Abstract
Total direct counts of bacterial abundance are central in assessing the biomass and bacteriological quality of water in ecological and industrial applications. Several factors have been identified that contribute to the variability in bacterial abundance counts when using fluorescent microscopy, the most significant of which is retaining an adequate number of cells per filter to ensure an acceptable level of statistical confidence in the resulting data. Previous studies that have assessed the components of total-direct-count methods that contribute to this variance have attempted to maintain a bacterial cell abundance value per filter of approximately 106 cells filter−1. In this study we have established the lower limit for the number of bacterial cells per filter at which the statistical reliability of the abundance estimate is no longer acceptable. Our results indicate that when the numbers of bacterial cells per filter were progressively reduced below 105, the microscopic methods increasingly overestimated the true bacterial abundance (range, 15.0 to 99.3%). The solid-phase cytometer only slightly overestimated the true bacterial abundances and was more consistent over the same range of bacterial abundances per filter (range, 8.9 to 12.5%). The solid-phase cytometer method for conducting total direct counts of bacteria was less biased and performed significantly better than any of the microscope methods. It was also found that microscopic count data from counting 5 fields on three separate filters were statistically equivalent to data from counting 20 fields on a single filter.
Bacteria play a central role in the cycling of carbon, organic and inorganic nutrients, minerals, and metals in aquatic systems. Their biomass is also a source of carbon and nutrients for higher organisms in these systems. Accordingly, accurate measurements of bacterial abundance and biomass are important factors to consider when assessing the role of bacteria in aquatic systems. Epifluorescent microscopy (EFM) has become a standard technique for estimating the abundance, size, biovolume, biomass, and physiological activity of bacteria recovered from aquatic systems and retained on membrane filters (7, 11, 16-18). Though this technique is relatively simple and easy to perform, several factors have been identified that, if not eliminated or minimized, can significantly increase the variability and therefore affect the reliability of the resulting data. When using membrane filtration to retain and concentrate bacteria, one of the more significant variables to consider is having a sufficient number of cells present on a filter's surface to count and provide a level of acceptable reliability to the resulting data (6).
As the abundance of bacteria in a sample decreases, increasing volumes of sample must be filtered to maintain the same level of confidence in the final direct-count estimates. However, increasing the volume of water filtered through a single membrane filter is not always a practical option, as nonbacterial biomass and debris clog the types of filters routinely used in total-direct-count methods. The retained nonbacterial material can nonspecifically bind the fluorescent stain, be autofluorescent, or physically cover the bacterial cells. The effect each of these situations has on total direct counts, either individually or in combination, is a false reduction in the abundance of bacteria per filter.
Another concern for calculating the final abundance of bacteria per filter is the low percentage of the effective filtration area that is subsampled by accepted microscopic techniques. For example, when a filtration funnel tower with an inside diameter of 17 mm (providing an effective filtration area of 2.27 × 108 μm2) is used to count a sample in which there are 25 cells grid−1 in an ocular reticle with an area of 1.00 × 104 μm2 and the microscopist counts 20 grids at a total magnification of ×1,250 to achieve a total cell count of 500 cells, only 0.09% of the effective filtration area has been subsampled. However, the time and effort needed to count the number of cells on a more representative percentage of a membrane filter's surface would be prohibitive in most laboratories.
A recently developed technology, solid-phase cytometry (SPC), avoids this problem as it is capable of scanning, detecting, and counting all fluorescently labeled bacteria on the entire surface area of a 25-mm-diameter membrane filter in less than 3 min (16). This technology has been used by several groups to assess bacterial abundances and viability in different types of water (2, 4, 5, 10, 15, 16, 21-23, 25, 26). SPC combines a rapid detection system with direct microscopic verification of each fluorescent event, allowing the microscopist to determine if a recorded fluorescent event truly represents a bacterium. Having a detection sensitivity of a single bacterium on a filter's surface minimizes the necessity of having to filter large volumes of water to achieve an abundance of cells per filter that will provide an acceptable level of confidence in the resulting data. Additionally, the SPC counts all bacteria on a membrane's surface, thereby minimizing the inherent errors associated with counting bacteria in a limited number of fields and subsequently estimating the total number of cells per filter.
In this study we compare traditional EFM techniques for counting fluorescently labeled beads and bacteria to the SPC method to establish the minimum number of cells needed per filter for reliable total-direct-count data. We compared the bias and overall error of EFM and SPC after counting a range of fluorescently labeled beads and bacteria retained on membrane filters. Additionally, we used these same methods to determine if random selection of microscope fields provides more reliable data than systematic selection of areas, within which random fields are counted, when counting 20 fields on a single filter or 5 fields on each of three filters.
MATERIALS AND METHODS
Fluorescent beads.
Fluorescent beads (Fluoresbrite plain YG; 0.485 ± 0.01 μm in diameter) were purchased from Polysciences, Inc. (Warrington, Pa.). The stock bead suspension (approximately 3.54 × 1011 beads ml−1) was stored at 4°C in the dark. Three replicate dilution series were prepared from the stock bead suspension in sterile reagent-grade water (Milli-Q UV Plus; Millipore Corp., Bedford, Mass.) immediately prior to each experiment for each of the respective bead abundances. The final volume for each bead suspension was 1.0 ml. Prior to filtering each bead suspension, 1.0 ml of sterile reagent-grade water was added to a glass filtration funnel, followed by the addition of the bead suspension and a second 1.0 ml of sterile reagent-grade water. The final filtration volume was 3.0 ml. This bead suspension was filtered onto black polycarbonate filters (25 mm diameter, 0.2 μm pore size; GTBP Isopore Membranes; Millipore Corp.), under which a support filter (25 mm diameter, 0.8 μm pore size; AAWP; Millipore Corp.) was placed. Vacuum pressures were maintained between 50 to 70 mm Hg for all samples. The filtration funnel was rinsed three times with 3.0-ml volumes of sterile reagent-grade water. The filter membrane was transferred to a microscope slide, where a 25-mm-diameter coverslip, on which 20 μl of phosphate-buffered saline (13.9 mM NaCl, 1.0 mM Na2HPO4, 0.2 mM KH2PO4, 0.3 mM KCl, pH 8.5) had been pipetted, was gently dropped into place on top of the filter. The same filtration funnel, with an inside diameter of 16.6 mm, and fritted glass support (25 mm; Millipore Corp.) were used with all samples to maintain uniformity in effective filtration area within and between experiments. The target bead abundances were 104, 104.5, 105.5, 105.7, and 106 beads filter−1.
Bacterial strain and culture and labeling conditions.
Escherichia coli O157:H7 strain 932 was kindly provided by the U.S. Environmental Protection Agency. Cultures of E. coli O157:H7, maintained at −80°C in dimethyl sulfoxide (3.5% final concentration), were used to inoculate 100 ml of YT broth (containing, per liter, 10 g of tryptone, 5 g of yeast extract, 5 g of glucose, and 5 g of sodium chloride, pH 7.2). Cultures were grown for 16 to 18 h at 25°C and 130 rpm. The primary cultures were diluted in sterile reagent-grade water to a final abundance of 108 cells ml−1, as determined by Klett-Summerson colorimeter values and intralaboratory standardized growth curve data for this strain (data not shown). The adjusted cultures were used to make three replicate dilution series at the appropriate abundances and were filtered as described for the fluorescent beads. The cells were subsequently labeled by placing the filters on sterile pads saturated with the fluorogenic esterase substrate, ChemChrome V3, which had been diluted 100-fold in ChemSol B1 (Chemunex, Paris, France). The filters were incubated in the dark for 30 min at 30°C. The target bacterial abundances were 104, 105, and 105.7 cells filter−1.
Microscopic counting of fluorescent beads and bacteria.
All microscopic counts were performed on a Nikon Optiphot epifluorescent microscope system, equipped with a 100-W mercury light source, ×10 oculars, Nikon filter cube B-2A (excitation wavelength, 450 to 490 nm; dichroic mirror, 500 nm; barrier filter, 515 nm), and an automated stage (Prior Scientific Instruments Ltd., Cambridge, United Kingdom) that could be controlled manually or automatically. The traditional epifluorescent microscopic counts (i.e., the subjective and objective counting methods described below) were performed with a ×100 (UV-F) objective (total magnification, ×1,250). The SPC counts were performed with the same microscope, except a ×60 (Plan 60 NCG) objective (total magnification, ×750) was used for verification, as required by the manufacturer. Each sample was processed and analyzed by all counting methods on the same day. The same person prepared the suspensions and performed all assays.
Three methods were used for counting fluorescent beads and bacteria on the respective filters.
(i) Subjective counting methods.
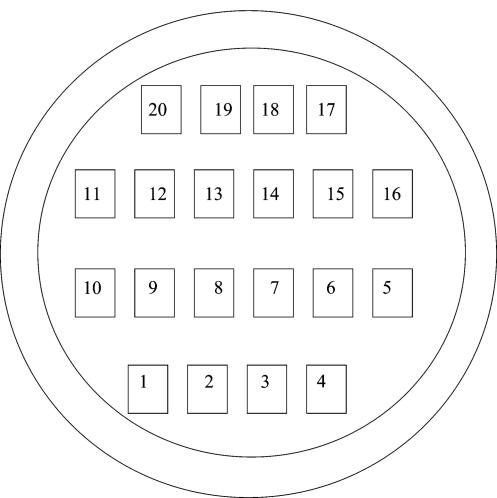
Three replicate filters were prepared, as described previously, for the respective fluorescent bead and bacterial abundances. Subjective method A (SubjA) systematically selected and counted 20 fields, as defined by a calibrated ocular grid reticle, on the first of three filters, following the pattern shown in Fig. 1. Subjective method B (SubjB) systematically selected five fields, from the areas numbered 5, 7, 10, 13, and 15 in Fig. 1, and were counted on the remaining two filters. The field counts from these same five areas determined by using the SubjA count method were included in the SubjB data sets to provide a triplicate set of data for the SubjB method. Preliminary experiments had shown that fluorescent beads and bacteria were retained on filters outside the measured inside diameter of the filtration funnel to approximately 17.0 mm (data not shown). To ensure this additional area was included in the random field selection, a filter diameter of 17.5 mm was used in the calculation of the effective filtration area.
FIG. 1.
General pattern used to select regions of the membrane filter from which microscope fields were selected when using the subjective microscope methods for counting beads and bacteria. The innermost area represents the effective filtration area of the filter membrane. The outer area represents the area of the membrane filter on which filtered beads or bacteria were not retained.
(ii) Objective counting method.
For the objective method, the software program that controls the automated stage was modified to randomly select nonoverlapping fields on each filter. The three filters used for the subjective counting methods were subsequently used for the objective counting methods, counting 20 fields on a single filter (objective method A [ObjA]) and counting 5 fields on three separate filters (objective method B [ObjB]). A filter diameter of 17.5 mm was used in the calculation of the effective filtration area.
(iii) SPC counting method.
The ScanRDI (Chemunex) is an SPC that is capable of detecting and counting fluorescent events (e.g., fluorescent beads and fluorescently labeled bacteria) that have been excited by a fixed-wavelength laser (488 nm) while scanning the surface of a 25-mm-diameter membrane (16, 23). The same fluorescent bead and bacterial suspensions used to inoculate the three filters for the subjective and objective count methods were diluted further to achieve a final abundance of approximately 200 beads or bacteria per filter. Each SPC filter was inoculated with 1.0 ml of the respective suspensions, following the same protocol as described for the microscope methods, except a coverslip was not placed on the filter prior to counting. Preliminary studies had shown 200 fluorescent events could be counted and verified by the SPC before the filter started to dehydrate (data not shown). The instrument was set to scan an area with a 20-mm diameter. After a successful scan each filter was transferred to the automated stage on the epifluorescent microscope, where each fluorescent event was positioned within the field of view by the automated stage. Each fluorescent event was visually verified as a bead, bacterium, or piece of debris.
Statistical analysis.
The total counts from each filter were multiplied by the appropriate microscope conversion and dilution factors to provide the final abundance of beads or cells per milliliter and were subsequently log10 transformed. Similarly, the SPC data were adjusted for the appropriate dilution factors to provide the bead and bacterial cell abundances per milliliter and were log10 transformed.
The bias associated with each of the methods was evaluated separately for each target abundance by using analysis of variance methods (20). The analysis of variance produced an estimate of the bias (B) represented by the following equation: B = log10(C/T), where C denotes the geometric mean of observed abundance value and T denotes the associated target abundance values. Bonferroni simultaneous t tests were used to determine whether each counting method had an estimated bias value that was statistically significantly different from zero at a 0.05 simultaneous level of significance (20). To make the bias estimates easier to interpret, they were transformed to the percentages by which the target abundance values were over- or underestimated (%T) by using the following equation: %T = (10B − 1) × 100.
RSD.
The precision of each method was assessed by using the repeatability standard deviation (RSD). The RSD is the standard deviation of the respective bead or bacterial abundance values (1).
RMSE.
The overall error of each counting method was assessed by using the root mean squared error (RMSE) (3), which was calculated with the formula 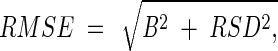 where B and RSD are the bias and RSD values associated with each method, respectively.
where B and RSD are the bias and RSD values associated with each method, respectively.
The RMSE values were partitioned into two separate analyses, one using data for the lower target abundances (104 to 105.5) and the other for the higher target abundances (105.7 and 106). A Dunnett's many-to-one simultaneous test was conducted to determine whether the RMSE for each microscope counting method was significantly different from that for the SPC at a simultaneous α of 0.05 (20).
All statistical analyses were performed with the Minitab release 13.3 software (Minitab, Inc., State College, Pa.).
RESULTS
Equality.
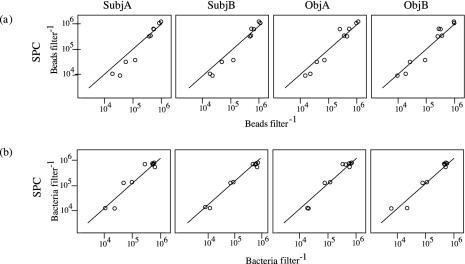
All of the methods were equivalent in estimating the number of beads filter−1 at the higher bead abundances of 105.7 to 106 (Fig. 2a). However, as bead abundances decreased, all microscopic methods showed a similar trend in overestimating the number of beads filter−1 relative to the SPC method. The bacterial data showed similar trends, but counts by the microscopic methods were closer to the corresponding SPC counts (Fig. 2b).
FIG. 2.
Bead (a) and bacterial (b) abundances on filter membranes, comparing the SubjA, SubjB, ObjA, and ObjB microscope methods to the SPC method. The solid line represents the line of equality.
Bias.
When counting fluorescently labeled beads, all microscope methods exhibited some degree of bias, with a general trend of increasing positive bias as bead abundances decreased (Table 1). These biases were most pronounced at bead abundances of <105.5 filter−1. The overestimations (%T) ranged from 34% (ObjB) to 156% (SubjA) at bead target abundances of 104 or 104.5 beads filter−1 (Table 2). At 105.5 beads filter−1, the SubjA, SubjB, and ObjA methods still overestimated the target abundance but by a much lower percentage, ranging from 24.8% (SubjA) to 56.5% (SubjB). The ObjB method exhibited a negative bias (Table 1) (i.e., underestimation of bead abundance [Table 2]) when the target abundance of beads was >104.5 filter−1.
TABLE 1.
Bias and RMSE valuesa
| Counting method | Bias values per filter (log10) for:
|
RMSE values per filter (log10) for:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beads
|
Bacteria
|
Beads
|
Bacteria
|
|||||||||||||
| 4.0 | 4.5 | 5.5 | 5.7 | 6.0 | 4.0 | 5.0 | 5.7 | 4.0 | 4.5 | 5.5 | 5.7 | 6.0 | 4.0 | 5.0 | 5.7 | |
| SubjA | 0.41* | 0.41* | 0.10 | 0.03 | −0.04 | 0.21 | −0.17 | 0.00 | 0.45** | 0.47** | 0.10** | 0.03 | 0.06 | 0.31* | 0.27* | 0.13 |
| SubjB | 0.31** | 0.39** | 0.19** | 0.08 | 0.02 | 0.11 | −0.11 | 0.08 | 0.32** | 0.48** | 0.20** | 0.10 | 0.03 | 0.13 | 0.14 | 0.10 |
| ObjA | 0.27 | 0.21 | 0.10 | −0.14 | 0.02 | 0.30 | −0.01 | 0.02 | 0.30** | 0.29** | 0.13** | 0.27 | 0.05 | 0.30* | 0.14* | 0.12 |
| ObjB | 0.13 | 0.19 | −0.02 | −0.22** | −0.02 | 0.06 | −0.03 | 0.00 | 0.24** | 0.42** | 0.11** | 0.23 | 0.02 | 0.41* | 0.12* | 0.04 |
| SPC | −0.01 | 0.02 | −0.01 | 0.06 | 0.02 | 0.04 | 0.04 | 0.05 | 0.04 | 0.05 | 0.02 | 0.06 | 0.04 | 0.04 | 0.04 | 0.08 |
*, Bias or RMSE values statistically significantly different from zero or RMSE for SPC, respectively (P < 0.05). **, Bias or RMSE values statistically significantly different from zero or RMSE for SPC, respectively (P < 0.01).
TABLE 2.
Percentage of over- and underestimations (%T) of target bead and bacteria abundancesa
| Counting method | No. of beads filter−1 (log10)
|
No. of bacteria filter−1 (log10)
|
||||||
|---|---|---|---|---|---|---|---|---|
| 4.0 | 4.5 | 5.5 | 5.7 | 6.0 | 4.0 | 5.0 | 5.7 | |
| SubjA | 155.1* | 156.0* | 24.8 | 7.1 | −8.5 | 60.9 | −31.7 | 0.6 |
| SubjB | 103.3** | 143.1** | 56.5* | 19.1 | 4.4 | 29.0 | −23.1 | 20.9 |
| ObjA | 86.2 | 63.5 | 26.9 | −27.0 | 4.2 | 99.3 | −2.7 | 5.5 |
| ObjB | 34.0 | 54.0 | −4.6 | −40.2** | −4.7 | 15.0 | −6.1 | 0.2 |
| SPC | −2.0 | 5.1 | −2.8 | 15.3 | 5.4 | 10.7 | 8.9 | 12.5 |
All values are expressed as percentages. Positive and negative values represent over- and underestimations of target values, respectively. *, Bias values statistically significantly different from zero (P < 0.05). **, Bias values statistically significantly different from zero (P < 0.01).
Among the microscope counting methods, the subjective methods of selecting fields to count were more positively biased, or overestimating, than the objective methods (Tables 1 and 2). Also, counting 20 fields on a single filter (method A) produced higher levels of positive bias at the lower target abundances than counting 5 fields on three separate filters (method B), regardless of the fields being systematically (subjective method) or randomly (objective method) selected. The SPC method displayed a relatively small bias over the range of bead abundances used in this study and had an under- and overestimation of the target bead abundance ranging from −2.8 to 15.3% (Tables 1 and 2).
The bacterial data show a trend similar to that of the bead data, with increasing bias as bacteria target abundances per filter decreased, though the magnitudes of the bias values were not as great (Table 1). The biases associated with the subjective and objective methods were greatest when counting 20 fields filter−1 relative to counting 5 fields on three filters at cell abundances of ≤105 (Table 1). However, as cell abundances increased above 105 cells filter−1, all microscopic methods accurately estimated the target abundances. The SPC method had a consistent and relatively small bias over the entire range of bacterial abundances counted and was similar to the bias values this method demonstrated with the bead count data.
The biases associated with bead and target abundance counts at <105 cells filter−1 were statistically significant from zero for the SubjA and SubjB methods. The negative bias for ObjB at the target abundance of 105.7 was the only significant bias value for any of the counting methods at bead or bacterial abundance counts of ≥105 cells filter−1. The biases associated with the SPC method for the ranges of bead and bacterial abundances used in this study were not statistically significant from zero.
RMSE.
The RMSE values for methods A and B exhibited an inverse relationship to bead abundances (Table 1). Also, the objective method had lower RMSE values than the subjective method, regardless of counting one or three filters, up to an abundance of approximately 105.5 beads filter−1. The SPC RMSE values were consistent over the range of target bead abundances counted and were lower than the values determined by any of the microscopic methods. For the set of lower target bead densities (104 to 105.5), all of the microscopic methods had significantly larger (P < 0.01) RMSE values than the SPC (Table 1), indicating that the SPC method was more proficient at counting beads in the lower abundance range than the microscopic methods. For the higher target bead abundance range (105.7 and 106), however, all of the microscopic methods were statistically equivalent (P > 0.76) to the SPC method.
For counting bacteria, the RMSE values for the SubjA, ObjA, and ObjB methods were similar to those for counting beads (Table 1). The performance of the SubjB method was consistent over the range of bacterial abundances counted, with relative RMSE values being lower with decreasing abundances of bacteria than those of the other microscopic methods but greater than that of the SPC. For the set of lower target bacterial abundances (104 and 105), the microscopic counting methods had a statistically significantly larger RMSE than did SPC (P < 0.03), except for SubjB (P = 0.08). Though there is a trend of increased overall performance of the microscopic methods when counting 5 fields on three filters as opposed to 20 fields on a single filter, there was no clear advantage of one method over the other.
DISCUSSION
Previous studies have listed and described aspects of sample processing and microscopic analyses that contribute to the overall variability associated with performing total direct counts of bacteria by using fluorescent microscopy. These include high vacuum pressures and filtration rates (24), not taking into account the spreading of cells outside the measured diameter of the filtration funnel (14), sample volume filtered (9), abundance of cells on a filter (12), suboptimal staining efficiency (11), counting too few microscope fields or bacteria per filter (6, 12), counting a single filter per sample as opposed to multiple filters (8, 12), and technician counting bias and fatigue (11, 13). We minimized the influence of several of these factors by consistently using the same low vacuum pressure, slow filtration rate, filtration funnel system, having the same person perform all of the analyses, and planning the experiments so the microscopist was not sitting at the microscope for extended periods of time.
Collectively the statistical methods of comparison used in this study indicate that as bead or bacterial abundances increased above approximately 105.5 filter−1 (20 to 25 beads or cells field−1), the variability between the different methods became progressively smaller and the methods were statistically equivalent. However, as bead or cell counts per filter decreased below this threshold, there was a concomitant decrease in the overall performance of all the microscopic methods and reliability of the resulting data, while the SPC performance remained consistently superior. Lemarchand et al. (15) determined that EFM and SPC were statistically equivalent when enumerating fluorescently labeled beads in the range of 1 × 104 to 5 × 105 beads filter−1. However, no statistical assessment of the reliability of the resulting count data was performed.
Kirchman et al. (12) observed a trend similar to that found in this study, as they found that when the number of bacteria was in the range of 30 to 50 cells field−1 there was no significant effect on the overall accuracy of the resulting data. However, they did not assess the influence of decreasing or lower cell abundances on data reliability.
Counting fewer fields on multiple filters that have been prepared from a single sample, as opposed to counting more fields on a single filter (thereby increasing the number of subsamples per individual sample), has been proposed as a method by which variability in total-direct-count data can be reduced (8, 12). In this study we compared abundance data from 20 fields on a single filter (method A) to abundance data from 5 fields on each of three filters (method B). However, there was no clear difference, for either method, between counting 5 fields on each of three filters and counting 20 fields on a single filter, regardless of whether we were counting beads or bacteria.
The data from the different microscope methods for counting bead abundances did not model the bacterial data, as we had originally hypothesized. We attribute this to the beads having a significantly greater density than the bacteria. The beads average 2.9 × 10−12 g bead−1 (per the manufacturer), which is approximately three times heavier than an average bacterium (9.5 × 10−13 g cell−1) (19). This difference in mass may have increased the bead-settling rate in the filtration tower during sample preparation and filtration and may have altered the distribution of the beads on the surface of the membrane filter relative to the bacterial samples.
Overall, the SPC method was consistently less biased, had a higher degree of repeatability, and had better overall performance than any of the microscope methods for counting beads or bacteria over the entire range of abundances used in this study. Also, and in contrast to the microscope methods, there was no significant difference in these statistical measurements of the SPC method regardless of counting filters with 104 or 106 beads or bacteria filter−1.
For performing total direct counts of bacteria using EFM, we recommend preparing two or three filter membranes per sample and that at least five truly randomly selected fields be counted per filter when the abundance of bacteria filter−1 is ≥105. However, when the abundance of bacteria per filter decreases to <105, the resulting total direct count data should be evaluated with caution.
Admittedly, increasing the number of experiments for each method and increasing the number of filters and fields counted per filter may produce results different from those presented here. However, we feel that the protocols described here more closely model those routinely used by researchers using total direct counts to measure and enumerate bacteria from water samples. Also, the general experimental designs used in this study have been recommended by others as being most appropriate for total direct counts (8, 11-13).
Kepner and Pratt (11) reviewed over 200 peer-reviewed publications in which total-direct-count methods were described and found that the number of publications providing adequate methodological detail on the total-direct-count method has been steadily decreasing. In an effort to provide some standardization of the types of information that should be included in publications, thus allowing interstudy comparisons, they provide a list of data types that should be included in any publication that presents total direct count data. One of these data types is the number of cells and microscope fields counted. However, the inclusion of data pertaining to the number of cells and microscope fields counted provides an estimate of the precision but not the accuracy of those data. Based on the results of this study, as the abundance of bacteria per filter decreases to <105 the precision of the data may be consistent, but it may be significantly over- or underestimating the true abundance. Accordingly, we recommend that the average number of bacteria per microscope field also be included in this list of required data types to allow the reader to determine the accuracy and overall reliability of the published total-direct-count data.
Acknowledgments
This work was partially supported by Cooperative Agreement EEC-8907039 between the U.S. National Science Foundation Engineering Research Centers Program and Montana State University, Bozeman.
REFERENCES
- 1.Association of Official Analytical Chemists. 1990. Official methods of the association of official analytical chemists, p. 681. Association of Official Analytical Chemists, Arlington, Va.
- 2.Baudart, J., J. Coallier, P. Laurent, and M. Prevost. 2002. Rapid and sensitive enumeration of viable diluted cells of members of the family Enterobacteriaceae in freshwater and drinking water. Appl. Environ. Microbiol. 68:5057-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, D., and B. Lindgren. 1990. Statistics: theory and methods. Brooks/Cole, Pacific Grove, Calif.
- 4.Catala, P., N. Parthuisot, L. Bernard, J. Baudart, K. Lemarchand, and P. Lebaron. 1999. Effectiveness of CSE to counterstain particles and dead bacterial cells with permeabilised membranes: application to viability assessment in waters. FEMS Microbiol. Lett. 178:219-226. [DOI] [PubMed] [Google Scholar]
- 5.D'Haese, E., and H. J. Nelis. 2000. Effect of antibiotics on viability staining of Escherichia coli in solid phase cytometry. J. Appl. Microbiol. 89:778-784. [DOI] [PubMed] [Google Scholar]
- 6.Fry, J. C. 1990. Direct methods and biomass estimation, p. 41-85. In R. Grigorova and J. R. Norris (ed.), Methods in microbiology. Academic Press, Ltd., Cambridge, United Kingdom.
- 7.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of nucleopore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, K., R. Lundman, and M. Hamilton. 1993. Efficient sampling designs for microbial processes: a case study. J. Microbiol. Methods 18:69-81. [Google Scholar]
- 9.Jones, J., and B. Simon. 1975. An investigation of errors in direct counts of aquatic bacteria by epifluorescence microscopy, with reference to a new method for dyeing membrane filters. J. Appl. Bacteriol. 39:317-329. [DOI] [PubMed] [Google Scholar]
- 10.Joux, F., and P. Lebaron. 2000. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microb. Infect. 2:1523-1535. [DOI] [PubMed] [Google Scholar]
- 11.Kepner, R. L., and J. R. Pratt. 1994. Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol. Rev. 58:603-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchman, D., J. Sigda, R. Kapuscinski, and R. Mitchell. 1982. Statistical analysis of the direct count method for enumerating bacteria. Appl. Environ. Microbiol. 44:376-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchman, D. L. 1993. Statistical analysis of direct counts of microbial abundances, p. 117-119. In P. Kemp, B. Sherr, E. Sherr, and J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 14.Lebaron, P., M. Troussellier, and P. Got. 1994. Accuracy of epifluorescence microscopy counts for direct estimates of bacterial numbers. J. Microbiol. Methods 19:89-94. [Google Scholar]
- 15.Lemarchand, K., N. Parthuisot, P. Catala, and P. Lebaron. 2001. Comparative assessment of epifluorescent microscopy, flow cytometry and solid-phase cytometry used in the enumeration of specific bacteria in water. Aquat. Microb. Ecol. 25:301-309. [Google Scholar]
- 16.Lisle, J., S. Broadaway, A. Prescott, B. Pyle, C. Fricker, and G. McFeters. 1998. Effects of starvation on physiological activity and chlorine disinfection resistance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4658-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisle, J., B. Pyle, and G. McFeters. 1999. The use of multiple indices of physiological activity to access viability in chlorine disinfected Escherichia coli O157:H7. Lett. Appl. Microbiol. 29:42-47. [DOI] [PubMed] [Google Scholar]
- 18.McFeters, G., B. Pyle, J. Lisle, and S. Broadaway. 1999. Rapid direct methods for enumeration of specific, active bacteria in water and biofilms. J. Appl. Microbiol. 85:193S-200S. [DOI] [PubMed] [Google Scholar]
- 19.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Inc., Sunderland, Md.
- 20.Neter, J., M. Kutner, C. J. Nachtsheim, and W. Wasserman. 1996. Applied linear statistical models. McGraw-Hill, Boston, Mass.
- 21.Parthuisot, N., P. Catala, K. Lemarchand, J. Baudart, and P. Lebaron. 2000. Evaluation of ChemChrome V6 for bacterial viability assessment in waters. J. Appl. Microbiol. 89:370-380. [DOI] [PubMed] [Google Scholar]
- 22.Pyle, B., S. Broadaway, and G. McFeters. 1999. Sensitive detection of Escherichia coli O157:H7 in food and water by immunomagnetic separation and solid-phase laser cytometry. Appl. Environ. Microbiol. 65:1966-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds, D., and C. Fricker. 1999. Application of laser scanning for the rapid and automated detection of bacteria in water samples. J. Appl. Microbiol. 86:785-795. [DOI] [PubMed] [Google Scholar]
- 24.Turley, C. M., and D. Hughes. 1992. Effects of storage on direct estimates of bacterial numbers of preserved seawater samples. Deep Sea Res. 39:375-394. [Google Scholar]
- 25.van Poucke, S. O., and H. J. Nelis. 2000. A 210-minute solid phase cytometry test for the enumeration of Escherichia coli in drinking water. J. Appl. Microbiol. 89:390-396. [DOI] [PubMed] [Google Scholar]
- 26.Vermis, K., P. A. R. Vandamme, and H. J. Nelis. 2002. Enumeration of viable anaerobic bacteria by solid phase cytometry under aerobic conditions. J. Microbiol. Methods 50:123-130. [DOI] [PubMed] [Google Scholar]