Abstract
Assessment of health risk associated with fecal pollution requires a reliable fecal indicator and a rapid quantification method. We report the development of a Taq nuclease assay for enumeration of 16S rRNA genes of Bacteroidetes. Sensitivity and correlation with standard fecal indicators provide experimental evidence for application of the assay in monitoring fecal pollution.
The enumeration of fecal indicators is the cornerstone of testing for fecal pollution in recreational, potable, and shellfish water. There is currently no single bacterial indicator used in all water systems. Total coliforms are the U.S. Environmental Protection Agency (EPA) standard indicators of pollution for drinking water (14), Escherichia coli and enterococci are approved for freshwater (12), enterococci are recommended for marine water (12), and fecal coliforms are used for shellfish waters (15). Traditional membrane filtration and most probable number (MPN) methods require 24 to 74 h for enumeration. Recent updates in methods include the Colilert-18 test (Idexx Laboratories, Westbrook, Maine), which has received U.S. EPA approval for use in ambient water testing (13). Colilert-18 uses a defined substrate medium to test colorimetrically for E. coli and total coliforms within 18 h. E. coli is considered a more specific fecal indicator than total or fecal coliforms, which are found in ambient water in the absence of fecal pollution (11). Epidemiological studies have established a correlation between standard fecal indicators and associated human health risks (5, 7, 12).
Fecal members of the class Bacteroidetes have distinct advantages over coliforms and E. coli as fecal indicators. They are more abundant in the feces of warm-blooded animals than E. coli (8). They are likely to predict recent fecal contamination because they are obligate anaerobes and are unlikely to survive long outside the intestinal tract (1, 8). Enterococci and E. coli are facultative anaerobes, and they can proliferate in soil, sand, and sediments (6, 10, 16, 17).
Bernhard and Field (3, 4) developed 16S rRNA gene (rDNA) markers from Bacteroidetes to detect fecal pollution and to distinguish between human and ruminant sources by PCR. Markers for additional host sources have been recently developed (L. K. Dick, A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field, unpublished data). PCR source identification is rapid, specific, and sensitive, and it does not require maintenance of databases or libraries of bacterial isolates.
Here we report a quantitative Taq nuclease assay (TNA) (2, 9) for general fecal pollution using a Bacteroidetes 16S rDNA marker. The TNA was compared with the Colilert-18 system for accuracy, range, and limits of quantification in serial dilutions of primary sewage influent.
A fluorogenic probe and primer set was designed for Bacteroidetes 16S rDNA by using the Primer Design function in the ARB software program (Ludwig and Strunk, Munich, Germany). The sequences were verified for use in a TNA with Primer Express software (PE Applied Biosystems, Foster City, Calif.). The primers did not bind to fecal bacteria outside the class Bacteroidetes when up to five mismatches were chosen by the ARB Probe Match program. They did amplify 16S rDNA of Bacteroidetes from human, cow, dog, cat, pig, elk, deer, and gull feces. Sequences used were AACGCTAGCTACAGGCTTAACA (3), ACGCTACTTGGCTGGTTCA (this study), and CAATATTCCTCACTGCTGCCTCCCGTA (this study), for the forward primer, reverse primer, and probe, respectively.
We collected 1 liter of primary influent from the Corvallis Wastewater Reclamation plant in Corvallis, Oreg. It was transported and stored in a sterile polypropylene container on ice. Six separate 10-fold serial dilutions to 10−10were made in 100-ml volumes in sterile glass containers with nanopure water. Colilert-18 tests were performed on three of the dilution sets. The other three sets were filtered, and the bacteria were extracted and used in the TNA. A nondiluted influent sample was not included in the experiment because it clogged the filter.
Three sets of 100-ml primary influent serial dilutions were filtered through 47-mm-diameter, 0.2-μm-pore-size filters (Supor-200 membrane disk filters; Pall Gelman Laboratory, Ann Arbor, Mich). The glass filtration apparatus was heat sterilized prior to use and was soaked for 3 min in 20% bleach and rinsed under distilled water between filtrations. The filters were placed in 500 μl of guanidine isothiocyanate buffer (5 M guanidine isothiocyanate, 100 mM EDTA [pH 8], 0.5% Sarkosyl) in 15-ml polypropylene tubes. DNA was extracted by using the DNeasy tissue kit (QIAGEN, Valencia, Calif.) with a slightly modified protocol. We omitted the proteinase K digestion, and 500 μl of QIAGEN AL buffer was added to the guanidine isothiocyanate filter and vortexed for 1 min. A second wash step was used to ensure a clean product, and the DNA was eluted in 200 μl of Tris-HCl buffer.
Amplifications were run on an ABI Prism 7700 (PE Applied Biosystems). The 25-μl PCR mixtures included 1× TaqMan buffer A (PE Applied Biosystems), 3.5 mM MgCl2, 400 μM dUTP, a 200 μM concentration of each remaining deoxynucleoside triphosphate, a 0.4 μM concentration of each primer, a 0.2 μM concentration of the fluorogenic probe, 0.06% bovine serum albumin, 0.25 U of uracil-N-glycosylase, 0.63 U of AmpliTaq Gold, and 2 μl of template DNA. Cycling parameters were 2 min at 50°C for uracil-N-glycosylase activation, 10 min at 95°C for denaturation, and 40 cycles of 15 s at 95°C, followed by 1 min at 60°C for annealing and extension. All of the reaction mixtures were run in triplicate, and a standard curve was created from serial dilutions of plasmid DNA containing known copy numbers of the template.
The Colilert-18 kit was used with the manufacturer's protocol. Briefly, medium was added to each 100-ml diluted sample, mixed, and transferred to 97-well trays (Quanti-Tray 2000; Idexx Laboratories). The trays were sealed and incubated at 35°C for 18 h. Wells that turned yellow were positive for coliforms, and wells that fluoresced under UV light were positive for E. coli. An MPN chart provided by the manufacturer was used for enumeration.
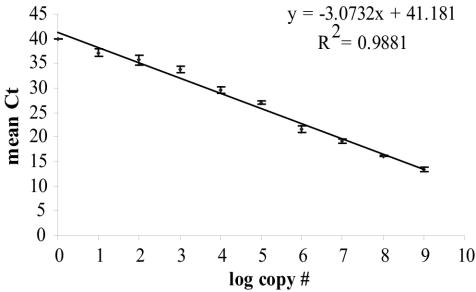
The threshold cycle method used in a TNA allows a wide, dynamic range for the calibration of copy numbers to standards. During 40 cycles of PCR, standards were quantified over 8 orders of magnitude, down to 10 copies (Fig. 1). The primers provided specificity, while the probe was more general. Becker et al. (2) found that using primers with specificity greater than or equal to that of the probe helped to reduce the PCR bias inherent when a background of complex DNA is present.
FIG. 1.
TNA dynamic range for enumeration of 16S rDNA of Bacteroidetes. Plasmid DNAs containing known template copy numbers were added in 10-fold serial dilutions to create the standard curve. Mean threshold cycle values (Ct) are plotted against corresponding DNA log concentrations of Bacteroidetes. Error bars represent the standard deviations among results of triplicate PCRs.
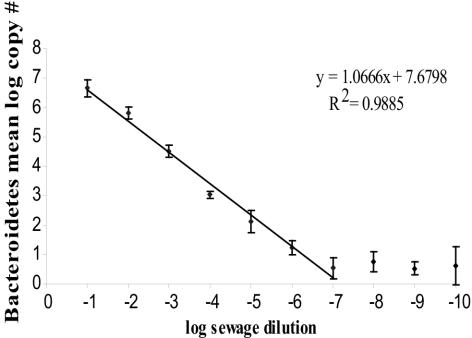
There was a linear decrease in copy numbers of 16S rDNA of Bacteroidetes as a function of serially diluting the sewage influent (Fig. 2). The decrease remained linear over 7 orders of magnitude. After the assay reached its minimum detection level of about 10 copies, it became inconsistent. The linearity of the dilution curve shows that neither PCR inhibitors nor the presence of large amounts of heterologous DNA inhibited amplification, even at the highest copy numbers.
FIG. 2.
TNA analysis of 16S rDNA copy numbers of Bacteroidetes in serial dilutions of sewage primary influent. Geometric mean copy numbers are plotted against log serial sewage dilutions. The error bars represent the standard deviations from results of three replicate serial dilutions of sewage.
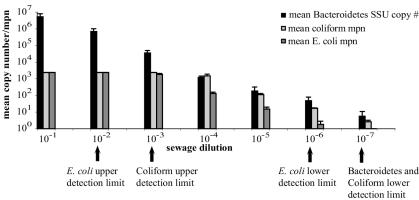
16S rDNA copy numbers of Bacteroidetes are compared with coliform and E. coli MPNs in sewage dilutions in Fig. 3. Geometric means were used, since they are the standard for coliform and E. coli data (12). The Colilert-18 upper limit of enumeration is 2,419 organisms for both coliforms and E. coli (Idexx Laboratories). This limit was reached or exceeded in the first three serial dilutions for coliforms and in the first two dilutions for E. coli. Copy numbers of Bacteroidetes were quantifiable in the first three dilutions. Lower limits of enumeration for Colilert-18 (one organism per 100 ml of dilution [Idexx Laboratories]) were reached at the 10−6 dilution for E. coli and the 10−7 dilution for coliforms. The limit for Bacteroidetes of 10 copies was also reached at the 10−7 dilution. Because cells of Bacteroidetes contain multiple copies of the 16S rRNA gene, 10 copies represents as few as one or two cells, and it suggests that the lower limits of enumeration for all three groups are approximately the same.
FIG. 3.
Comparison of copy numbers of 16S rDNA of Bacteroidetes with coliform and E. coli MPNs in 10-fold primary-influent dilutions. Equivalent sewage volumes (100 ml) were used for the TNAs and the Colilert assays, but only 1 μl of the total filtered, extracted DNA was used in the TNA. The MPN and copy number data were log transformed. The error bars represent 1 standard deviation based on geometric means of results for triplicate sewage dilutions.
Simple linear correlation analysis of the 10−4 to 10−7 dilutions showed that mean concentrations of Bacteroidetes were highly and positively correlated with both coliform and E. coli mean concentrations (r values of 0.999 for both comparisons). This dilution range contains the U.S. EPA threshold concentration for E. coli in ambient water above which the health risk from waterborne illness is deemed unacceptably high (5-day geometric mean of 126 organisms/100 ml) (12).
The TNA for 16S rDNA of Bacteroidetes is rapid, sensitive, and reproducible in sewage dilutions. It correlates with present standard indicators but takes only 3 to 4 h to complete. Recent advances in rapid thermocycling allow 40 cycles of PCR to be completed in 30 min, potentially reducing the assay time even more. The quantitative range spans 8 orders of magnitude, compared with approximately 4 orders for E. coli enumeration. However, it will be necessary to validate the temporal and spatial application of the assay by obtaining data from additional sites and over several seasons.
The quantitative assay described here provides a framework for expanding the use of PCR indicators for Bacteroidetes beyond fecal source tracking and into a health risk-based analysis of fecal pollution.
ADDENDUM IN PROOF
When tested on seawater samples, the quantitative assay reported here amplified nonfecal Bacteroidetes species, including Cytophaga fermentans. New quantitative primers GCTCAGGATGAACGCTAGCT (forward) and CCGTCATCCTTCACGCTACT (reverse), specific for sequences adjacent to and partially overlapping those specified by the original primers, amplified fecal Bacteroidetes only but otherwise had quantitative characteristics identical to those of the original primers.
Acknowledgments
This work was partially supported by grant R82-7639 from the U.S. Environmental Protection Agency, grant 00-S1130-9818 from the U.S. Department of Agriculture, and grant NA76RG0476 (project no. R/ECO-04) from the National Oceanic and Atmospheric Administration to the Oregon Sea Grant College Program.
REFERENCES
- 1.Allsop, K., and D. J. Stickler. 1985. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J. Appl. Bacteriol. 58:95-99. [DOI] [PubMed] [Google Scholar]
- 2.Becker, S., P. Boger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabelli, V. J., A. P. Dufour, M. A. Levin, and L. J. McCage. 1982. Swimming-associated gastroenteritis and water quality. Am. J. Epidemiol. 115:606-616. [DOI] [PubMed] [Google Scholar]
- 6.Desmarais, T. R., H. M. Solo-Gabriele, and C. J. Palmer. 2001. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dufour, A. P. 1984. Health effects criteria for fresh recreational waters. EPA/600/R-84/004. U.S. Environmental Protection Agency. Cincinnati, Ohio.
- 8.Fiksdal, L., J. S. Maki, S. J. LaCroix, and J. T. Staley. 1984. Survival and detection of Bacteroides spp., prospective indicator bacteria. Appl. Environ. Microbiol. 49:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 10.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 1999. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toranzos, G. A., and G. A. McFeters. 1997. Detection of indicator microorganisms in environmental freshwaters and drinking waters, p. 184-194. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. American Society for Microbiology, Washington, D.C.
- 12.U.S. Environmental Protection Agency. 2003. Bacterial water quality standards for recreational waters (freshwater and marine waters). EPA/823/R-03/008. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 13.U.S. Environmental Protection Agency. 2003. Guidelines establishing test procedures for the analysis of pollutants: analytical methods for biological pollutants in ambient water, final rule. Fed. Regist. 68:43272-43283. [Google Scholar]
- 14.U.S. Environmental Protection Agency. 2001. Total coliform rule: a quick reference guide. EPA/816/F-01/035. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 15.U.S. Food and Drug Administration. 2000. National Shellfish Sanitation Program model ordinance. Office of Seafood, USDA Center for Food Safety and Applied Nutrition, Rockville, Md.
- 16.Wheeler, A. E., J. Burke, and A. Spain. 2003. Fecal indicator bacteria are abundant in wet sand at freshwater beaches. Water Res. 37:3978-3982. [DOI] [PubMed] [Google Scholar]
- 17.Whitman, R. L., D. A. Shively, H. Pawlik, M. B. Nevers, and M. N. Byappanahalli. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69:4714-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]