Abstract
The onset of streptothricin (ST) biosynthesis in Streptomyces rochei F20 was studied by using reverse transcription-PCR (RT-PCR) to detect transcripts of ST genes during growth in liquid medium, soil, and the rhizosphere. In situ results correlated with those obtained in vitro, illustrating the growth phase-dependent manner of ST production by F20. Maximal transcription of ST resistance (sttR) and biosynthesis (sttA) genes occurred during the transition between the exponential and stationary phases of growth, when the specific growth rate (μ) started to decline. A higher level of gene expression of sttR versus sttA was observed in all experiments. In liquid culture, maximal transcript accumulation of the sttA gene was only ca. 40% that of the sttR gene. sttA and sttR mRNAs were detected in soil containing approximately 106 CFU of growing cells g of soil−1. sttR mRNA was detected in sterile and nonsterile rhizosphere colonized with growing mycelium of F20 at 1.2 × 106 and 4.0 × 105 CFU g of soil−1, respectively. However, neither sttR nor sttA transcripts were detected by RT-PCR in the rhizoplane, which supported a lower population density of F20 than the rhizosphere.
The involvement of antibiotics in biological control has been reported (3, 14, 24, 27). However, antibiotic production in natural environments such as soil is difficult to demonstrate, because low levels of nutrients limit growth and production (32). Detection of antibiotic production in soil could be achieved only in the presence of nutrient sources, such as roots, seeds, or straw fragments. In addition, many antibiotics are adsorbed to clay particles and are difficult to detect by extraction (22, 23). Only a few studies using conventional extraction methods have provided direct evidence supporting the crucial role of antibiotics in disease suppression by biological control agents in situ. By use of a bioassay technique, geldanamycin was detected at 88 μg g of soil−1 in pea rhizosphere where Streptomyces hygroscopicus var. geldanus was used to control Rhizoctonia solani infection (24). Levels of thiostrepton production in sterile soil were reported by Wellington et al. (31) within the range of 30 to 50 ng g of soil−1 following extraction and bioassay via a specific thiostrepton-inducible promoter coupled to a resistance gene. A similar method using a highly sensitive inducible promoter driving gfp expression was used to assay oxytetracycline production by Streptomyces rimosus in soil microcosms (11). Fluorescence-activated cell sorter analysis was used to detect fluorescing cells in soil extracts. Extraction of phenazine from wheat rhizosphere inoculated with Pseudomonas fluorescens 2-79 and Pseudomonas aureofaciens 30-84 and the use of mutants with phenazine blocks unequivocally proved the significance of antibiotic production in control of take-all disease caused by Gaeumannomyces graminis var. tritici (27). Other studies have demonstrated the positive correlation between production of antibiotics using fluorescent pseudomonads and control of soil-borne fungal disease in plants (5, 14, 16).
Detection of mRNA can provide evidence for bacterial gene expression in situ (20). The expression of several genes in sediments (18, 20), and soil (2, 7, 9, 19) has been investigated by using reverse transcription-PCR (RT-PCR). The expression of manganese peroxidase (mnP) genes during removal of polycyclic aromatic hydrocarbons from a culture of Phanerochaete chrysosporium grown in presterilized soil was studied by using competitive RT-PCR to quantify the mRNA extracts (2). The high mnp transcript levels were shown to correlate with the manganese peroxidase enzyme activities observed. Thus far, no study has reported detection of mRNA involving antibiotic biosynthesis in soil by using the RT-PCR technique. The present study aimed to establish a reliable molecular method for monitoring the production by Streptomyces rochei F20 of streptothricin (ST), a possible candidate for control of soil-borne fungal plant pathogens (6), in soil and rhizosphere. The expression of the ST biosynthesis and resistance genes sttA and sttR in F20 was determined. Transcripts of these genes were monitored semiquantitatively as an indication of antibiotic production throughout the growth cycle of F20 in submerged culture compared with that in soil microcosms and the rhizosphere of wheat.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. rochei F20 (6), the ST-producing strain, was kindly supplied by Francisco Malpartida (National Center of Biotechnology, CSIC, Madrid, Spain). Streptomyces lividans 1326 (13) was used for the bioassay of ST. In liquid medium, the strains were grown in SMMS broth (26) at 28°C and 200 rpm. Reduced arginine-starch-salts (RASS) agar (12) or oatmeal (OM) agar was used for the preparation of streptomycete spore suspensions.
In vitro production of ST.
An F20 spore suspension (1010 spores ml−1) was inoculated into SMMS broth to an optical density at 450 nm (OD450) of 0.05 (approximate concentration, 107 CFU ml−1). The specific growth rate (μ) was derived from the OD measurement. The culture growth was monitored at OD450 at 0, 10, 20, 30, and 50 h. At each time point the culture was also subjected to RNA extraction and an ST antibiotic activity assay.
ST antibiotic activity assay.
The culture was centrifuged at 14,926 × g and 4°C for 10 min to remove cells, and the supernatant was filtered through a 0.45-μm-pore-size membrane. A 25-μl volume of the filtrate was dried onto a 6-mm-diameter disk (Whatman). The disk was placed on a plate overlaid with 3 ml of soft medium (0.6% agar) seeded with 106 CFU of S. lividans ml−1 and was incubated at 30°C overnight. The inhibition zone was measured. The specificity of the bioassay was confirmed by using the inactivation technique described by Fernandez-Moreno et al. (6). The cell extract was obtained from F20 mycelia by sonication, and the acetyltransferase in the cell extract was bound to ST and inactivated it, as demonstrated by a bioassay. This is to confirm that the inhibitory action observed on the plate assay resulted from ST only and not from other compounds that might be produced by F20. The concentration of ST was quantified by using a rectilinear dose-response curve with S. lividans and a commercially available ST designated nourseothricin. This antibiotic from Streptomyces noursei is a mixture of ST-F (identical to ST) and ST-D (Werner Bioagents, Jena, Germany). The calibration produced a coefficient of determination (R2) of 0.97.
Sterile soil microcosms.
Experiments were carried out in triplicate with an uninoculated soil as a control. Cryfield wheat rhizosphere soil, University of Warwick, was used; the composition is given by Wellington et al. (30). Soil microcosms were set up as described by Wellington et al. (30). Air-dried and sieved (pore size, 2 mm) soil was amended with 1% (wt/wt) chitin (Sigma) and 1% (wt/wt) starch). Ten grams of amended soil was then added to a 25-ml Falcon tube and sterilized by autoclaving twice (24 h apart) at 121°C and 15 lb/in2 for 15 min. Three Falcon tubes were set for each day of sampling. Soil was inoculated with a spore suspension of F20 at 107 CFU g of soil−1, and the moisture content in the soil was adjusted to 15% by adding sterile distilled water. Soil microcosms were then incubated at 28°C for 5 days. Three samples were taken each day for viable (spore and mycelium) and spore counts, RNA extraction, and antibiotic extraction. Viable and spore counts were performed by the method of Herron and Wellington (12).
Extraction of ST from soil.
A 10-g soil sample was resuspended in 20 ml of 10 mM N-tris(hydroxymethyl) methyl-2-aminoethanesulfonic acid (TES) buffer (pH 7.2). The suspension was shaken for 30 min and centrifuged at 1,600 × g for 15 min. The supernatant was removed and transferred to a fresh 20-ml Sarstedt screw-capped plastic bottle, while the pellet was resuspended in TES buffer and extracted again by shaking. The pooled supernatants were filtered through a 0.45-μm-pore-size membrane. The filtrate was lyophilized overnight and then dissolved in 2 ml of water. The bioassay of ST was performed. If ST activity was detected, water was added to bring the total volume to 50 ml. The solution was purified by ion-exchange chromatography (IRC50 resins and CM-Biogel A columns) as for the protocol for ST extraction from liquid-grown culture (Francisco Malpartida, personal communication). The elution condition for the first column (IRC50) was a 200-ml linear gradient within the range from 0 to 1 M HCl. For the second column (CM-Biogel), 200 ml of a 0 to 2 M NaCl linear gradient was used.
Rhizosphere microcosms.
Approximately 50 seeds of spring wheat (Triticum aestivum var. amtrack) were surface sterilized in 50 ml of 70% (vol/vol) ethanol in a 250-ml sterile conical flask by shaking gently at room temperature for 15 min. The ethanol was decanted. Seeds soaked with 50 ml of 25% (vol/vol) sodium hypochlorite were shaken gently at room temperature for 30 min before the liquid was discarded. This procedure was repeated once for 15 min with a fresh solution. Washes with 100 ml of sterile distilled water were performed five times, for 1 min each time. The seeds were then placed on 0.7% (wt/vol) agar (Difco), covered with sterile moistened Whatman no. 1 filter papers, and left to germinate in the dark at room temperature for 24 h. Contamination was examined by placing seedlings on nutrient agar (Oxoid, Basingstoke, United Kingdom) and incubating overnight at 30°C. Inoculation was done by immersion of 50 sterile seedlings in 25 ml of an F20 spore suspension (108 to 109 spores ml−1 resuspended in 0.05% [vol/vol] Tween 80) for 20 min, which gives a covering of 106 to 107 spores seedling−1 as estimated by a viable count. Seedlings immersed in sterile distilled water were used as controls. Two seedlings were sown at a 1-cm depth in 20 g of amended (1% chitin, 1% starch) sterile or nonsterile soil wetted with sterile distilled water to 15% (vol/wt) moisture content (−67 kPa) in a 50-ml glass tube. The plants were grown at 23°C in a chamber providing 12 h of darkness and 12 h of light until examination. There was no watering during the incubation. All tubes were wrapped with sterile or nonsterile plastic bags in order to keep the plants under sterile conditions and prevent evaporation of water in the soil. A 0.5-mm-diameter hole was made in the plastic bags and covered with 3M microperforated tape to allow respiration of plants.
Rhizosphere and rhizoplane sample preparation.
Rhizosphere and rhizoplane samples were examined at days 3, 5, 7, 14, and 21. The plants were carefully removed from the soil. The roots were tapped to remove the loose adhering soil and weighed. To obtain the rhizosphere sample, the tightly attached soil on the root surface was washed off with 10 ml of sterile phosphate-buffered saline (PBS) by vigorous agitation for 1 min. To prepare rhizoplane samples, the washed roots were removed from the rhizosphere soil suspension and ground in 1 ml of sterile PBS in a 1.5-ml glass tissue grinder (Bio-Rad). The samples were then ready for plate counting and DNA extraction. For RNA extraction, the samples were prepared separately under RNase-free conditions, and an extraction buffer (9) was used instead of PBS during sample preparations. Viable counts of F20 and indigenous soil microflora were performed by a dilution plate counting technique using selective agar supplemented with appropriate antibiotics (Table 1). The plates were incubated at 30°C until the colonies could be counted.
TABLE 1.
Selective media for viable counts of F20 and indigenous bacteria and fungi in nonsterile wheat rhizosphere and rhizoplane
| Target organism | Medium | Antibiotic supplement (μg ml−1) |
|---|---|---|
| S. rochei F20 | RASS | Nystatin (50) |
| Nourseothricin (10) | ||
| Bacteria | Nutrient agar | Nystatin (50) |
| Fungi | DOX's agara | Polymyxin A (50) |
| Thiostrepton (50) | ||
| Rifampin (10) |
In grams per liter: sucrose, 20.0; NaNO3, 2.0; K2HPO4, 1.0; MgSO4·7H2O, 0.5; Kcl, 0.5; agar, 15.0.
Extraction of total RNA from streptomycetes grown in liquid medium, soil, or rhizosphere.
RNA was extracted by the method of Griffiths et al. (9) with the following modifications. The rhizosphere samples were prepared under RNase-free conditions by washing the tightly adhered soil on roots excised from wheat plants (0.5 g [wet weight]) with RNase-free 5% hexadecyltrimethylammonium bromide (CTAB; Sigma, Poole, United kingdom)-120 mM potassium phosphate buffer (pH 8.0). The root-washed soil suspension was used as a rhizosphere sample. The washed roots were ground in the same buffer by using a 1.5-ml glass tissue grinder (Bio-Rad). The retrieved suspension was used as a rhizoplane sample. RNA was extracted from F20 grown in liquid culture by using 0.1 to 0.2 g (wet weight) of mycelia from 2 ml of a pure culture of F20. Cells were lysed using a ribolyser (FastPrep FP120 bead beating system; Bio-101, Vista, Calif.) at 4.0 for 15 s. The RNA extracts were quantified by agarose gel electrophoresis.
Detection of mRNA transcripts by RT-PCR.
Primers STR-F (5′-ACG CCG AGG CCA TCG AG-3′) and STR-R (5′-CAG GGC ATG CTC ATG TAG A-3′), designed from ST resistance genes of three ST producers, Streptomyces lavendulae sta (NCBI accession no. M16183), S. noursei snnat1 (NCBI accession no. X73149), and S. rochei F20 sttR (NCBI accession no. Y10293), were used for detection of sttR mRNA (514 bp). Primers STA-F (5′-CGC GAC CTG TAC GGC ATC GG-3′) and STA-R (5′-CCC GTT GGC GGA CAG TGG C-3′), designed from an S. rochei F20 ST biosynthesis gene coding for peptide synthetase (sttA [NCBI accession no. Y10293]), were used for detection of sttA mRNA (475 bp). The cDNA was synthesized from STR-R or STA-R primers by using Expand reverse transcriptase (Roche) according to the manufacturer's instructions. Amplification of cDNA was carried out by using a Hybaid PCR Express thermal cycler with the following steps: (i) 1 cycle of 94°C for 4 min; (ii) 30 cycles of 94°C for 30 s, 52°C (for sttR) or 56°C (for sttA) for 50 s, and 72°C for 1 min; and (iii) 1 cycle of 72°C for 5 min. A 50-μl PCR mixture contained 0.2 μM reverse and forward primers, 1× buffer, 5% dimethyl sulfoxide, 0.1 mM deoxynucleoside triphosphates, 2 mM MgCl2, 60 μg of bovine serum albumin ml−1, and 1.5 U of Taq polymerase (GIBCO BRL) μl−1. DNA from a pure culture of F20 and sterile distilled water were used as positive and negative controls, respectively, during the cDNA amplification step. RT-PCR products were analyzed by gel electrophoresis at 80 V (0.9% [wt/vol] agarose plus 0.5 mg of ethidium bromide ml−1 in Tris-borate-EDTA buffer). A 1-kb DNA marker was used as a molecular weight standard. No DNA contamination of RNA extracts was observed in any experimental controls.
Southern blot hybridization.
RT-PCR products were transferred from gels to Hybond N+ (Amersham) nylon membranes by blotting (25). Probes were prepared by using PCR products of F20 pure culture (which have 99% identities with the sttR and sttA genes) randomly labeled with [α-32P]dGTP (30 μCi) by using a random-primed DNA labeling kit (Boehringer Mannheim). The hybridization was carried out at 65°C for 18 h according to the manufacturer's recommendations (Amersham). The blots were washed twice (15 min each time) with 2×SSC (1×SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% (wt/vol) sodium dodecyl sulfate and once (10 min) with 0.5× SSC-0.1% (wt/vol) sodium dodecyl sulfate at 65°C. The hybridized probe was detected by autoradiography (25) using Fuji XR X-ray film.
Quantification of RT-PCR products by image analysis.
Image analysis of one-dimensional gel electrophoresis by use of Quantity One Software (version 4.1.1) provides relative quantification of RT-PCR products. Images of samples on electrophoresis gels were captured by using GelDoc 2000 (Bio-Rad). Once an image had been acquired, optimization to reduce noise or background was performed. Relative amounts of amplified products determined obtained by calculation of the average signal intensity (OD) of each DNA band in relation to that of a known quantity of the standard.
Statistical analysis.
All statistical analysis in this study was carried out using the Microsoft Excel 97 analysis toolpack. All data were calculated from at least three replicates. Data point standard errors were calculated for graphs. Multiple comparisons were carried out by using analysis of variance. Significant differences among sample means were determined by using the Tukey test. The minimum significant difference (MSD) was calculated by using the range (Q) adjusted by the Student t test at a P value of 0.05. Two means are declared significantly different only if their difference equals or exceeds the MSD.
RESULTS
Growth and ST production in liquid culture.
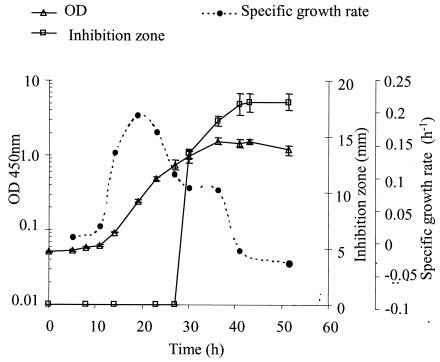
F20 had a doubling time of 4.1 h (μ = 0.17 h−1) in the exponential phase and a lag phase of 10 h (Fig. 1). Exponential growth occurred from 11 to 23 h, and peak cell production occurred at 36 h, with a stationary phase from 40 to 50 h. ST production was detected at the interface between the exponential and stationary phases of growth (Fig. 1) at 27 h after inoculation. The maximal amount of ST was detected at 43 h in the stationary-growth phase.
FIG. 1.
Growth and ST production profiles of F20 in liquid culture, obtained by monitoring the OD450 (open triangles), inhibition zones from the bioassay (open squares), and the specific growth rate (solid circles). Error bars, standard deviations (n = 3).
In vitro expression of sttR and sttA genes in liquid culture.
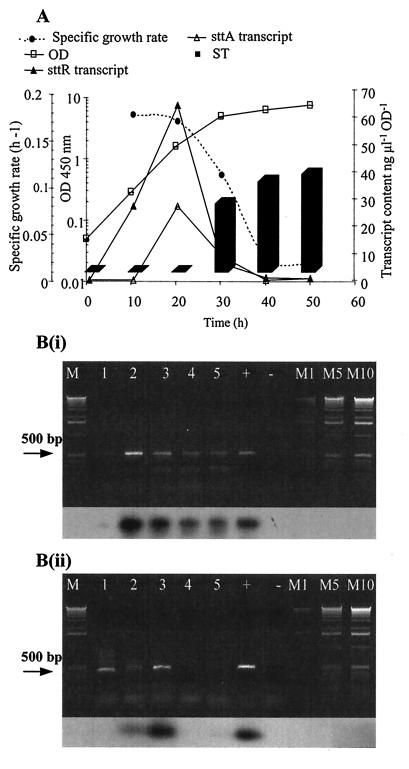
Transcripts of the sttR gene were detected in the lag phase and over a wide time range from 10 to 40 h, covering both the exponential and stationary phases of growth [Fig. 2A and B(i)]. The lag phase is not shown, because sampling was done at 10 h. The level of sttR mRNA was maximal at 20 h (transcript content, 64.23 ng μl−1 OD unit−1), twice that found at 10 h (27.19 ng μl−1 OD unit−1). Transcripts of the sttA gene were detected over a narrow time range, with a peak at 20 h (transcript content, 27.09 ng μl−1 OD unit−1) [Fig. 2A and B(ii)]. Levels of sttA transcripts were therefore approximately half those of sttR at 20 h. During the stationary phase, transcript levels of both genes were low while ST production increased (Fig. 2A). Both transcripts were maximal at the transition between the exponential and stationary phases.
FIG. 2.
Expression of ST genes and ST production in liquid culture. (A) Transcript contents of sttR (solid triangles) and sttA (open triangles), bioassay of ST production (solid bars), and specific growth rate (solid circles) monitored throughout the growth of F20 (OD450 [open squares]). Error bars (standard deviations, (n = 3) less than symbols used in graphs. (B) Gel electrophoresis and Southern blot hybridization of RT-PCR products of sttR (i) and sttA (ii). Lanes 1 through 5, 10, 20, 30, 40, and 50 h after inoculation, respectively; +, positive control; −, negative control; M1, M5, and M10, 1, 5, and 10 μl of the 1-kb DNA marker, respectively.
In situ expression of the sttR and sttA genes in soil.
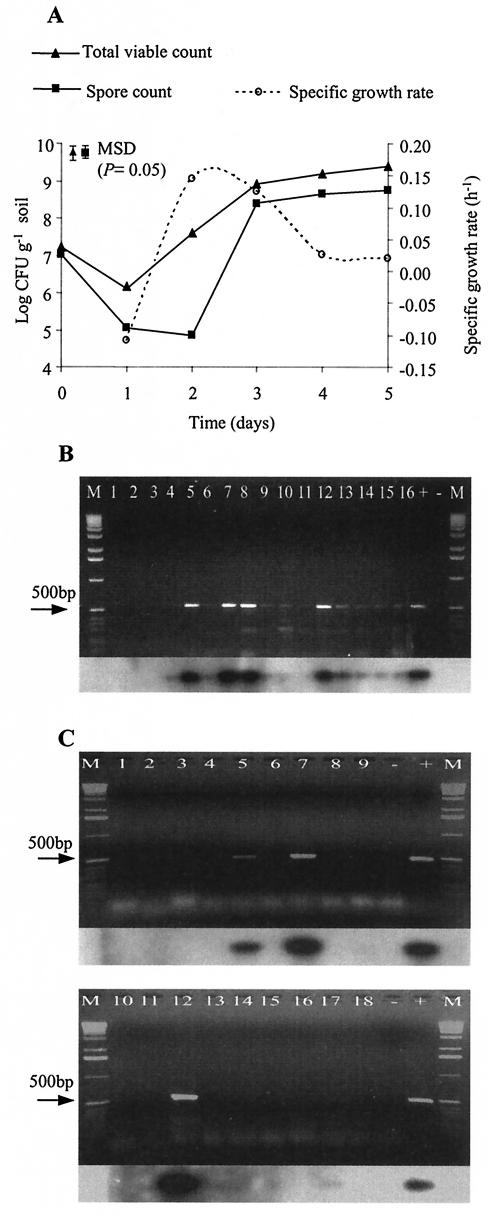
A drop in F20 viable counts was observed between days 0 and 1 (Fig. 3A). The spore count indicated that approximately 99% of spores underwent germination between days 1 and 2. Exponential growth ranged from day 1 to day 3, with a doubling time of 5.1 h (μ = 0.14 h−1). From day 3 to day 5, the ratio of the total viable count to the spore count decreased, indicating an increase in sporulation, suggesting that cellular differentiation of F20 in soil occurred during this period.
FIG. 3.
Growth and expression of ST genes in sterile amended soil. (A) Total viable counts (solid triangles), spore counts (solid squares), and specific growth rate (open circles). Symbols with error bars represent MSDs (P = 0.05; n = 3). (B and C) Gel electrophoresis and Southern blot hybridization of sttR (B) and sttA (C) RT-PCR products. Lanes: 1 to 3, day 0; 4 to 6, day 1; 7 to 9, day 2; 10 to 12, day 3; 13 to 15, day 4; 16, day 5; 17 and 18, day 5 RT-PCR products; +, positive control; −, negative control; M, 1-kb DNA marker.
Attempts to extract ST from soil failed. RT-PCR was used to detect ST biosynthesis in soil, and the results correlated with in vitro expression of ST genes in liquid culture. Expression of the sttR gene was not detected at day 0 but occurred at day 1 (Fig. 3B). Despite variability in replicate samples, higher signal intensity of sttR mRNA was observed at day 2 and day 3 at the transition between the exponential and the stationary phase. sttA mRNA was detected from day 1, and a high transcript level was observed at day 2 and day 3, although detection was achieved for only one sample each day (Fig. 3C). These observations confirmed that expression of the sttR and sttA genes of F20 grown in soil was dependent on the growth phase, a finding that correlated with the in vitro results.
Growth and in situ expression of sttR and sttA genes in the rhizosphere and rhizoplane of wheat in sterile soil.
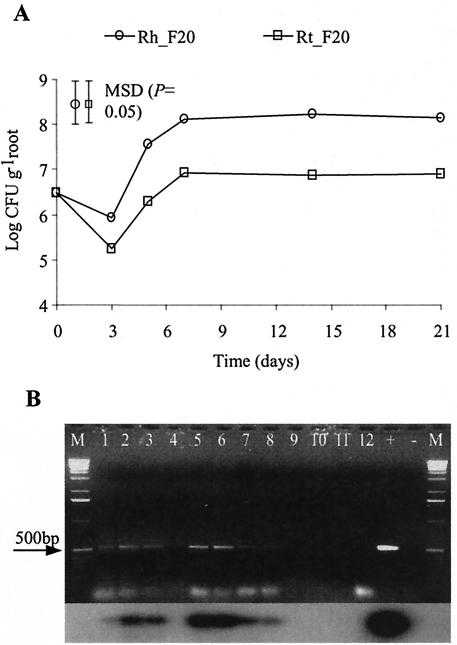
The growth of F20 in the rhizosphere and rhizoplane is shown in Fig. 4A. F20 attained a significantly higher population in the rhizosphere than in the rhizoplane. The results showed that growth and sporulation of F20 in the rhizosphere and rhizoplane occurred from day 3 to day 7. In the rhizosphere, sttR mRNA was detected from day 3 until day 14 (Fig. 4B). Viable growth profiles were different from those observed for soil microcosms; this is due to the inoculation of seedlings rather than dispersion of the inoculum throughout the soil. sttR mRNA was not detected in the rhizoplane, and sttA mRNA could not be detected in any rhizosphere or rhizoplane samples (data not shown).
FIG. 4.
Growth and expression of ST genes in the rhizosphere and rhizoplane of wheat in sterile soil. (A) Total viable counts (CFU per gram of soil) of F20 populations from rhizosphere (Rh) (circles) and rhizoplane (Rt) (squares) samples. Symbols with error bars represent MSDs (P = 0.05; n = 3). (B) Analysis of rhizosphere samples. Lanes: 1 to 3, day 3; 4 to 6, day 5; 7 to 9, day 14; 10 to 12, day 21; +, positive control; −, negative control; M, 1-kb DNA marker.
Growth and in situ expression of sttR and sttA genes in the rhizosphere and rhizoplane of wheat in nonsterile soil.
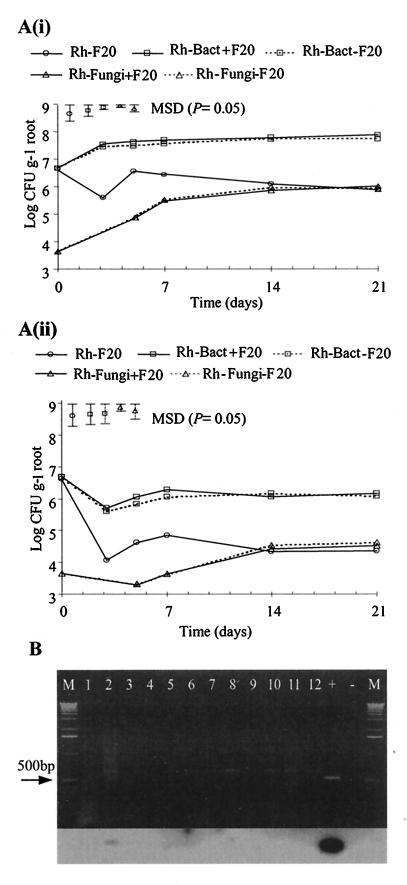
The population dynamics of F20 and of indigenous soil fungi and bacteria in the rhizosphere and rhizoplane are shown in Fig. 5A(i) and (ii), respectively. There was no significant difference between counts of the indigenous soil microflora in the presence of F20 and counts in the absence of F20 either in the rhizosphere or in the rhizoplane. Although viable counts of indigenous bacteria in the rhizosphere in the presence of F20 were higher than those in the absence of F20, this difference was not significant at a P value of 0.037. This showed that the F20 introduced had no effect on the growth of indigenous microflora in soil. The colonizing ability of F20 in nonsterile soil was lower than that in sterile soil, since F20 counts in nonsterile soil were lower by more than 2 orders of magnitude [see Fig. 4A and 5A(i) and (ii)]. In the rhizosphere, growth and sporulation of F20 occurred from day 3 to day 5 [Fig. 5A(i)]. The same trend was observed in the rhizoplane [Fig. 5A(ii)], where the count at day 7 was not significantly different from that at day 5. sttR mRNA was detected at day 3 only (Fig. 5B, lane 2) in the rhizosphere but was not detected in the rhizoplane (data not shown). Transcription of sttA was not detectable in either the rhizosphere or the rhizoplane (data not shown).
FIG. 5.
Growth and expression of ST genes in the rhizosphere and rhizoplane of wheat in nonsterile soil. (A) Population dynamics of indigenous bacteria and fungi in the presence (+F20) and absence (−F20) of F20 in the rhizosphere (Rh) (i) and the rhizoplane (Rt) (ii). Symbols with error bars represent MSDs (P = 0.05; n = 3). (B) Gel electrophoresis and Southern blot hybridization of sttR RT-PCR products. Lanes: 1 to 3, day 3; 4 to 6, day 5; 7 to 9, day 14; 10 to 12, day 21; +, positive control; −, negative control; M, 1-kb DNA marker.
DISCUSSION
ST production in liquid culture was detected first during the transition between the exponential and stationary growth phases, showing the growth phase-dependent manner of ST production in F20. This correlates with the general view of antibiotic production in streptomycetes, that production coincides with morphological differentiation and occurs after vegetative growth has ceased (1). Regulation of ST production was at the level of transcription, as observed for other antibiotics (oxytetracycline [17], streptomycin [21], and actinorhodin [8]).
In both soil and liquid culture, resistance gene expression proceeded biosynthesis. McDowall et al. (17) showed similar expression patterns for oxytetracycline biosynthesis (otcC, otcX, otcZ) and resistance (otrA) genes in S. rimosus culture by using S1 nuclease protection experiments. During oxytetracycline production, a significant drop in levels of otrA transcripts driven by otrAp1 was observed, while those transcribed from otcCp1 increased. In the present study, high levels of sttR transcripts were detected at the transition between exponential and stationary growth phases, but transcript levels declined markedly (>90%) later, when ST production was detected. Lower expression levels of sttA compared with sttR were possibly due to differences in promoter strength or transcript stability and reflect the importance of the sttR resistance gene product, ST acetyltransferase, for inactivation of any ST within the cell. The biosynthesis gene sttA encodes a peptide synthetase, a large protein with 481 amino acids, which is present at low levels in the cell, and gene dosage is obviously not limiting production initially. The sttR gene encodes a small enzyme, ST acetyltransferase (190 amino acids), which may in part explain the lower level of expression of sttA, which encodes a larger protein of 481 amino acids (6).
We report evidence for in situ antibiotic production in soil and the rhizosphere. Although it was not possible to extract ST from soil, the correlation of gene expression in bulk soil with that in liquid culture proved that ST production occurred in situ. Transcription of ST genes occurred in the same growth phase in bulk soil as in liquid culture. A drop of viable count at day 1 during F20 growth in soil follows the usual growth pattern for streptomycete growth and may be due to the death phase of young germlings (4). The lack of detection of biosynthesis gene transcripts under nonsterile conditions is probably due to the lower population level, although resistance gene expression was observed. Nevertheless, detection of sttR mRNA does imply ST biosynthesis in the rhizosphere, due to the fact that the sttR gene is clustered with other ST biosynthetic genes, and the acetyltransferase encoded by this gene is highly specific for ST produced by F20 (6). ST resistance in indigenous bacteria was detected in the soil used, but there was no significant increase in ST resistance to high ST levels (20 μg ml−1) due to the introduction of F20 during the experiments (29).
The inability to extract ST from soil is related to a number of extraction constraints. ST is soluble in water and insoluble in organic solvents (10), making extraction and concentration from soil more difficult when very small amounts of the antibiotic are produced in soil (31, 33). In addition, binding of antibiotics to clay particles is also likely to cause extraction problems in the soil used, which has a high clay content (31, 32). Basic antibiotics such as streptomycin and neomycin, which are aminoglycosides, and the peptide viomycin were studied for their behavior in soil and appear to be strongly adsorbed (22, 23). ST is a peptide antibiotic with a heterocyclic β-amino acid, streptolidine, an amino sugar, 4-carbamido-d-gulosamine, and one or more β-lysine residues attached by amide linkage at the C-2 position of the glucosamine moiety. The basic amino groups of ST may interact with organic matter in the soil, but clays have also been implicated in adsorption of aminoglycoside antibiotics such as streptomycin (22, 23, 32). Montmorillonite was found to have the greatest capacity for binding streptomycin compared to other clays with lower cation-exchange capacities, including vermiculite, illite, and kaolinite.
Further work is needed to elucidate the exact nature of interactions between ST and soil particles, particularly because soil is a complex, heterogeneous environment. Bound antibiotics may not be biologically inactivated; studies with streptomycin bound to soil proved that it was still capable of inhibiting salmonellae in situ in soil when the activity of the salmonellae was determined by an in situ assay using a lux-marked strain (15). Previous studies of the antagonistic activity of streptomycin-producing streptomyctes in soil showed that Streptomyces bikiniensis, a streptomycin producer, was capable of having a significant negative effect on the survival in soil of Salmonella enterica serovar Dusseldorf compared to that with a non-antibiotic-producing Streptomyces species (28). The ST producer employed in the present study, S. rochei, failed to exert a negative impact on indigenous bacteria; this may result from necrotrophic growth of resistant bacteria in the presence of killed sensitive groups. The overall effect would be to maintain similar population levels. It is likely that some susceptible soil bacteria have been inhibited. In addition, production may be highly localized in soil and thus cause significant inhibition of bacteria in a restricted area, but the overall impact on the population may be very small. The increase in total viable bacterial counts in the presence of S. rochei may be due to the stimulatory effect of killing some members of the indigenous microflora, thus providing nutrients for others.
Acknowledgments
We are grateful to the Development and Promotion of Science and Technology Talent Project (DPST) of Thailand for supporting this work. E. M. H. Wellington acknowledges financial support from European Community grant QLRT-2000-01783.
We thank F. Malpartida, National Centre of Biotechnology, CSIC, Spain, for providing S. rochei F20 and helpful advice on ST extraction.
REFERENCES
- 1.Bibb, M. J. 1996. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142:1335-1344. [DOI] [PubMed] [Google Scholar]
- 2.Bogan, B. W., B. Schoenike, R. T. Lamar, and D. Cullen. 1996. Manganese peroxide mRNA and enzyme activity levels during bioremedation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl. Environ. Microbiol. 62:2381-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brisbrane, P. G., and A. D. Rovira. 1988. Mechanisms of inhibition of Gaeumannomyces graminis var. tritici by fluorescent pseudomonads. Plant Pathol. 37:104-111. [Google Scholar]
- 4.Burroughs, N. J., P. Marsh, and E. M. H. Wellington. 2000. The growth and interaction dynamics of streptomycetes and phage in soil. Appl. Environ. Microbiol. 66:3868-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buyer, J. S., M. G. Kratzke, and L. J. Sikora. 1993. A method for detection of pseudobactin, the siderophore produced by a plant growth-promoting Pseudomonas strain, in the barley rhizosphere. Appl. Environ. Microbiol. 59:677-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Moreno, G., C. Vallin, and F. Malpartida. 1997. Streptothricin biosynthesis is characterized by enzymes related to nonribosomal peptide bond formation. J. Bacteriol. 179:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming, J. T., J. Sanseverino, and G. S. Sayler. 1993. Quantitative relationship between naphthalene catabolic gene frequency and expression in predicting PAH degradation in soils at town gas manufacturing sites. Environ. Sci. Technol. 27:1068-1074. [Google Scholar]
- 8.Gramajo, H. C., E. Takano, and M. J. Bibb. 1993. Stationary-phase production of antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7:837-845. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for extraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and RNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glasby, J. S. 1979. Encyclopedia of antibiotics, 2nd ed., p. 327-328. Wiley, Chichester, United Kingdom.
- 11.Hansen, L. H., B. Ferrari, A. H. Sørensen, D. Veal, and S. J. Sorensen. 2001. Detection of oxytetracycline production by Streptomyces rimosus in soil microcosms by combining whole-cell biosensors and flow cytometry. Appl. Environ. Microbiol. 67:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herron, P. R., and E. M. H. Wellington. 1990. New method for the extraction of streptomycete spores from soil and application to the study of lysogeny in sterile amended and nonsterile soil. Appl. Environ. Microbiol. 56:1406-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopwood, D. A., M. J. Bibb, K. F. Charter, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 14.Howie, W. J., and T. V. Suslow. 1991. Role of antibiotic biosynthesis in the inhibition of Pythium ultimum in cotton spermosphere and rhizosphere by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 4:393-399. [Google Scholar]
- 15.Huddleston. A. S. 1995. The distribution and diversity of streptomycin-producing streptomycetes in Brazilian soil. Ph.D. thesis. University of Warwick, Coventry, United Kingdom.
- 16.Lemanceau, P., P. A. H. M. Bakker, W. Jan de Kogel, C. Alabouvette, and B. Schippers. 1992. Effect of pseudobactin 358 production by Pseudomonas putida WC538 on suppression of Fusarium wilt of carnations by nonpathogenic Fusarium oxysporum Fo47. Appl. Environ. Microbiol. 58:2978-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDowall, K. J., A. Thamchaipenet, and I. J. Hunter. 1999. Phosphate control of oxytetracycline production by Streptomyces rimosus is at the level of transcription from promoters overlapped by tandem repeats similar to those of the DNA-binding site of the OmpR family. J. Bacteriol. 181:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meckenstock, R., P. Steinle, J. R. van der Meer, and M. Snozzi. 1998. Quantification of bacterial mRNA involved in degradation of 1,2,4-trichlorobenzene by Pseudomonas sp. strain P51 from liquid culture and from river sediment by reverse transcriptase PCR (RT/PCR). FEMS Microbiol. Lett. 167:123-129. [DOI] [PubMed] [Google Scholar]
- 19.Mendum, T. A., R. E. Sockett, and P. R. Hirsch. 1998. The detection of Gram-negative bacterial mRNA from soil by RT-PCR. FEMS Microbiol. Lett. 164:369-373. [Google Scholar]
- 20.Miskin, I. P., P. Farrimond, and I. M. Head. 1999. Identification of novel bacterial lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology 145:1977-1987. [DOI] [PubMed] [Google Scholar]
- 21.Piepersberg, W. 1997. Molecular biology, biochemistry, and fermentation of aminoglycoside antibiotics, p. 81-161. In W. R. Strohl (ed.), Biotechnology of industrial antibiotics, 2nd ed. Marcel Dekker Inc., New York, N.Y.
- 22.Pinck, L. A., D. A. Soulides, and F. E. Allison. 1961. Antibiotics in soils. I. Physicochemical studies of antibiotics-clay complexes. Soil Sci. 91:22-28. [Google Scholar]
- 23.Pinck, L. A., D. A. Soulides, and F. E. Allison. 1962. Antibiotics in soils. IV. Polypeptides and macrolides. Soil. Sci. 94:129-131. [Google Scholar]
- 24.Rothrock, C. S., and D. Gottleib. 1984. Role of antibiosis in antagonism of Streptomyces hygroscopicus var. geldanus to Rhizoctonia solani in soil. Can. J. Microbiol. 30:1440-1447. [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Takano, E., H. C. Gramajo, E. Strauch, N. Andres, J. White, and M. J. Bibb. 1992. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 6:2797-2804. [DOI] [PubMed] [Google Scholar]
- 27.Thomashow, L. S., D. M. Weller, R. F. Bonsall, and L. S. Pierson III. 1990. Production of antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl. Environ. Microbiol. 56:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turpin, P. E., V. K., Dhir, K. A. Maycroft, C. Rowlands, and E. M. H. Wellington. 1992. The effect of Streptomyces species on the survival of Salmonella in soil. FEMS Microbiol. Ecol. 101:271-280. [Google Scholar]
- 29.Watyam, U. 2003. The use of molecular techniques for in situ study of ST production in soil and rhizosphere. Ph.D. thesis. University of Warwick, Coventry, United Kingdom.
- 30.Wellington, E. M. H., N. Cresswell, and V. A. Saunders. 1990. Growth and survival of streptomycete inoculants and extent of plasmid transfer in sterile and nonsterile soil. Appl. Environ. Microbiol. 56:1413-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wellington, E. M. H., P. Marsh, I. Toth, N. Cresswell, L. Huddleston, and M. B. Schihabel. 1993. The selective effects of antibiotics in soils, p. 331-336. In R. Guerrero and C. Pedros-Alio (ed.), Trends in microbial ecology. Spanish Society for Microbiology, Barcelona, Spain.
- 32.Williams, S. T., and M. R. Khan. 1974. Antibiotics—a soil microbiologist's viewpoint. Postepy. Hig. Med. Dosw. 28:395-407. [PubMed] [Google Scholar]
- 33.Williams, S. T. 1982. Are antibiotics produced in soil? Pedobiologia 23:427-435. [Google Scholar]