Abstract
The metagenomic approach provides direct access to diverse unexplored genomes, especially from uncultivated bacteria in a given environment. This diversity can conceal many new biosynthetic pathways. Type I polyketide synthases (PKSI) are modular enzymes involved in the biosynthesis of many natural products of industrial interest. Among the PKSI domains, the ketosynthase domain (KS) was used to screen a large soil metagenomic library containing more than 100,000 clones to detect those containing PKS genes. Over 60,000 clones were screened, and 139 clones containing KS domains were detected. A 700-bp fragment of the KS domain was sequenced for 40 of 139 randomly chosen clones. None of the 40 protein sequences were identical to those found in public databases, and nucleic sequences were not redundant. Phylogenetic analyses were performed on the protein sequences of three metagenomic clones to select the clones which one can predict to produce new compounds. Two PKS-positive clones do not belong to any of the 23 published PKSI included in the analysis, encouraging further analyses on these two clones identified by the selection process.
Natural compounds produced by bacteria have been extensively used to develop most of the antibacterial drugs proposed by pharmaceutical companies over the last 50 years. However, new antibiotics from natural origins have become more difficult to detect in spite of efforts to increase both screening capacities and the number of bacteria tested (32). From 40,000 microbial cultures screened in 10 years, only three usable antibiotics were discovered (8). However, these efforts have concentrated on bacterial isolates capable of growth in vitro, thus missing the 99% of bacteria that remain uncultivated (2, 11). The production capacities of the microbial world are, therefore, far from being explored with these classical approaches.
Based on physiological studies, cultivated bacterium numbers will increase significantly in the near future, thus providing new bacterial isolates for screening tests (17). However, to avoid culture limits, another approach has been developed consisting of the screening of recombinant bacteria that could express genes from the metagenome, defined as all bacterial genomes of a given environment (6, 19, 21, 24). The interest of this metagenomic approach has already been demonstrated with the analysis of clones containing ribosomal genes. Phylogenetic studies of 16S rRNA genes indicate that metagenomic DNA encompasses a large bacterial diversity including uncultivated bacteria and even unknown bacterial phyla (4, 19, 29).
Beyond the descriptive analysis of diversity, the metagenome has been shown to provide the functional identification of bacterial genes that encode bioactive compounds (11), new polyketide synthases (6, 21), and even new functions like a membrane-associated proteolytic system (4). The functional analysis of metagenomic clones requires their genes, operons, or biosynthetic pathway to be entirely cloned and then transferred into an adapted host for heterologous expression. The construction of metagenomic libraries leads to such a genetic manipulation. The screening of large libraries for biosynthetic genes was demonstrated to detect numerous potentially interesting clones (6). Due to the technical difficulties encountered with heterologous expression, production of the expressed compound, and chemical analysis of the compound, the choice of clones to study is crucial. Type I polyketide synthases (PKSI) synthesize natural products of therapeutic interest such as erythromycin, rapamycin, or epothilone, and their organization provides facility in the selection of promising clones.
Properties of PKSI make them particularly well suited for the metagenomic DNA library approach. These large multienzymes are composed of a succession of modules. A loading module loads and activates the first substrate. Then each extender module catalyzes an elongation step with condensation of extender units onto the growing polyketide chain (28). A minimal extender module is composed of three domains: a ketosynthase (KS) domain for decarboxylative condensation of the extender unit onto the growing chain, an acyl transferase (AT) domain for substrate selection, activation, and transfer, and an acyl carrier protein (ACP), which loads the growing chain. Each substrate can be reductively tailored by additional domains, ketoreductase, enoylreductase, and dehydratase (14, 27). A thioesterase domain is often localized after the PKSI extender modules and catalyzes the release of the completed polyketide chain.
The method for selecting the promising clones is supplied by phylogenetic analysis of PKSI domains. Phylogenies of the protein sequences of KS and AT domains led to (i) the determination of the taxonomic position of the donor DNA, since most of KS domains from the order Actinomycetales are monophyletic (20, 21), (ii) the identification of unusual KS domain functions, such as KS from loading modules and from hybrid nonribosomal peptide synthases (NRPS)/PKS (20, 21), (iii) the prediction of the incorporated substrate of each module by AT phylogeny (13, 15, 26, 34), and (iv) the prediction of the polyketide novelty. Indeed, one can observe with phylogeny analysis that KS domains, with a usual function, from a PKSI operon tend to cluster (11a, 20). This clustering can be linked with the synthesized polyketide. To select the promising clones containing PKSI genes, their KS domains have to be analyzed though KS phylogeny to predict their functional novelty, which is defined as the novelty of their potentially synthesized polyketide. In others words, the branching of uncharacterized KS domains within clusters of published PKSI operons can lead to the exclusion of these uncharacterized PKSI from further experiments.
Among PKSI domains, the KS domain is the most conserved (3, 20, 21). Thus, we designed PCR primers in the conserved regions of the KS domain to detect PKSI genes in recombinant clones from a large metagenomic soil library. Three of 139 detected positive PKSI clones were entirely sequenced. The KS and AT domains of these three metagenomic clones were analyzed and compared to the domains from 23 previously published PKSI. This method led to the detection of numerous PKSI-positive clones and to the selection of two promising clones for the potential production of new compounds.
MATERIALS AND METHODS
DNA extraction from soil.
The soil was sampled in an area located 50 km east of Lyon, France (Montrond). The soil sample was collected during autumn 2001 from a depth range of 10 to 20 cm. The soil (clay loam sandy type) has a high level of organic matter (47 g/kg). The fresh soil was dried at room temperature for 24 h before being sieved through 4-mm mesh and then 2-mm mesh.
DNA extraction was performed by using a centrifugation-based separation of bacteria from soil particles, followed by the incorporation of bacteria in agarose before a gentle bacterial lysis as described by Nalin et al. (R. Nalin, P. Robe, and V. Tran Van, 11 January 2001, Method for indirectly extracting noncultivable DNA organisms and DNA by said method, French Patent Office). The Nycodenz-mediated extraction of bacteria from the soil matrix was achieved as previously described (5). The bacterial pellets were resuspended in a 50 mM Tris (pH 8.0), 100 mM EDTA buffer, mixed with an equal volume of molten 1.6% Incert agarose (BMA), and then transferred into disposable plug molds (Bio-Rad). The lysis of the soil bacteria was then performed in agarose. Agarose plugs were first transferred in 45 ml of LA lysis buffer (50 mM Tris [pH 8.0], 100 mM EDTA, 5 mg of lysozyme/ml, 0.5 mg of achromopeptidase/ml) and incubated at 37°C for 6 h. The agarose plugs were then incubated in 45 ml of SP lysis buffer (50 mM Tris [pH 8.0], 100 mM EDTA, 1% lauryl sarcosyl, 2 mg of proteinase K/ml) at 55°C for 24 h. An additional incubation for 24 h was performed with fresh SP buffer. Agarose plugs were finally equilibrated in a 10 mM Tris (pH 8.0), 1 mM EDTA storage buffer.
Construction of the metagenomic library.
High-molecular-weight bacterial DNA trapped in agarose plugs was immediately inserted into the wells of an 0.8% low-melting-temperature gel (Bio-Rad) and separated for 18 h by pulsed-field gel electrophoresis at 4.5 V/cm with 5- to 40-s pulse times with a CHEF-DRIII apparatus (Bio-Rad). DNA fragments ranging between 35 and 48 kbp were isolated and then recovered from the gel with GELase (Epicentre Technologies). Metagenomic DNA was then cloned into fosmids by using the EpiFos fosmid library production kit (Epicentre Technologies) as recommended by the manufacturer. Recombinant colonies were transferred to 96-well microtiter plates containing freezing medium (Luria-Bertani, 20% glycerol complemented with 12.5 μg of chloramphenicol/ml). After growing at 37°C for 22 h, the plates were stored at −20°C.
PCR screening of clones for PKSI genes.
Overnight cultures of 1 ml per well in 96-deepwell plates (22 h, 37°C, 250 rpm shaking) were pooled and purified with the Nucleobond PC100 kit (Macherey Nagel) by following the instructions of the manufacturer. Purified DNA from these 96 pools (100 to 500 ng) was used as a template. Primers KSLF (5′-CCSCAGSAGCGCSTSYTSCTSGA-3′) and KSLR (5′-GTSCCSGTSCCGTGSGYSTCSA-3′) were designed based on the conserved KS domain motifs. The specific fragment amplified with KSLF-KSLR is about 700 bp in length. PCR amplification on DNA 96-well pools was performed with recombinant Taq DNA polymerase (Sigma) as follows: a denaturation step at 96°C for 5 min; 7 cycles consisting of 1 min at 96°C, 1 min at 65°C (annealing temperature lowering 1°C per cycle), and 1 min at 72°C; 40 cycles consisting of 1 min at 96°C, 1 min at 58°C, and 1 min at 72°C; and a final extension for 7 min at 72°C.
Localization of PKSI-positive clones.
The microtiter plate positions of PKSI-positive clones were determined by colony hybridization. A PCR fragment obtained with degenerate PKSI primers set from four positive pools was used to generate the probe. The four PCR products were mixed to hybridize the four respective microtiter plates. This probe mixing minimized the number of labeling reactions. The mixed probes were labeled with [α-33P]dCTP by using the random priming DNA labeling kit (Roche) in accordance with the manufacturer's protocol. Transformants were spotted onto GeneScreen Plus (NEF988) nylon membranes previously laid onto Luria-Bertani agar plates and then incubated at 37°C for 18 h. Colonies were lysed by incubating the membranes for 15 min on a sheet of 3M paper (Whatman) saturated with 0.5 M NaOH-1.5 M NaCl. The membranes were then neutralized by incubation for 15 min on a sheet of 3M paper (Whatman) saturated with 1.5 M NaCl-1 M Tris (pH 7.5). After drying at room temperature for 20 min, immobilization of DNA on membranes was performed by the UV cross-linking technique (312 nm for 4 min). A prehybridization was realized for 2 h with conditions as follows. Hybridization with the probe was performed for 16 h at 68°C with a 1% sodium dodecyl sulfate-5× Denhardt's-1 M NaCl solution. Membranes were washed sequentially at 68°C in (i) 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 10 min, (ii) 2× SSC-0.1% sodium dodecyl sulfate for 20 min, and (iii) 1× SSC for 10 min. The hybridization signals were visualized after exposure for 4 h by using a PhosphorImager (Bio-Rad GS-525).
Sequencing.
Fosmid inserts were sequenced by using both transposon-mediated and shotgun subcloning approaches. Transposition was performed by using the transposition kit (Epicentre) according to the manufacturer's instructions. For subcloning, the purified fosmid DNA was partially restricted with Sau3AI. Restriction fragments ranging from 1 to 3 kbp were size selected by standard gel electrophoresis and then cloned into the pBC SK (+/−) vector (Stratagene) (25). In addition, PCR products of about 700 bp were purified from an agarose gel with a gel extraction kit (Qiagen) and then cloned by using the Topo PCR II kit (Invitrogen). Recombinant plasmids were purified by using the QIAprep plasmid extraction kit (Qiagen) and sequenced with forward and reverse M13 primers. Sequencing reactions were performed with the DTCS cycle sequencing kit (Beckman Coulter) as recommended by the supplier. Sequencing reactions were run on a CEQ 2000 sequencer (Beckman Coulter). Then the overlaps of at least four independent clones were assembled.
Phylogenetic analysis.
The protein sequences of KS and AT domains detected in the metagenomic library were aligned with a large set of published sequences (supplemental table available at http://web.libragen.com/Phylogeny/sup_table.html). This set contains 23 PKSI clusters representing 203 KS domains and 207 AT domains. The KS and AT domains from three clones detected in the metagenomic library, named Lib4, Lib7, and Lib10, were included and aligned with respective published sequences by using DbClustal (30). Alignments were manually corrected by using Seaview (10). Phylogenetic reconstructions were performed with Phylo_win (10) for the distance method by using neighbor joining (NJ) and the PAM matrix. The program PhyML (12) was used for the maximum-likelihood (ML) method by using BIONJ with the JTT model of substitution (16). All trees were built with 500 bootstrap replicates. For the ML reconstruction, the 500 data sets were generated by using SEQBOOT from the PHYLIP, version 3.57c, package (7). A tree was built for each replicate with PhyML, and then bootstraps were computed with CONSENSE. All trees were drawn with NJplot (22). The trees were rooted by a fatty acid synthase (mas gene, Uniprot/Swiss-Prot database accession number M95808).
Nucleotide sequence accession number.
The three nucleotide sequences Lib4, Lib7, and Lib10 encoding the KS and AT domain regions have been assigned the accession numbers AJ639921, AJ639922, and AJ639923, respectively, in the EMBL database. The accession numbers for published PKSI sequences are reported in the supplemental table (http://web.libragen.com/Phylogeny/sup_table.html).
RESULTS AND DISCUSSION
Metagenomic DNA library construction.
A large metagenomic library of more than 100,000 fosmid clones was constructed, with a high cloning efficiency of 3 × 105 clones/μg of metagenomic DNA. Clones were randomly chosen to estimate the insert size. All were shown to contain inserts ranging from approximately 30 to 40 kbp, in agreement with the encapsidation-mediated selection of inserts. The library was organized on microtiter plates for high-throughput screening. The cumulated size of inserts encompassed 3.5 Gbp, corresponding approximately to 850 times the size of the Escherichia coli genome (4.1 × 106 bp). If DNA redundancy (overrepresentation of DNA from some bacteria because of population size or extraction and cloning biases) remained low, this available metagenomic library encompasses a little bit more than 5% of the estimated soil bacterial diversity based on the 10,000 different bacterial types detected in pristine soils and sediments (31). Even without constructing an exhaustive metagenomic library of more than 2,000,000 clones, our 100,000-clone library already provides access to bacterial diversity 24 times greater than traditional cultivation methods.
PCR-based metagenomic library screening for PKSI genes.
Recombinant fosmid DNA from 60,000 of the 100,000 clones of the soil metagenomic library was extracted for use as a template for PCR screening according to the protocol described in Materials and Methods. Primers were designed based on the most conserved DNA regions of the KS domain and gave a positive PCR response for the 139 clones. Further analysis was restricted to 40 randomly chosen fosmids, which were subsequently cloned and sequenced. All 40 DNA sequences were unique. The 40 deduced protein sequences never exceeded 67% similarity to published sequences (maximum of 141 of 210 amino acid identities) according to BLAST analysis (1). These results confirm that the diversity level in the metagenomic DNA library was relatively high, as described for a previous 5,000-clone-rich library in which 11 different KS domain sequences were detected (6).
Phylogenetic analysis of the detected PKSI genes.
An active KS domain site encoding an open reading frame was detected in each of the 39 DNA sequences amplified from the metagenomic DNA library clones. Among the 39 sequences, 11 sequences displayed a unique pattern N(DE)KD 22 amino acids upstream from the cysteine active site in the KS domain and the conserved pattern VDTACSSS was replaced by VQTACSTS (amino acid modifications are shown in boldface type). These two patterns were shown to identify KS domains belonging to hybrids between NRPS and PKSI (21) and, more precisely, KS domains preceded by an NRPS, thus acting on an amino acid chain (11a).
For further analysis, complete insert sequences were obtained for 3 of the 39 clones, Lib4, Lib7, and Lib10. Lib10 was selected as a representative of the 11-clone group that could exhibit the presence of an NRPS domain upstream of the KS domain. Lib4 and Lib7 were chosen because the extremities of their fosmid inserts did not contain any PKS genes, and thus, their complete biosynthetic pathway genes were expected to be contained in the cloned DNA inserts.
Analysis of KS domains.
Only one KS domain was found in Lib4, Lib7, and Lib10 clone sequences, indicating that the potentially synthesized compounds would not exhibit a typical linear polyketide structure. Phylogenetic analyses were carried out with the objective of locating these metagenomic KS domains inside the phylogenetic tree built with the KS domains from 23 PKSI published sequences. Both reconstruction methods, distance and ML, provided the same tree topologies, suggesting that observed groups do not result from computational artifacts.
The KS domain sequences of the Lib4, Lib7, and Lib10 clones exhibited low similarity values to those available in GenBank (58, 61, and 58% BLASTP identities with their closest neighbors, respectively) and were not identical to each other.
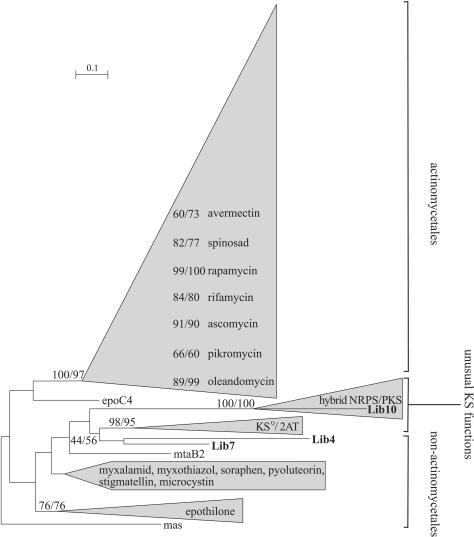
None of the KS domains from the metagenomic clones Lib4, Lib7, and Lib10 were clustered within the Actinomycetales group (bootstrap values, 100 [ML] and 97 [NJ]), suggesting that these clones probably do not belong to this order (Fig. 1). However, such a hypothesis cannot be totally supported considering that functional constraints could have led some genes to evolve differently than the other genes that encode typical KS domains. For instance, KS domains that do not catalyze a usual incoming acyl chain but an amino acid chain were found to cluster together (called a hybrid group) and separately from the other KS domains (20, 21), independently of their taxonomic positions. An NRPS module systematically precedes these unusual KS domains. Interestingly, an NRPS gene was also detected upstream of the KS domain in the Lib10 clone that strongly clustered in the hybrid group (Fig. 1).
FIG. 1.
Phylogenetic analysis of KS domains. The reconstruction was computed for 207 protein sequences on 413 sites by the ML method (BIONJ, JTT matrix) and the distance method (NJ, PAM matrix) with 500 bootstrap replicates. The group named hybrid NRPS/PKS concerns KS domains preceded by an NRPS. KSQ domains and KS domains from loading modules with the organization ACP-KS-AT-AT-ACP are clustered and are called the KSQ/2AT group. As the tree topologies obtained with both methods are similar, bootstrap values are indicated as ML/NJ. The strongly clustered KS domains of a PKSI are given with the name of the produced polyketide and their respective bootstrap values. The number of amino acid substitutions is proportional to the length of the scale. Sequences obtained from the metagenomic library are given in boldface type. The entire tree is available upon request.
As most KS domains within a PKSI are clustered (11a, 20), the KS domains from Lib4 and Lib7 clones would not belong to any of the 23 PKSI computed in this study (Fig. 1). Thus, the 2 polyketides synthesized by the metagenomic PKSI are predicted to be different from these 23 polyketides. Since the KS domain from Lib10 does not present an usual KS function, it cannot be used as a predictive domain for the functional novelty of its PKSI.
Analysis of the complete sequences confirmed that the Lib4 and Lib7 inserts contained all of the genes coding for a complete biosynthetic pathway. The two KS domains from Lib4 and Lib7 belong to the loading module of their respective PKSI. They did not exhibit the active-site mutation specific to KSQ domains in which cysteine is replaced by glutamine. These KSQ domains have lost their condensation activity but still decarboxylate the ACP-bound dicarbocylic acid, giving rise to the initial substrate (33). Moreover, as these modules contain only one AT domain, they cannot be classified in the starting module group that presents the organization ACP-KS-AT-AT-ACP (18). As expected, distance and ML phylogenetic methods did not include the KS domains from Lib4 and Lib7 in the KSQ/2AT, group although they were closed (Fig. 1). Thus, the KS domains from Lib4 and Lib7 may have a usual function and are reliable for novelty prediction.
Substrate specificity prediction of AT domains.
The chemical structure of the final compound is dictated by (i) the incorporated substrate recognized by AT domains, (ii) the degree of the reduction cycle catalyzed by additional domains of each module (14), and (iii) the number of modules and their succession. Substrate recognition is a major factor influencing polyketide structure and diversity. In most cases, the incorporated substrates are predicted by phylogenetic analyses of AT domains (13, 15, 26, 34).
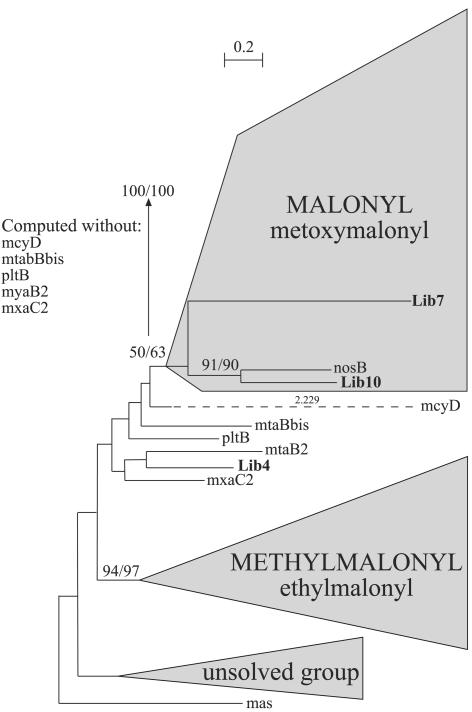
AT domains specific to malonyl and methylmalonyl were found to cluster in two separate groups. However, the malonyl node is not supported by high bootstrap values when five AT domains (pltB, mtaBbis, mtaB2, mcyD, and mxaC2) were included in the reconstruction. A second phylogenetic reconstruction performed without these five protein sequences showed that the typical malonyl incorporation coding sequences clustered together with bootstrap values of 100 (ML) and 100 (NJ) (Fig. 2). The two AT domains from Lib7 and Lib10 are clustered in the real malonyl group and, therefore, must incorporate malonyl (Fig. 2).
FIG. 2.
Phylogenetic analyses of AT domains. The mtaBbis domain corresponds to the second AT domain in the unusual organization ACP-KS-AT-AT-ACP. The reconstruction was computed for 210 protein sequences on 169 sites by the ML method (BIONJ, JTT matrix) and the distance method (NJ, PAM matrix) with 500 bootstrap replicates. As the tree topologies obtained with both methods are similar, bootstrap values are indicated as ML/NJ. The AT domains located outside the two clusters for malonyl and methylmalonyl remain unsolved by phylogenetic analysis. The number of amino acid substitutions is proportional to the length of the scale except for mcyD, which has its branch length written on the dashed line. Sequences obtained from the metagenomic library are given in boldface type. The entire tree is available upon request.
The study of the active-site signatures (23, 34) confirmed a malonyl substrate incorporation for the Lib7 AT domain (Tables 1 and 2). The AT domain from Lib10 also displayed a malonyl-specific signature, even if among the 25 first substrates predicted by the SEARCHPKS tool (35) one differed from malonyl (Table 2). In the case of AT domains from Lib7 and Lib10, both signature and phylogeny methods provided the same conclusion for malonyl incorporation.
TABLE 1.
Reeves signature model and associated predicted substrates for metagenomic AT domainsa
| Substrate or AT domain | Conserved pattern sequences | Predicted substrate(s) | ||
|---|---|---|---|---|
| Substratesb | ||||
| M | (Q/R/D/E)TX(Y/F) (T/A)Q | GHS(L/V/I)GE | H(A/G)FH | |
| mM | R(V/A/I)(D/E)VVQ | GHS(Q/M)GE | YASH | |
| eM | RVDVV(Q/H) | GHSQGE | TAGH | |
| AT domains | ||||
| Lib4 | RTEIAQ | GHSVGE | YAFH | M, mM |
| Lib7 | ETIYTQ | GHSIGE | RAFH | M |
| Lib10 | APSVGL | GHSMGE | VAAH | M, mM |
Reeves signature model (23). The consensus patterns for malonyl, methylmalonyl, and ethylmalonyl are given with all possibilities (indicated in parentheses). Signatures of the metagenomic AT domains were extracted and reported with the deduced substrate(s).
M, malonyl; mM, methylmalonyl; eM, ethylmalonyl.
TABLE 2.
Yadav signature model and associated predicted substrates for metagenomic AT domainsa
| AT domain | No. of occurrences of substrate in first 25 predicted substrates
|
|||||||
|---|---|---|---|---|---|---|---|---|
| M | mM | M/mM | eM | 2mB | 3mB | G | B | |
| Lib4b | 1 | 19* | 2 | 2 | 1 | |||
| Lib7 | 24* | 1 | ||||||
| Lib10 | 24* | 1 | ||||||
Yadav signature model (34). The distribution of the first 25 results for substrate prediction by the SEARCHPKS tool (35) is shown. The first predicted substrate for each domain is indicated by an asterisk. Abbreviations: M, malonyl; mM, methylmalonyl; eM, ethylmalonyl; 2mB, 2-methylbutyryl; 3mB, 3-methylbutyryl; G, glyceryl; B, benzoyl.
Described as a loading domain.
The Reeves signature model (23) was based on 71 AT domain sequences and did not include substrates other than malonyl and methylmalonyl. Moreover, 57 of 71 (80%) AT domains belonged to the Actinomycetales, suggesting this model to be more adapted to this order. Our results indicate that the Reeves model could not lead to a reliable substrate prediction for the Lib4 AT domain, supporting the results of KS domain phylogeny, i.e., this clone could not have come from an Actinomycetales species.
The majority of AT domains from loading modules of non-Actinomycetales fall in the unsolved group (Fig. 2). AT domains from loading modules can accept different loading units with a lower specificity, maybe to adapt to the substrate availability in the cell (9). The AT domain from Lib4 did not cluster within any of the malonyl or methylmalonyl reliable groups (Fig. 2); thus, any prediction for substrate incorporation based on phylogenetic analysis is excluded. Moreover, the Yadav prediction for the Lib4 AT domain provided similar expect values (5 e−70 to 3 e−58) for various substrates, including 3-methylbutyryl, which ranked at the first position (Table 2). Since, this AT domain belongs to a loading domain, which is also predicted by the Yadav model, it may recognize several substrates.
The organization of clone libraries containing large fragments of metagenomic DNA provides access to a wide diversity of uncharacterized genes, operons, and biosynthetic pathways. By targeting PKSI genes, our results and those of a previous report (6) indicate that one can expect 0.23% (139 PCR hits for 60,000 clones screened) of PKSI genes from metagenomic libraries. This frequency and the absence of redundancy observed in this study confirmed the potential and the quality of this soil library. Since large and homogeneous libraries of more than 100,000 clones are available, sorting the numerous detected clones is an important challenge. Indeed, the transfer into an adapted host and the identification of conditions for their heterologous expression are fastidious to perform. Moreover, even when this heterologous expression is achieved, chemical analysis of natural compounds is often difficult. Our study demonstrates that complete KS and AT sequences from PKSI provide enough fundamental information to select the promising clones. This information includes the prediction of novelty of the potentially synthesized compound and the incorporated substrates. These useful predictions will help the chemical characterization of the polyketide. Thus, this method will enable investigators to decide which clones deserve to be studied further. In this study, we detected and selected Lib4 and Lib7 as the most pertinent clones for potentially novel active compounds.
Acknowledgments
We are indebted to the Pôle Bioinformatique Lyonnais for kind access to databanks and information resources. We gratefully acknowledge Philippe Normand for helpful discussions regarding the manuscript.
This work is part of the project “Développement et exploitation de librairies d'ADN metagénomique,” which was funded by Région Rhône-Alpes (Thématiques Prioritaires, Sciences Analytiques Appliquées) and was supported by the Agence Nationale de Valorisation de la Recherche.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio, J. F., R. Fouces, M. V. Mendes, N. Olivera, and J. F. Martin. 2000. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem. Biol. 7:895-905. [DOI] [PubMed] [Google Scholar]
- 4.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. [DOI] [PubMed] [Google Scholar]
- 5.Courtois, S., A. Frostegard, P. Goransson, G. Depret, P. Jeannin, and P. Simonet. 2001. Quantification of bacterial subgroups in soil: comparison of DNA extracted directly from soil or from cells previously released by density gradient centrifugation. Environ. Microbiol. 3:431-439. [DOI] [PubMed] [Google Scholar]
- 6.Courtois, S., C. M. Cappellano, M. Ball, F. X. Francou, P. Normand, G. Helynck, A. Martinez, S. J. Kolvek, J. Hopke, M. S. Osburne, P. R. August, R. Nalin, M. Guerineau, P. Jeannin, P. Simonet, and J. L. Pernodet. 2003. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl. Environ. Microbiol. 69:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 8.Firn, R. D., and C. G. Jones. 2000. The evolution of secondary metabolism—a unifying model. Mol. Microbiol. 37:989-994. [DOI] [PubMed] [Google Scholar]
- 9.Gaitatzis, N., B. Silakowski, B. Kunze, G. Nordsiek, H. Blocker, G. Hofle, and R. Muller. 2002. The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J. Biol. Chem. 277:13082-13090. [DOI] [PubMed] [Google Scholar]
- 10.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie, D. E., S. F. Brady, A. D. Bettermann, N. P. Cianciotto, M. R. Liles, M. R. Rondon, J. Clardy, R. M. Goodman, and J. Handelsman. 2002. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Ginolhac, A., C. Jarrin, P. Robe, G. Perrière, T. M. Vogel, P. Simonet, and R. Nalin. J. Mol. Evol., in press. [DOI] [PubMed]
- 12.Guindon, S., and O. Gascuel. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 13.Haydock, S. F., J. F. Aparicio, I. Molnar, T. Schwecke, L. E. Khaw, A. Konig, A. F. Marsden, I. S. Galloway, J. Staunton, and P. F. Leadlay. 1995. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Microbiol. Lett. 374:246-248. [DOI] [PubMed] [Google Scholar]
- 14.Hopwood, D. A. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465-2497. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda, H., T. Nonomiya, M. Usami, T. Ohta, and S. Omura. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 96:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biol. Sci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 17.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ligon, J., S. Hill, J. Beck, R. Zirkle, I. Molnar, J. Zawodny, S. Money, and T. Schupp. 2002. Characterization of the biosynthetic gene cluster for the antifungal polyketide soraphen A from Sorangium cellulosum So ce26. Gene 285:257-267. [DOI] [PubMed] [Google Scholar]
- 19.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez, J. V. 2003. Naturally mosaic operons for secondary metabolite biosynthesis: variability and putative horizontal transfer of discrete catalytic domains of the epothilone polyketide synthase locus. Mol. Genet. Genomics 270:420-431. [DOI] [PubMed] [Google Scholar]
- 21.Moffitt, M. C., and B. A. Neilan. 2003. Evolutionary affiliations within the superfamily of ketosynthases reflect complex pathway associations. J. Mol. Evol. 56:446-457. [DOI] [PubMed] [Google Scholar]
- 22.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 23.Reeves, C. D., S. Murli, G. W. Ashley, M. Piagentini, C. R. Hutchinson, and R. McDaniel. 2001. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry 40:15464-15470. [DOI] [PubMed] [Google Scholar]
- 24.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schwecke, T., J. F. Aparicio, I. Molnar, A. Konig, L. E. Khaw, S. F. Haydock, M. Oliynyk, P. Caffrey, J. Cortes, J. B. Lester, G. A. Bohm, J. Stauton, and P. F. Leadlay. 1995. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. USA 92:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen, B. 2003. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 7:285-295. [DOI] [PubMed] [Google Scholar]
- 28.Staunton, J., and K. J. Weissman. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380-416. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki, M. T., O. Béjà, L. T. Taylor, and E. F. Delong. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3:323-331. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., F. Plewniak, J.-C. Thierry, and O. Poch. 2000. DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res. 28:2919-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torsvik, V., L. Ovreas, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 32.Watve, M. G., R. Tickoo, M. M. Jog, and B. D. Bhole. 2001. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176:386-390. [DOI] [PubMed] [Google Scholar]
- 33.Weinig, S., H. J. Hecht, T. Mahmud, and R. Muller. 2003. Melithiazol biosynthesis: further insights into myxobacterial PKS/NRPS systems and evidence for a new subclass of methyl transferases. Chem. Biol. 10:939-952. [DOI] [PubMed] [Google Scholar]
- 34.Yadav, G., R. S. Gokhale, and D. Mohanty. 2003. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328:335-363. [DOI] [PubMed] [Google Scholar]
- 35.Yadav, G., R. S. Gokhale, and D. Mohanty. 2003. SEARCHPKS: a program for detection and analysis of polyketide synthase domains. Nucleic Acids Res. 31:3654-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]