Abstract
A method for rapid detection and identification of hyperthermophilic archaea of the family Thermococcaceae based on PCR amplification of 16S rRNA gene fragments with primers TcPc 173F (5′-TCCCCCATAGGYCTGRGGTACTGGAAGGTC-3′) and TcPc 589R (5′-GCCGTGRGATTTCGCCAGGGACTTACGGGC-3′) was developed and used for identification of new isolates.
The family Thermococcaceae (order Thermococcales, kingdom Euryarchaeota, domain Archaea [14]) includes the phenotypically close genera Thermococcus (17), comprising 20 valid species, and Pyrococcus (6), comprising 4 valid species. Thermococcaceae have attracted consistent attention from researchers because of their evolutionary significance (1, 16), as well as their biotechnological potential, connected with production of thermostable enzymes. The present work focused on elaboration of a method for rapid identification of Thermococcaceae based on PCR amplification of 16S rRNA gene fragments with specific oligonucleotide primers.
BlastN (2) of the National Center for Biotechnology Information site (http://www.ncbi.nih.gov/BLAST/) showed that GenBank contained 142 sequences of 16S rRNA genes with 95 to 100% similarity with the 16S rRNA gene of Thermococcus celer (including sequences pertaining to valid species of Thermococcus and Pyrococcus). Other GenBank sequences had less than 91% similarity with the 16S rRNA gene of T. celer as a query. The 142 sequences of 16S rRNA genes related to Thermococcaceae were retrieved from GenBank and aligned with the MultAlin program (5; http://prodes.toulouse.inra.fr/multalin/). Consensus sequences of Archaea, Bacteria, and Eucarya were deduced, by using a specially written program with a straightforward algorithm, from aligned small-subunit (SSU) rRNA sequences available from RDP, release 8.0 (9), at the website http://rdp.cme.msu.edu/html/ in files SSU_Prok.gb and SSU_Euk.gb (1,107 archaeal sequences apart from Thermococcaceae, 15,104 bacterial sequences, and 2,054 eukaryotic sequences). The MultAlin-generated consensus sequences of Thermococcaceae were aligned, again with the use of MultAlin, with consensus sequences of Archaea, Bacteria, and Eucarya and examined for sites that are conserved among Thermococcaceae and that at the same time exhibit strong signatures at and/or close to the extending (3′) ends of prospective primers. The sites corresponding to primers TcPc 173F and TcPc 589R were finally chosen for this work. As seen from Fig. 1, most of the nontarget sequences belonging to the groups whose rRNA genes could be aligned with Thermococcaceae at the site of interest exhibit mismatches with primer TcPc 173F in positions 1, 2, and 4, counting from the 3′ end of the primer, and mismatches in positions 1 and 4 with primer TcPc 589R; this should be sufficient for reliable discrimination. However, in higher-level consensus sequences of Archaea and Bacteria, the above-mentioned alignment positions were strongly degenerated (occupied by Ns), making final conclusions on the primer specificity impossible. Therefore, assessment of the affinity of candidate primers for individual sequences of SSU rRNA genes was performed with individual 16S rRNA gene sequences of Thermococcaceae and individual SSU rRNA sequences available from RDP, release 8.0, in the aforementioned files SSU_Prok.gb and SSU_Euk.gb. The candidate primers were applied to all sites (taken with a 1-nucleotide step) of all sequences (with the help of a specially written program with a straightforward algorithm). The affinity of a primer for a particular sequence was expressed in terms of the number of mismatches and of the total weight of mismatches recorded for the sequence site exhibiting the highest affinity. Weighting of mismatches was undertaken because the discriminating ability of a mismatch in a 3′-to-5′ exonuclease-free reaction mixture is higher the closer the mismatch is to the extending (3′) end of the primer (see, e.g., references 7 and 8). It was performed with the geometric progression formula wi = w1 × q(i−1), where wi is the weight of a given mismatch, w1 is the weight of the 3′-terminal mismatch, i is the position number of the mismatched nucleotide (counting from the 3′ end of the primer), and q is a coefficient specifying the decrease in the mismatch weight with each step from the 3′ primer end to the 5′ primer end. The value of w1 was chosen in such a way as to make the average value of the weights of all possible mismatches equal to 1. This condition is fulfilled if the weight of the 3′-terminal mismatch is specified as w1 = N × (q − 1)/(qN − 1), where N is the total number of nucleotides in the primer. Thus, the formula used was as follows:
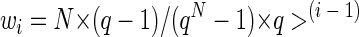 |
(1) |
Three variants of formula 1 were used, differing in the q value (0.95, 0.9, and 0.85). In silico testing of a number of candidate primers led us to choose the primer pair TcPc 173F-TcPc 589R, since these primers exhibited low affinity for nontarget sequences in terms of both unweighted mismatches and mismatches weighted by any of the three variants of the geometric progression formula. Apart from a few environmental clones of 16S rRNA genes, the nontarget organisms exhibiting the highest affinity for the forward primer were Archaeoglobus fulgidus and Acidianus infernus; Acidilobus aceticus was the organism showing the highest affinity for the reverse primer, and Archaeoglobus fulgidus exhibited the highest affinity for the primer pair (Fig. 2). These cultures were used along with other negative controls during in vitro verification of primer specificity.
FIG. 1.
Primers TcPc 173F and TcPc 589R and the corresponding sites of consensus sequences of 16S rRNA genes of organisms related to Thermococcaceae and of consensus sequences deduced for Archaea (without Thermococcaceae) and Bacteria. 173 and 589 refer to the locations of the sites in the 16S rRNA molecules (Escherichia coli numbering). The percentages show the consensus levels. + and −, plus and minus DNA strands, respectively. Mismatches are in lowercase. The site 173 to 193 is highly variable among Bacteria and Eucarya, including variations in size; therefore, the consensus for Thermococcaceae in this region could be compared only with the archaeal consensus. The site 589 to 618 is highly variable among Eucarya, including variations in length, making comparison with Eucarya at this site impossible.
FIG. 2.
Negative controls that were deduced to be the most appropriate for the primers TcPc 173F and TcPc 589R. + and −, plus and minus DNA strands, respectively. Mismatches are in lowercase. The numbers of mismatches and, in parentheses, the total weights of mismatches according to formula 1 at q = 0.9 are specified on the right.
Tests of the primer specificity used DNAs of the following archaeal and bacterial strains: positive controls Thermococcus celer DSM 2476T, Thermococcus pacificus DSM 10394T, Thermococcus litoralis DSM 5473T, Thermococcus sibiricus DSM 12597T, Thermococcus gorgonarius DSM 10395T, Thermococcus peptonophilus DSM 10343T, Thermococcus alcaliphilus DSM 10322T, Thermococcus profundus DSM 9503T, Thermococcus chitonophagus DSM 10152T, and Pyrococcus woesei DSM 3773Tand negative controls Acidianus infernus DSM3191T, Haloferax mediterranei DSM 1411T, Methanopyrus kandleri DSM6324T, Methanococcus jannaschii DSM2661T, Sulfolobus solfataricus DSM1616T, Archaeoglobus fulgidus DSM 4304T, Methanosarcina barkeri DSM 800T, Desulfurococcus amylolyticus DSM 3822T, Acidilobus aceticus DSM 11585T, Thermoanaerobacter wiegelii DSM 10319T, Thermotoga maritima DSM 3109T, Thermoterrabacterium ferrireducens DSM 11255T, and Moorella glycerini DSM 11254T. The strains were grown on media and under conditions recommended by the Deutsche Sammlung von Mikroorganismen und Zellkulturen. DNA was isolated by a standard procedure (10).
PCR was run in a reaction mixture (20 μl) containing 1 μl of template DNA, primers (0.1 μM each), 2 μl of 10× PCR buffer, 1.0 mM MgCl2, deoxyribonucleotide triphosphates (200 μM each), and 1.5 U of Taq DNA polymerase (set #K0163; Fermentas, Vilnius, Lithuania) on a Tertsik multichannel thermal cycler (DNK-Tekhnologiya, Moscow, Russia). The temperature profile was as follows: initial DNA denaturation for 3 min at 94°C, 25 to 30 cycles of denaturation at 92°C for 20 s and primer annealing and extension at 72°C for 1 min 40 s, and final extension for 5 min at 72°C. Amplification products (10 μl) were visualized in agarose gel according to standard protocols (3).
In specificity tests, the annealing temperature was varied from 65 to 75°C. A single PCR product of the expected size (about 400 bp) was produced on the DNA of all Thermococcaceae strains at all annealing temperatures tested except 75°C, at which no amplification occurred. No products were formed on the DNAs of negative controls in the entire range of the annealing temperatures tested. For further work, the annealing temperature of 72°C was chosen as providing a safety margin for primer specificity.
The proposed primer pair TcPc 173F-TcPc 589R was used for identification of hyperthermophilic isolates maintained at the laboratory culture collection and morphologically resembling Thermococcaceae (Table 1). A specific amplification product was obtained for seven strains, four of which had been isolated and cultivated under conditions unusual for Thermococcaceae: strains SN531 and SM402 used Fe(III) as the electron acceptor, and strains 515 and AM4 used inorganic energy sources, H2 and CO, respectively (M. L. Miroshnichenko, unpublished data; T. G. Sokolova, J. M. Gonzalez, N. A. Kostrikina, N. A. Chernyh, T. P. Tourova, E. A. Bonch-Osmolovskaya, and F. T. Robb, Abstr. 4th Int. Congr. Extremophiles, abstr. P275, p. 417, 2002). Identification of strain SB611, isolated from a North Sea oil well, as a representative of Thermococcaceae provides additional evidence of the presence of these microorganisms in high-temperature oil reservoirs (15). The sequence of the strain SB611 amplification product (GenBank accession no. AY591754) showed 99% similarity (BlastN) with that of T. litoralis.
TABLE 1.
Identification of new isolates by PCR with the primer pair TcPc 173F-TcPc 589R, specific for Thermococcaceae
| Strain | Isolation source | Cultivation conditionsa
|
Amplification resultb | |||
|---|---|---|---|---|---|---|
| Mineralization of the medium | Electron donor | Electron acceptor | Referencec | |||
| SN531 | Hydrothermal vent chimney, East Pacific Rise | Marine | Peptone | Fe(III) | 12 | + |
| 515 | Hydrothermal vent chimney, East Pacific Rise | Marine | H2 | S0 | 11 | + |
| AM4 | Hydrothermal vent chimney, East Pacific Rise | Marine | CO | None | 13 | + |
| AM8 | Hydrothermal vent chimney, East Pacific Rise | Marine | Peptone | S0 | 11 | + |
| SB611 | Oil well, North Sea | Marine | Peptone | None | 12 | + |
| SM402 | Shallow-water marine hydrothermal sediment, Kuril Islands | Marine | Peptone | Fe(III) | 12 | + |
| SM4022 | Shallow-water marine hydrothermal sediment, Kuril Islands | Marine | Peptone | Fe(III) | 12 | + |
| 204 | Continental hot spring, Kuril Islands | Freshwater | Peptone | S0 | 4 | − |
| 313 | Continental hot spring, Kamchatka | Freshwater | Peptone | S0 | 4 | − |
In all cases, the media contained 200 mg of yeast extract (Sigma)/liter, the medium pH was 6.5, and the incubation temperature was 85°C.
+, single band of the expected size; −, absence of any amplification products.
References are sources for medium compositions.
Acknowledgments
We thank E. Stackebrandt (DSMZ, Braunschweig, Germany) for providing reference strains and C. Schleper (Technical University of Darmstadt, Darmstadt, Germany) for providing DNAs of several archaeal cultures.
This research was supported by the Program on Physicochemical Biology of the Russian Academy of Sciences.
REFERENCES
- 1.Achenbach-Richter, L., R. Gupta, W. Zillig, and C. R. Woese. 1988. Rooting the archaebacterial tree: the pivotal role of Thermococcus celer in archaebacterial evolution. Syst. Appl. Microbiol. 10:231-240. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D Moore, G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Bonch-Osmolovskaya, E. A., A. I. Slesarev, M. L. Miroshnichenko, T. P. Svetlichnaya, and V. A. Alekseev. 1988. Characteristics of Desulfurococcus amylolyticus n.sp.—a new extremely thermophilic archaebacterium isolated from thermal springs of Kamchatka and Kunashir Island. Mikrobiologiya 57:78-84. [Google Scholar]
- 5.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 7.Huang, M. M., N. Arnheim, and M. F. Goodman. 1992. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 20:4567-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ignatov, K. B., V. M. Kramarov, O. L. Uznadze, and A. I. Miroshnikov. 1997. Tth DNA polymerase-mediated amplification of DNA fragments using primers with mismatches in the 3′-region. Bioorg. Khim. 32:817-822. [PubMed] [Google Scholar]
- 9.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marmur, J. 1961. A procedure for the isolation of desoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 11.Miroshnichenko, M. L., G. M. Gongadze, F. A. Rainey, A. S. Kostyukova, A. M. Lysenko, N. A. Chernyh, and E. A. Bonch-Osmolovskaya. 1998. Thermococcus gorgonarius sp. nov. and Thermococcus pacificus sp. nov.: heterotrophic extremely thermophilic archaea from New Zealand submarine hot vents. Int. J. Syst. Bacteriol. 48:23-29. [DOI] [PubMed] [Google Scholar]
- 12.Slobodkin, A. I., B. Campbell, S. C. Cary, E. A. Bonch-Osmolovskaya, and C. Jeanthon. 2001. Evidence for the presence of thermophilic Fe(III)-reducing microorganisms in deep-sea hydrothermal vents at 13°N (East Pacific Rise). FEMS Microbiol. Ecol. 36:235-243. [DOI] [PubMed] [Google Scholar]
- 13.Sokolova, T. G., J. M. Gonzalez, N. A. Kostrikina, N. A. Chernyh, T. P. Tourova, C. Kato, E. A. Bonch-Osmolovskaya, and F. T. Robb. 2001. Carboxydobrachium pacificum gen. nov., sp. nov., a new anaerobic, thermophilic, CO-utilizing marine bacterium from Okinawa Trough. Int. J. Syst. Bacteriol. 51:141-149. [DOI] [PubMed] [Google Scholar]
- 14.Stetter, K. O. 1996. Hyperthermophilic procaryotes. FEMS Microbiol. Rev. 18:149-158. [Google Scholar]
- 15.Stetter, K. O., R. Huber, E. Blochl, M. Kurr, R. D. Eden, M. Fielder, H. Cash, and I. Vance. 1993. Hyperthermophilic archaea are thriving in deep North Sea and Alaskan oil reservoirs. Nature (London) 365:743-745. [Google Scholar]
- 16.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zillig, W., I. Holtz, D. Janekovic, W. Schafer, and W. D. Reiter. 1983. The archaebacterium Thermococcus celer represents a novel genus within the thermophilic branch of the Archaebacteria. Syst. Appl. Microbiol. 4:88-94. [DOI] [PubMed] [Google Scholar]




