn. While one can only speculate on the possible mechanism of transfer, a transposase gene was identified directly upstream of the cld gene in Pseudomonas sp. strain PK (data not shown). Other genes involved in the perchlorate reduction pathway were also identified in the direct proximity of the cld gene in D. agitata and “D. aromatica,” indicating that this metabolism may have been conferred through the action of a mobile genetic element. While phylogenetic comparisons of the cld gene and the 16S rDNA gene indicate that horizontal transfer is involved in the evolution of (per)chlorate metabolism, an interesting question still remains regarding the progenitor of (per)chlorate reduction and the selective advantage for retaining this metabolic machinery given that (per)chlorate has been widespread in the environment only in the last 50 years and that many DPRB are found in pristine areas.
Acknowledgments
This work was supported by grant number DACA72-00-C-0016 from the Department of Defense to J.D.C. and L.A.A.
Special thanks to D. Bazylinski, G. Gilbert, M. Madigan, L. Karr, and J. Pollock for generously providing cultures and environmental samples used within this study.
REFERENCES
- 1.Achenbach, L. A., U. Michaelidou, R. A. Bruce, J. Fryman, and J. D. Coates. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. E vol. Microbiol. 51:527-533. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and B. J. Schileifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, K. S., S. M. O'Connor, R. Chakraborty, J. D. Coates, and L. A. Achenbach. 2002. Sequencing and transcriptional analysis of the chlorite dismutase gene of Dechloromonas agitata and its use as a metabolic probe. Appl. Environ. Microbiol. 68:4820-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce, R. A., L. A. Achenbach, and J. D. Coates. 1999. Reduction of (per)chlorate by a novel organism isolated from paper mill waste. Environ. Microbiol. 1:319-329. [DOI] [PubMed] [Google Scholar]
- 6.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunbar, J., S. White, and L. J. Forney. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert, D. G. 1993. SeqApp 1.9a157 ed. Biocomputing Office, Biology Dept., Indiana University, Bloomington.
- 9.Herrick, J. B., K. G. Stuart-Keil, W. C. Ghiorse, and E. L. Madsen. 1997. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl. Environ. Microbiol. 63:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter, W. J. 2002. Bioremediation of chlorate or perchlorate contaminated water using permeable barriers containing vegetable oil. Curr. Microbiol. 45:287-292. [DOI] [PubMed] [Google Scholar]
- 11.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating ‘uncultivable’ microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 12.Karr, E. A., W. M. Sattley, D. O. Jung, M. T. Madigan, and L. A. Achenbach. 2003. Remarkable diversity of phototrophic purple bacteria in a permanently frozen Antarctic lake. Appl. Environ. Microbiol. 69:4910-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, K., and B. E. Logan. 2001. Microbial reduction of perchlorate in pure and mixed culture packed-bed bioreactors. Water Res. 35:3071-3076. [DOI] [PubMed] [Google Scholar]
- 14.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logan, B. E., H. Zhang, P. Mulvaney, M. G. Milner, I. M. Head, and R. F. Unz. 2001. Kinetics of perchlorate- and chlorate-respiring bacteria. Appl. Environ. Microbiol. 67:2499-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mesearch, M. B., C. H. Nakatsu, and L. Nies. 2000. Development of catechol 2,3-dioxygenase-specific primers for monitoring bioremediation by competitive quantitative PCR. Appl. Environ. Microbiol. 66:678-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michaelidou, U., L. A. Achenbach, and J. D. Coates. 2000. Isolation and characterization of two novel (per)chlorate-reducing bacteria from swine waste lagoons, p. 271-283. In E. D. Urbansky (ed.), Perchlorate in the environment. Kluwer Academic/Plenum, New York, N.Y.
- 19.O'Connor, S. M., and J. D. Coates. 2002. Universal immunoprobe for (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 68:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution: a ribosomal approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 21.Rambaut, A. 2002. Sequence Alignment Editor v2.0. Department of Zoology, University of Oxford, Oxford, United Kingdom.
- 22.Renner, R. 1998. Perchlorate-tainted wells spur government action. Environ. Sci. Technol. News 32:210A. [DOI] [PubMed] [Google Scholar]
- 23.Rikken, G. B., A. G. M. Kroon, and C. G. van Ginkel. 1996. Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45:420-426. [Google Scholar]
- 24.Smets, B. F., S. D. Siciliano, and W. Verstraete. 2002. Natural attenuation: extant microbial activity forever and ever? Environ. Microbiol. 4:315-317. [DOI] [PubMed] [Google Scholar]
- 25.Swofford, D. L., G. J. Olsen, P. J. Waddell, and D. M. Hollis. 1996. Phylogenetic inference, p. 407-514. In D. M. Hollis, C. Moritz, and B. K. Mable (ed.), Molecular Systematics, 2nd ed. Sinauer Associates, Inc., Sunderland, Mass.
- 26.Swofford, D. L. 1999. PAUP*: Phylogenetic analysis using parsimony (and other methods). Version 4.0. Sinauer Associates, Sunderland, Mass.
- 27.Tipton, D. K., D. E. Rolston, and K. M. Scow. 2003. Transport and biodegradation of perchlorate in soils. J. Environ. Qual. 32:40-46. [DOI] [PubMed] [Google Scholar]
- 28.Urbansky, E. T. 1998. Perchlorate chemistry: implications for analysis and remediation. Biorem. J. 2:81-95. [Google Scholar]
- 29.U.S. Environmental Protection Agency. 2003. National Academy of Sciences' review of EPA's draft perchlorate environmental contamination: toxicological review and risk characterization. [Online.] http://cfpub.epa.gov/ncea/cfm/recordisplay.cfm?deid=72117.
- 30.van Ginkel, C. G., G. B. Rikken, A. G. M. Kroon, and S. W. M. Kengen. 1996. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch. Microbiol. 166:321-326. [DOI] [PubMed] [Google Scholar]
- 31.Wallace, W., T. Ward, A. Breen, and H. Attaway. 1996. Identification of an anaerobic bacterium which reduces perchlorate and chlorate as Wolinella succinogenes. J. Ind. Microbiol. 16:68-72. [Google Scholar]