Abstract
Two genes encoding EngB endoglucanase and mini-CbpA1 scaffolding protein of Clostridium cellulovorans were constructed and coexpressed in Bacillus subtilis WB800. The resulting minicellulosomes were isolated by gel filtration chromatography and characterized. Biochemical and immunological evidence indicated that fraction II contained minicellulosomes consisting of mini-CbpA1 and EngB. The in vivo synthesis of minicellulosomes suggests that it will be possible in the future to insert into B. subtilis cellulosomal genes that will allow growth on cellulosic materials and the production of various designer cellulosomes with specific functions.
Clostridium cellulovorans, an anaerobic, mesophilic, spore-forming bacterium (14), produces a cellulosome with a molecular weight of about 1 million and in which several cellulases are bound tightly to a scaffolding protein, CbpA (3). Dockerin domains of cellulosomal enzyme subunits bind to hydrophobic domains termed “cohesins” (20, 27), which are repeated nine times in CbpA. So far, eight cellulosomal cellulase genes (engB, engE, engH, engK, engL, engM, engY, and exgS) (19) and the gene for the scaffolding protein CbpA (13) have been cloned and sequenced from C. cellulovorans.
It is very important to make recombinant cellulosomes, or designer cellulosomes, with specific functions for understanding the hydrolytic mechanism of cellulosomes and for industrial processes. There has been interest in constructing designer minicellulosomes of C. cellulovorans for several purposes (2, 8, 9, 10), such as for synergy studies between various cellulosomal enzymes and for improving the efficiency of cellulosomes. Production of intact C. cellulovorans endoglucanase, EngB, in Bacillus subtilis WB800 and the interaction with mini-CbpA produced in Escherichia coli allowed the successful construction of minicellulosomes in vitro (10). These studies indicated that a minicellulosome containing mini-scaffolding protein and cellulosomal enzyme subunits could be created and could be functional in vitro (4, 10). Also, there have been reports of the production of heterologous and chimeric scaffoldins in Clostridium acetobutylicum (11) and the in vivo production of recombinant minicellulosomes in C. acetobutylicum (12). These works showed the possibility of assembling in vivo a recombinant minicellulosome by overexpressing a mini-scaffolding protein.
We have used B. subtilis, which is an aerobic, mesophilic bacterium and an attractive host for the heterologous production of secretory proteins, as an expression host (22, 24). B. subtilis strain WB800, which is a strain deficient in eight extracellular proteases, was constructed by Wong and colleagues (25, 26, 28) to overcome the problem of protein degradation by extracellular B. subtilis proteases (1). To prepare recombinant cellulosomes, it is necessary to express intact cellulosomal subunits which retain their dockerin domains.
In this study, we report the construction of expression plasmids carrying genes encoding EngB endoglucanase and mini-CbpA1 scaffolding protein of C. cellulovorans and demonstrate the expression of these genes and the in vivo assembly of minicellulosomes in B. subtilis.
Construction of pWB980-mini-cbpA1-engB expression plasmid.
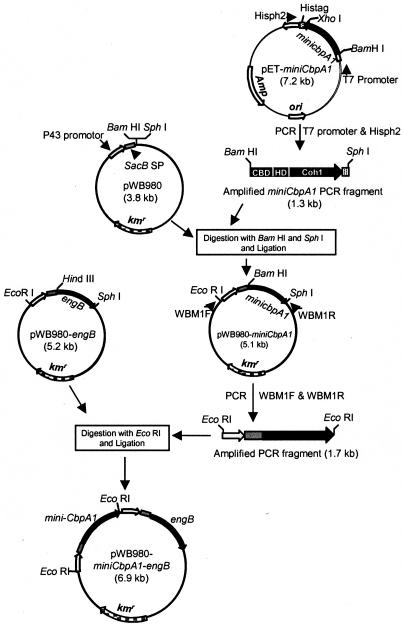
We have expressed C. cellulovorans cellulosomal enzyme subunit genes in E. coli successfully; however, these expressed subunits were partially degraded by E. coli proteases during the culture period. Therefore, the use of B. subtilis WB800, deficient in eight extracellular proteases, was essential for the production of intact EngB and minicellulosomes (26, 28). For B. subtilis expression, we chose the construction of a small mini-CbpA (i.e., mini-CbpA1), which consisted of a cellulose binding domain, a hydrophilic domain (HLD1), and one cohesin domain (Coh1) of the scaffolding protein, CbpA (17). The DNA fragment coding for mini-CbpA1 was amplified from pET-mini-cbpA1, which was designed for E. coli expression, by using primers T7 promoter (5′-TAATACGACTCACTATAGGG) and Hisph2 (5′-CGGGCTTTGTTAGCAGCGCATGCTCAGTGGTG) (Fig. 1). The Hisph2 primer was designed to allow an in-frame fusion at the C-terminal end with a His tag from pET-22b (Novagen). This amplified 1.3-kb fragment was introduced between the BamHI and SphI sites of the pWB980 Bacillus expression vector that carried the constitutively expressed P43 promoter (21); the engineered levansucrase signal sequence, sacB signal peptide (SP) (23); and the kanamycin resistance marker to generate pWB980-mini-cbpA1. The P43 promoter is a strong and constitutively expressed promoter and was used to direct the transcription of mini-cbpA1 and engB during growth. The sacB signal sequence allowed the recombinant proteins to be secreted into the culture broth. B. subtilis WB800 harboring pWB980-mini-cbpA1 produced intact mini-CbpA1 in B. subtilis and could be purified by a Ni2+-nitrilotriacetic acid agarose column (QIAGEN). For the two-gene construct of engB and mini-cbpA1, the pWB980-mini-cbpA1 plasmid was amplified with a pair of primers, WBM1F (5′-GGTTTTATGGTTTTGGTCGGCACTGAATTC) and WBM1R (5′-CGGGGCGAATTCGCATGCTCAGTGGTGGTG), which allowed the amplification of the mini-cbpA1 gene containing a promoter and signal sequence. The amplified 1.7-kb PCR fragment was digested with EcoRI, inserted into pWB980-engB (10), and digested with the same restriction enzyme to generate pWB980-mini-cbpA1-engB (Fig. 1). We chose engB for this experiment because it was the smallest cellulosomal cellulase gene of C. cellulovorans (19). Standard DNA transformation was performed with B. subtilis by the competent-cell method (15). The clone was selected on an agar plate containing, per liter, 10 g of peptone, 5 g of tryptone, and 5 g of NaCl supplemented with kanamycin (50 μg/ml).
FIG. 1.
Construction of the pWB980-mini-cbpA1-engB expression plasmid. For B. subtilis expression, we chose a small mini-CbpA, i.e., mini-CbpA1 consisting of a cellulose binding domain, a hydrophilic domain (HLD1), and one cohesin domain (Coh1). Plasmid pWB980, which is a Bacillus expression vector, carries both the constitutively expressed P43 promoter and sacB SP, the engineered levansucrase signal sequence for heterologous protein expression in B. subtilis (23). The sequences coding for the kanamycin resistance marker and levansucrase signal sequence are represented by kmr and sacB SP, respectively. Arrows show the transcription directions for the genes.
Expression and purification of minicellulosomes from B. subtilis.
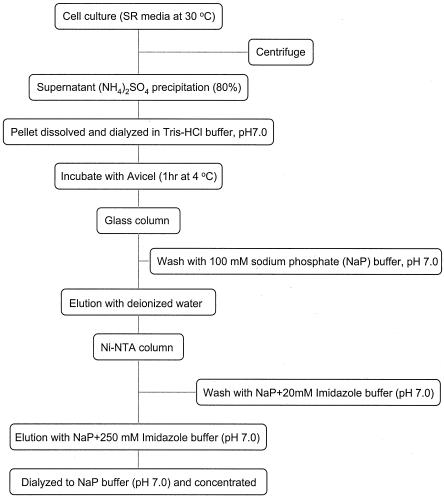
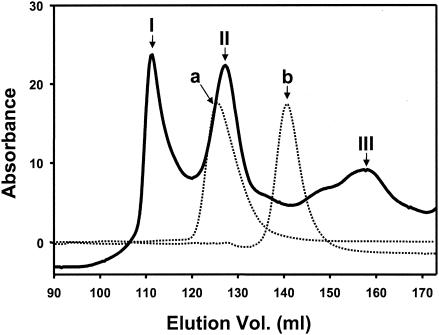
B. subtilis WB800 harboring pWB980-mini-cbpA1-engB was cultivated in 2,000 ml of superrich medium (containing, per liter, 25 g of yeast extract, 15 g of tryptone, and 3 g of K2HPO4 supplemented with kanamycin [50 μg/ml]) for 20 h at 30°C. Figure 2 shows the purification procedure for minicellulosomes. The culture was centrifuged to remove cells. Culture supernatant was collected, precipitated with ammonium sulfate at 80% saturation, and dialyzed. This enzyme solution was mixed with Avicel, which resulted in the binding of the minicellulosomes and free mini-CbpA1. After incubation for 1 h at 4°C, the suspension was poured into a glass column and the column was washed with 3 volumes of 100 mM sodium phosphate buffer (pH 7.0). The Avicel-bound fraction was eluted with deionized water and concentrated. This Avicel-bound enzyme solution was then applied to a Ni2+-nitrilotriacetic acid agarose column (QIAGEN) and washed with 2 column volumes of 50 mM sodium phosphate buffer with 20 mM imidazole (pH 7.0). The bound fraction was eluted with 250 mM imidazole in 50 mM sodium phosphate buffer (pH 7.0), dialyzed against 50 mM sodium phosphate buffer (pH 7.0), and concentrated with Vivaspin 20 mini-spin column (10-kDa cutoff) (Viva Science). The apparent size and homogeneity of the cellulosome preparation were determined by gel filtration on a Superdex 75 preparation-grade column (Amersham Pharmacia Biotech) with 50 mM sodium phosphate buffer as the elution buffer (Fig. 3). Three fractions (I, II, and III) were isolated. Fraction I corresponded to an apparent molecular mass of about 300 kDa, fraction II corresponded to an apparent molecular mass of about 120 kDa, and fraction III corresponded to an apparent molecular mass of about 50 kDa. These values were calculated from the calibration curve obtained with the standards used, i.e., aldolase (158 kDa) and bovine serum albumin (67 kDa). Based on the molecular mass, fraction II appeared to be the minicellulosome fraction. However, the molecular mass from the gel filtration result (120 kDa) was larger than the theoretical molecular mass of 94 kDa (mini-CbpA1, 46 kDa; EngB, 48 kDa). The difference in size seemed likely to be due to the resulting mini-CbpA1-EngB complex being an elongated molecule rather than a globular molecule. Since both the shape and the mass of the molecule determine the apparent molecular mass of this molecule by the gel filtration method, the deviation from the expected molecular mass in this case may be attributed to the nonglobular shape of the complex.
FIG. 2.
Purification procedure for minicellulosomes from B. subtilis WB800 harboring pWB980-mini-cbpA1-engB. SR, superrich; NTA, nitrilotriacetic acid.
FIG. 3.
Gel filtration chromatography of the water-eluted and Ni2+-nitrilotriacetic acid agarose-purified cellulosome fraction on a Superdex 75 preparation-grade column. Fractions I, II, and III are shown as solid lines, and standards are shown as dotted lines; (a, aldolase [158 kDa]; b, bovine serum albumin [67 kDa]).
Characterization of minicellulosomes assembled in B. subtilis.
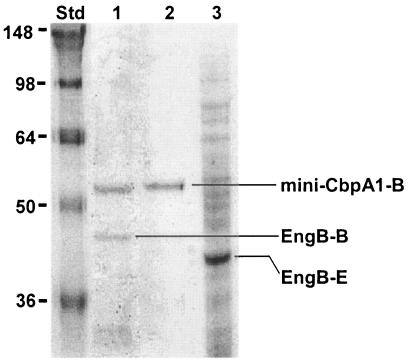
Two fractions from the gel filtration analysis were collected and characterized. Based on the estimated molecular mass from gel filtration analysis, fraction II was considered to be the minicellulosome fraction. We subjected fraction II to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to identify the subunit composition (Fig. 4, lane 1). Purified mini-CbpA1 from B. subtilis (Fig. 4, lane 2) and the crude extract from E. coli harboring the engB gene (Fig. 4, lane 3) were also analyzed as standards. From the SDS-PAGE gel, fraction II (lane 1) showed two bands. The upper band exactly matched the purified mini-CbpA1 from B. subtilis (lane 2), although the molecular mass was larger than the theoretical value (46 kDa). This result showed the difference between the theoretical and experimental molecular mass determinations from SDS-PAGE and gel filtration chromatography. The lower band visible in Fig. 4, lane 1, is EngB, as demonstrated later by Western blotting and zymogram analysis. The band visible in Fig. 4, lane 3, was larger than the EngB produced from E. coli, because E. coli produces a truncated EngB without dockerin (5). From these SDS-PAGE results, we could see that fraction II (Fig. 4, lane 1) consisted mainly of mini-CbpA1 and EngB.
FIG. 4.
SDS-PAGE analysis of fraction II stained with Coomassie blue. Lane 1, fraction II (mini-CbpA1-B and EngB-B); lane 2, purified mini-CbpA1 from B. subtilis WB800 (mini-CbpA1-B); lane 3, crude extract from E. coli harboring the engB gene (EngB-E). Lane Std, molecular weight standards (indicated in thousands).
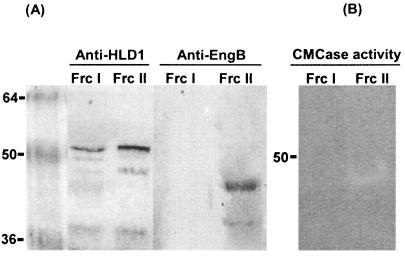
To verify the SDS-PAGE results, we performed a Western blot analysis. The samples were subjected to SDS-10% PAGE and blotted onto nitrocellulose membranes for Western blot analysis (Fig. 5A). We used two kinds of antibodies for detection. We have defined anti-HLD as the antibody made against one of the hydrophilic domains of CbpA, and thus anti-HLD1 is specific for hydrophilic domain 1 of CbpA. Anti-EngB is the specific antibody for EngB. The membrane incubated with the HLD1-specific antibody revealed bands corresponding to mini-CbpA1, showing that both fractions I and II contained mini-CbpA1. The membrane incubated with EngB-specific antibody revealed that only fraction II contained a band corresponding to EngB. These results confirmed that fraction II consisted of mini-CbpA1 and EngB, which had been assembled into minicellulosomes in vivo. Fraction III contained only free mini-CbpA and a small amount of free EngB, as shown by Western blot analysis (data not shown).
FIG. 5.
Characterization of fractions I and II. (A) Western blot showing detection with antisera raised against HLD1 and EngB, respectively. Anti-HLD1 was made against the hydrophilic domain 1 of CbpA, and anti-EngB was made against EngB. (B) Zymogram analysis with 0.1% carboxymethyl cellulase (CMCase) incorporated into the polyacrylamide. Lane Frc I, fraction I; lane Frc II, fraction II. Molecular weights are indicated on the left in thousands.
As an additional method for confirming the nature of the minicellulosome, zymograms were created by using a 0.1% (wt/vol) concentration of carboxymethyl cellulose incorporated into the polyacrylamide gel. After electrophoresis, the gel was incubated at 30°C for 2 h in 50 mM sodium acetate buffer (pH 6.0). The enzymatic activity was visualized after staining with 0.25% Congo red and destaining with 1 M NaCl (6). As a result, strong EngB activity was detected in fraction II, which corresponded to a molecular weight of about 48 kDa, and no activity was detected in fraction I. However, this minicellulosome could not degrade Avicel. From the immunological and biochemical evidence, we concluded that fraction II contained minicellulosomes consisting of mini-CbpA1 and EngB. The yield of minicellulosomes in fraction II was approximately 0.2 mg/liter of culture.
Until now, the C. cellulovorans cellulosomal engB and mini-cbpA genes had been expressed in E. coli (5, 7, 16, 17, 18, 19) and an in vitro interaction had been shown between EngB and mini-CbpA. This is the first report that a cellulosomal cellulase gene and a scaffolding gene could be coexpressed and could form a minicellulosome in B. subtilis in vivo. Our goal is to express minicellulosomes and a variety of designer cellulosomes in vivo in high yields. The in vivo construction of minicellulosomes suggests that it will be possible in the future to insert into B. subtilis cellulosomal genes that will produce various designer cellulosomes with specific functions. Moreover, with further development of this system, B. subtilis may be able to grow on cellulose and on plant cell wall materials and produce valuable products from inexpensive substrates.
Acknowledgments
We thank Helen Chan for skillful technical assistance.
This research was supported in part by a grant from the Research Institute of Innovative Technology for the Earth and by U.S. Department of Energy grant DDF03-92ER20069.
REFERENCES
- 1.Aminov, R. I., N. P. Golovchenko, and K. Ohmiya. 1995. Expression of a celE gene from Clostridium thermocellum in Bacillus. J. Ferment. Bioeng. 79:530-537. [Google Scholar]
- 2.Doi, R. H., A. Kosugi, K. Murashima, Y. Tamaru, and S. O. Han. 2003. Cellulosomes from mesophilic bacteria. J. Bacteriol. 185:5907-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doi, R. H., and Y. Tamaru. 2001. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem. Rec. 1:24-32. [DOI] [PubMed] [Google Scholar]
- 4.Doi, R. H., J. S. Park, C. C. Liu, L. M. Malburg, Y. Tamaru, A. Ichiishi, and A. Ibrahim. 1998. Cellulosome and noncellulosomal cellulases of Clostridium cellulovorans. Extremophiles 2:53-60. [DOI] [PubMed] [Google Scholar]
- 5.Foong, F., T. Hamamoto, O. Shoseyov, and R. H. Doi. 1991. Nucleotide sequence and characteristics of endoglucanase gene engB from Clostridium cellulovorans. J. Gen. Microbiol. 137:1729-1736. [DOI] [PubMed] [Google Scholar]
- 6.Kosugi, A., K. Murashima, and R. H. Doi. 2001. Characterization of xylanolytic enzymes in Clostridium cellulovorans: expression of xylanase activity dependent on growth substrates. J. Bacteriol. 183:7037-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu, C. C., and R. H. Doi. 1998. Properties of exgS, a gene for a major subunit of the Clostridium cellulovorans cellulosome. Gene 211:39-47. [DOI] [PubMed] [Google Scholar]
- 8.Murashima, K., A. Kosugi, and R. H. Doi. 2003. Synergistic effects of cellulosomal xylanase and cellulases from Clostridium cellulovorans on plant cell wall degradation. J. Bacteriol. 185:1518-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Synergistic effects on crystalline cellulose degradation between cellulosomal cellulases from Clostridium cellulovorans. J. Bacteriol. 184:5088-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murashima, K., C.-L. Chen, A. Kosugi, Y. Tamaru, R. H. Doi, and S.-L. Wong. 2002. Heterologous production of Clostridium cellulovorans engB, using protease-deficient Bacillus subtilis, and preparation of active recombinant cellulosomes. J. Bacteriol. 184:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perret, S., L. Casalot, H.-P. Fierobe, C. Tardif, F. Sabathé, J.-P. Belaich, and A. Belaich. 2004. Production of heterologous and chimeric scaffoldins by Clostridium acetobutylicum ATCC 824. J. Bacteriol. 186:253-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabathé, F., and P. Soucaille. 2003. Characterization of the CipA scaffolding protein and in vivo production of a minicellulosome in Clostridium acetobutylicum. J. Bacteriol. 185:1092-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoseyov, O., M. Takagi, M. A. Goldstein, and R. H. Doi. 1992. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc. Natl. Acad. Sci. USA 89:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sleat, R., R. A. Mah, and R. Robinson. 1984. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium cellulovorans sp. nov. Appl. Environ. Microbiol. 48:88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleotide. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamaru, Y., and R. H. Doi. 2000. The engL gene cluster of Clostridium cellulovorans contains a gene for cellulosomal ManA. J. Bacteriol. 182:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamaru, Y., and R. H. Doi. 2001. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. Proc. Natl. Acad. Sci. USA 98:4125-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamaru, Y., and R. H. Doi. 1999. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J. Bacteriol. 181:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokatlidis, K., P. Dhurjati, and P. Beguin. 1993. Properties conferred on Clostridium thermocellum endoglucanase CelC by grafting the duplicated segment of endoglucanase CelD. Protein Eng. 6:947-952. [DOI] [PubMed] [Google Scholar]
- 21.Wang, P. Z., and R. H. Doi. 1984. Overlapping promoters transcribed by Bacillus subtilis sigma 55 and sigma 37 RNA polymerase holoenzymes during growth and stationary phases. J. Biol. Chem. 259:8619-8625. [PubMed] [Google Scholar]
- 22.Wong, S. L. 1995. Advances in the use of Bacillus subtilis for the expression and secretion of heterologous proteins. Curr. Opin. Biotechnol. 6:517-522. [DOI] [PubMed] [Google Scholar]
- 23.Wong, S. L. 1989. Development of an inducible and enhancible expression and secretion system in Bacillus subtilis. Gene 83:215-223. [DOI] [PubMed] [Google Scholar]
- 24.Wong, S.-L., R. Ye, and S. Nathoo. 1994. Engineering and production of streptokinase in a Bacillus subtilis expression-secretion system. Appl. Environ. Microbiol. 60:517-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, S.-C., J. C. Yeung, Y. Duan, R. Ye, S. J. Szarka, H. R. Habibi, and S.-L. Wong. 2002. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl. Environ. Microbiol. 68:3261-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, X.-C., W. Lee, L. Tran, and S.-L. Wong. 1991. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J. Bacteriol. 173:4952-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaron, S., E. Morag, E. A. Bayer, R. Lamed, and Y. Shoham. 1995. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 360:121-124. [DOI] [PubMed] [Google Scholar]
- 28.Ye, R., L. P. Yang, and S.-L. Wong. 1996. Construction of protease-deficient Bacillus subtilis strains for expression studies: inactivation of seven extracellular proteases and the intracellular LonA protease, p. 160-169. In S. K. Hong and W. J. Lim (ed.), Proceedings of the International Symposium on Recent Advances in Bioindustry. The Korean Society of Applied Microbiology, Seoul, Korea.