Abstract
In response to reports that the contamination of food can occur during the on-farm primary phase of food production, we report data that describes a possible cost-effective intervention measure. The effect of time before soil incorporation of livestock wastes spread to land on the rate of decline of zoonotic agents present in the waste was investigated. Fresh livestock wastes were inoculated with laboratory-cultured Salmonella, Listeria, and Campylobacter spp. and Escherichia coli O157 before they were spead onto soil. Incorporation of the spread wastes was either immediate, delayed for 1 week, or did not occur at all. Bacterial decline was monitored over time and found to be significantly more rapid for all waste types when they were left on the soil surface. There were no significant differences in initial bacterial decline rates when wastes were spread in summer or winter. Our results indicate that not incorporating contaminated livestock wastes into soil is a potential intervention measure that may help to limit the spread of zoonotic agents further up the food chain. The implications of these findings are discussed in relation to current advice for livestock waste disposal.
Food can become contaminated with pathogenic microorganisms at all stages of manufacture and processing (13). However, there is a recognized potential for the on-farm transfer of pathogens to food during primary production (32). Livestock infected with zoonotic agents can excrete pathogens into their feces, and animal wastes have been implicated as a source of infection in a number of cases of human food-borne illness (2, 7). Since livestock wastes are routinely disposed of by spreading to agricultural land used for food production, the practice of waste spreading is an obvious consideration for any integrated pathogen-spread prevention-control strategy (5, 17, 18).
Over the last decade, there has been an increase in the awareness of British farmers on the best practices for storage and disposal of livestock wastes (33). The publication of specific management guidance (26, 27) was driven largely by the need to control chemical pollution from wastes, including nitrate contamination of watercourses and airborne ammonia emissions (35). The effects of these chemical pollutants are immediate and obvious and overshadow more subtle environmental damage such as the dissemination of bacterial pathogens. Evaluation of current guidance, which has been targeted toward the control of chemical pollutants, suggested that it may increase the length of time that pathogens present in the waste could survive in the environment (18). Of particular concern is a move toward immediate solid waste incorporation and band spreading or direct injection of liquid wastes into soil. Such practices are likely to decrease the rate of waste drying, the levels of UV irradiation, and the daily range of temperatures experienced by pathogens present in the waste, potentially extending their survival (6). However, there is currently no experimentally derived information that supports or disproves this hypothesis.
The purpose of the present study was to determine whether the survival of bacteria present in livestock wastes are affected by the manner in which the wastes are disposed of to soil. Specifically, we investigated the effect that the amount of time before incorporation into soil influenced the decline of bacteria present in the waste.
MATERIALS AND METHODS
Microorganisms and culture conditions.
Zoonotic bacteria used for these studies were all recent livestock waste isolates. Strains of Salmonella enterica serotype Typhimurium DT104 isolated from cattle slurry (S8118/99), serovar Typhimurium DT104 from pig slurry (S10570/99) and S. enterica serovar Enteritidis PT4 from poultry wastes (S8167/99) were used for each respective waste type. Campylobacter jejuni (strain 20001424), Listeria monocytogenes (strain 20001532), and a non-verotoxin-producing Escherichia coli O157 (strain 20001383) were isolated from cattle waste during a British manure surveillance exercise (17).
E. coli O157 and S. enterica were propagated in Luria-Bertani broth, C. jejuni was grown in modified Park-Sanders broth supplemented with 2% (vol/vol) water-lysed fresh human blood, and L. monocytogenes was cultured in Listeria selective enrichment broth. No medium supplements inhibitory to other bacteria were used for Listeria selective enrichment broth and Park-Sanders broth. All broths were supplemented with 3% (wt/vol) ammonium chloride and 1% sodium chloride. Cultures were grown without agitation or aeration at 37°C (E. coli, S. enterica, and C. jejuni) or 25°C (L. monocytogenes). Headspace in Campylobacter culture vessels was filled by using a custom formulated mixture of 8% (vol/vol) carbon dioxide, 7% (vol/vol) oxygen, and 85% (vol/vol) nitrogen (British Oxygen Company, Guilford, United Kingdom).
Wastes and inoculation of bacterial pathogens.
Solid farmyard manure (FYM) and/or liquid wastes (slurries or dirty waters) from dairy cattle, laying chickens, poultry broilers, or breeder pigs were investigated as part of the present study. Pathogen declines were measured in studies commencing during summer and winter so that seasonal differences could be compared. All wastes were fresh (<72 h postdeposition) and collected from commercial farms. Bacterial pathogens (cultured as described above) were introduced directly into the wastes and distributed through the material by either vigorous agitation (slurries and dirty waters) or tumbling in a concrete mixer for 5 min. Bacteria were added to the wastes to give, as close as possible, a theoretical concentration of 107 CFU g of waste−1. Care was taken to ensure that the volume of culture added did not exceed 5% of the mass of the waste. The amounts of waste used for each 3-m2 field plot were 25 liters of cattle slurry, 12.5 kg of cattle FYM, 10.7 kg of pig FYM, 18.7 liters of pig slurry, and 2.6 kg of broiler litter. The waste masses were calculated from an average nitrogen content for each livestock waste type (derived from analyses undertaken by this laboratory between 1995 and 2000 [n = 16,322; results not shown]) and a target concentration of 200 kg of total nitrogen Ha−1 (the maximum manure application rate advised by the government of the United Kingdom) (26). The nitrogen values used for the calculations were 3 g of N liter−1 at 6% dry matter for cattle slurry, 6 g of N kg−1 for cattle FYM, 4 g of N liter−1 at 4% dry matter for pig slurry, 7 g of N kg−1 for pig FYM, and 30 g of N kg−1 for poultry litter.
Field spreading.
A week before commencing the experiment, three soil samples each of 5-kg mass were collected by taking handfuls of soil randomly from the field site. These samples were sent to an external laboratory (Direct Laboratories Analytical Chemistry) for analyses of their physicochemical properties (Table 1). Three replicated field plots were generated for each treatment. Incorporation treatments done were as follows: <2 h after spreading (immediate), 7 days after spreading (delayed), or no incorporation into the soil. Manures were incorporated into the soil by using a customized spading machine attached to the back of a tractor. The spading machine was designed to mimic manual digging with an action specifically designed to minimize the movement of waste from the plots. The incorporation depth was 10 to 15 cm. Controls that were not spread with any manure or pathogen were included for each treatment. Experiments were conducted during spring and summer 2001 (summer treatments) and also during autumn and winter (winter treatments) 2002.
TABLE 1.
Physicochemical profile of the soil used for this study
| Analysis | Mean result (n = 3) |
|---|---|
| Parameters | |
| Moisture (%) | 12.1 |
| Chloroform N (μg g−1) | 8.53 |
| Biomass N (μg g−1) dry matter | 16 |
| Biomass C (μg g−1) | 39.7 |
| pH | 6.9 |
| Magnesium (mg liter−1) | 129 |
| Total nitrogen (% mm) | 0.12 |
| Organic matter (% mm) | 1.82 |
| Total potassium (% mm) | 0.126 |
| Total phosphorus (% mm) | 0.07 |
| Chloroform C (μg g−1) | 17.9 |
| Particle size distribution (%) | |
| 2,000-600 μm (coarse sand) | 3 (wt/wt) |
| 599-212 μm (medium sand) | 43 (wt/wt) |
| 211-63 μm (fine sand) | 23 (wt/wt) |
| 62-20 μm (coarse silt) | 11 (wt/wt) |
| 19-2 μm (fine silt) | 12 (wt/wt) |
| <2 μm (clay) | 8 (wt/vol) |
Sample collection and transit.
Samples from each replicated field plot were collected over a 9-month period and analyzed independently. Samples of wastes that had been immediately incorporated were collected 0, 2, 4, 8, 16, 34, 64, 121, and 278 days after waste spreading. Wastes that had delayed incorporation or were not incorporated were sampled on days 0, 2, 8, 16, 64, and 121 days after waste spreading. Each sample was ∼500 g of soil generated from a minimum of 20 combined subsamples collected to a depth of 15 cm by using a sterilized soil auger. Samples were refrigerated at 2°C and shipped from the farm site to laboratories, where analyses were commenced within 16 h of sampling.
Environmental temperatures and rainfall.
Soil temperature was recorded at a depth of 5 cm by using a tiny talk data logger (Gemini Data Loggers, Chichester, United Kingdom). Precipitation was collected in a rain gauge located ca. 300 meters from the plots and recorded daily.
Determination of bacterial levels.
E. coli O157, Salmonella, Campylobacter, and Listeria organisms were enumerated by using a general filter resuscitation method with different selective media (Table 2) used for each organism. Briefly, subsamples (25 g) of wastes were mixed with 225 ml of initial diluent. Samples were homogenized in mesh filter bags (6041/STR; Seward, Thetford, United Kingdom) in a stomacher (Colworth 400; Seward) for 1 min before centrifugation (300 × g, 5 min) and filtration of the supernatant through a glass fiber filter (Sartorius 13430-0475) to remove granular particulates. Filtrates were diluted decimally, 10−1 to 10−5 in initial diluent. Each dilution was vacuum filtered through a cellulose nitrate filter with an organism-specific pore size (Table 2). Filters containing bacteria were placed on felt pads soaked in resuscitation medium at 37°C for different periods depending on organism (Table 2). Filters were transferred to counting medium plates and incubated as shown (Table 2). Confirmation of presumptive-positive, colored colonies was as described in (Table 2). Colony counts were converted to CFU g of waste−1 according to the criteria specified by the International Organization for Standardization (19, 20).
TABLE 2.
Selective media and organism-specific variations used for enumeration of bacterial zoonotic agents from livestock wastesa
| Parameter | Selective media and organism-specific variation(s) for:
|
|||
|---|---|---|---|---|
| E. coli O157 | Salmonella sp. | Listeria sp. | Campylobacter sp. | |
| Initial diluent | mTSB, 80 μg of novobiocin ml−1 | mTSB, 40 μg of novobiocin ml−1 | LSEB | PSW (boiled to remove O2) |
| Cellulose nitrate filter pore size (μm) | 0.45 | 0.45 | 0.45 | 0.1 |
| Resuscitation medium | mTSB, 40 μg of novobiocin ml−1 | mTSB, 20 μg of novobiocin ml−1; 1% (wt/vol) iodine | LSEB | BFEB (microaerophilic conditions) |
| Resuscitation time (h) | 6 | 16 | 24 | 24 |
| Counting medium | CHROMagar O157 (Becton Dickson 264105) | Rambach agar | BCM listeria agar (Biosynth C0608 and C0610) | CCDA microaerophilic conditions |
| Incubation conditions prior to counting | 16 h at 41°C | 28 h at 37°C | 28 h at 37°C | 28 h at 37°C |
| Color/morphology of pre- sumptive positive colonies | Purple | Pinkish red | Blue/convex | Gray/moist/flat |
| Confirmation method or apparatus | Latex agglutination (Oxoid DR120M) | API 20E (BioMérieux) | Listeria confirmation agar (Biosynth C0612) | API Campylobacter (BioMérieux) |
All media were from Oxoid (Basingstoke, United Kingdom) unless otherwise specified. mTSB, modified tryptone soy broth; LSEB, Listeria selective enrichment broth; PSW, peptone salt water; BFEB, blood-free enrichment broth; CCDA, Campylobacter agar with charcoal and deoxycholate.
Chemical methods.
The pH and conductivity of liquid or solid wastes were determined directly on samples without dilution (slurry) or with decimal dilution (FYM), respectively, by using conductivity (Philips PW9526) and pH meters (WPA model CD620). Dry matter was assessed by weighing the wastes and drying them in an oven at 100°C for 16 h; remaining weight was expressed as a percentage of the initial weight. Ammonium-N (the N ions contributed by NH4 in the waste) was extracted from wastes with 2 M KCl. The pH of an aliquot of each extract was raised by the addition of NaOH, and the released gaseous ammonia recovered by distillation and condensation. The concentration of ammonia was determined by titration with 0.05 M sulfuric acid with methyl red-bromocresol green as indicators.
Analyses of results.
Log averages and associated standard deviations (SDs) from each set of three replicates were calculated for each sample time by using Excel 2000 (Microsoft). R2 values were determined by the least-squares method, and coefficients of variation (CVs) were calculated by dividing the means by the SD for each sample time. Groups of CVs were compared by using the Mann-Whitney U-test for nonparametric data (SPSS 11.5; SPSS, Inc., Chicago, Ill.). D values (the number of days required for a 1-log decline in bacterial numbers) were calculated from data generated during the first 16 days immediately after waste spreading. Groups of D values were compared by using one-way analysis of variance (ANOVA) with Tukey's post-hoc test (SPSS).
RESULTS AND DISCUSSION
Bacteria were added to the livestock wastes at a theoretical concentration of 107 CFU g of waste−1. However, the measured levels of each pathogen were typically ca. 106 CFU g−1, indicating either significant bacterial death occurred upon introduction to the wastes or that organisms were lost or irreversibly bound to organic material in the wastes. A count of 106 CFU g−1, however, was still typical of the levels of zoonotic agents observed during an on-farm survey of fresh livestock wastes (17).
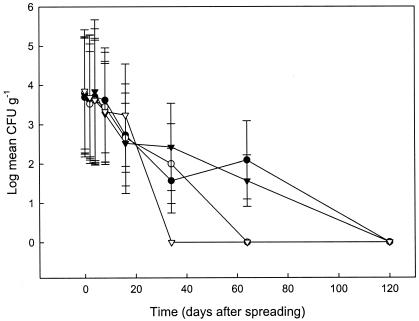
Bacterial decline in plots spread with dairy cattle slurry that had been immediately incorporated into the soil is depicted in Fig. 1. The declines shown in Fig. 1 are typical of those observed in the other plots that were spread with different wastes. Declines measured over the first 16 days were linear. However, in laboratory-based pilot studies we observed a significant range of up to 4 logs in the levels of zoonotic agents isolated from single handfuls of inoculated FYM and up to 2 logs for slurries. Thus, it is likely that the distribution of bacteria through the waste before spreading was not uniform. However, longer or more vigorous agitation of wastes may have resulted in aeration of the material which is an effective control strategy for reducing bacterial numbers (30, 34). Furthermore, single soil plots into which pathogen-inoculated FYM and slurry was incorporated by using a spade showed a range of almost 3 logs for both waste types when single core samples taken from the plots were analyzed (results not shown). Consequently, and although multiple soil cores were combined to generate samples, the average bacterial counts obtained from the replicate plots had large SDs (Fig. 1). However, the experimental setup was typical of that encountered on a working farm, and it is likely that relative hotspots of bacterial population are generated during commercial waste spreading. Generally, CVs for slurries were significantly lower than those calculated for solid wastes, which had higher dry-matter contents (P < 0.05 [Mann-Whitney]). Similarly high variation has been observed previously for E. coli in wastes spread to soil or pasture (6, 37). In the present study, Listeria levels were generally much less variable, and consequently their decline graphs usually had higher R2 values than those calculated for the other bacteria (data not shown). It is uncertain why this should be when all pathogens were introduced into the manures and spread in an identical manner. The analysis methods used for the present study are labor-intensive, and it was not possible to determine native populations of each pathogen in the uninoculated manures. L. monoctyogenes is, however, very commonly isolated from livestock wastes (18a). It is possible that a native Listeria population was more evenly distributed through the material. If this were the case, it would likely result in a more even decline. Control plots, which did not contain manure, exclude the possibility of native soil populations being implicated. Molecular typing of bacteria was not undertaken as part of the study, and thus we were unable to differentiate between native and inoculated listeriae.
FIG. 1.
Decline of inoculated S. enterica (•), E. coli O157 (○), L, monotytogenes (▾), and C. jejuni (▿) introduced into fresh dairy cattle slurry and spread onto a sandy loam soil in late spring. Livestock waste was incorporated into the soil within 2 h of spreading. Each datum point is the log average of three independent replicates. Error bars indicate log SDs. A value of 1 CFU g of soil−1 was used for samples in which no zoonotic agents were detected to enable plotting of log data.
D values calculated from the first 16 days of decline during summer and winter are shown in Tables 3 and 4, respectively. For the experiments which monitored bacterial decline over the summer, a significant difference between the incorporation treatments was detected (P < 0.05 [ANOVA]). Further analyses (P < 0.05 [Tukey's post-hoc test]) revealed that rates of decline for pathogens in waste incorporated immediately were significantly slower than when the waste was left on the soil surface. There was no significant difference in pathogen decline rates between the delayed and unincorporated treatments.
TABLE 3.
Effect of interval before soil incorporation on decimal reduction values (D values) for zoonotic agents present in livestock wastes spread onto an arable sandy loam soil in late spring 2001a
| Waste type | Treatment | D value (days) for:
|
Mean | |||
|---|---|---|---|---|---|---|
| Salmonella sp. | E. coli O157 | Listeria sp. | Campylobacter sp. | |||
| Dairy cattle FYM | Immediate | 2.18 | 2.31 | 2.94 | 2.91 | 2.59 |
| Delayed | 1.04 | 5.06 | 2.59 | 0.98 | 2.42 | |
| Unincorporated | 1.12 | 1.04 | 3.44 | 1.13 | 1.68 | |
| Dairy cattle slurry | Immediate | 3.98 | 1.62 | 1.37 | 4.03 | 2.75 |
| Delayed | 1.11 | 0.89 | 0.88 | 0.86 | 0.94 | |
| Unincorporated | 0.89 | 0.99 | 0.88 | 0.91 | 0.92 | |
| Poultry broiler litter | Immediate | 1.28 | 1.05 | 1.00 | 1.11 | 1.11 |
| Delayed | 0.76 | 0.66 | 0.81 | 1.15 | 0.84 | |
| Unincorporated | 1.66 | 0.75 | 0.97 | 0.79 | 1.04 | |
| Pig FYM | Immediate | 2.90 | 1.00 | 1.32 | 1.58 | 1.70 |
| Delayed | 1.28 | 0.95 | 0.78 | 0.74 | 0.94 | |
| Unincorporated | 1.33 | 0.67 | 0.93 | 0.79 | 0.93 | |
| Pig slurry | Immediate | 2.44 | 4.89 | 1.13 | 1.03 | 2.37 |
| Delayed | 1.89 | 0.97 | 0.66 | 0.63 | 1.04 | |
| Unincorporated | 0.79 | 1.76 | 0.84 | 2.26 | 1.41 | |
Values were calculated from measurements of bacterial decline over the 16 days immediately after waste spreading. FYM is a mixture of fecal material and bedding.
TABLE 4.
Effect of interval before soil incorporation on decimal reduction times (D values) for zoonotic agents present in livestock wastes spread onto an arable sandy loam soil in early winter 2002a
| Waste type | Treatment | D value (days) for:
|
Mean | |||
|---|---|---|---|---|---|---|
| Salmonella sp. | E. coli O157 | Listeria sp. | Campylobacter sp. | |||
| Dairy cattle FYM | Immediate | — | — | 2.31 | 2.15 | 2.23 |
| Unincorporated | 1.72 | 2.16 | 1.10 | 1.65 | 1.66 | |
| Dairy cattle slurry | Immediate | — | — | 1.09 | 2.55 | 1.82 |
| Unincorporated | 1.06 | 3.77 | 0.76 | 1.59 | 1.80 | |
| Dirty water | Immediate | 1.26 | 1.75 | 1.05 | 2.50 | 1.64 |
| Unincorporated | 0.95 | — | 0.66 | 0.38 | 0.66 | |
| Poultry layer FYM | Immediate | 0.87 | 1.86 | 0.99 | 1.25 | 1.24 |
| Unincorporated | — | 0.93 | 0.92 | 0.83 | 0.89 | |
| Poultry broiler litter | Immediate | 1.10 | 1.88 | 2.22 | 2.63 | 1.96 |
| Unincorporated | 1.10 | 1.96 | 2.02 | 0.64 | 1.43 | |
| Pig FYM | Immediate | 2.50 | 2.75 | 3.04 | 3.90 | 3.05 |
| Unincorporated | 1.46 | 2.52 | 2.92 | 1.64 | 2.14 | |
| Pig slurry | Immediate | 5.20 | 1.86 | 1.71 | 1.14 | 2.48 |
| Unincorporated | 2.00 | 1.54 | 1.21 | 0.92 | 1.42 | |
Values were calculated from measurements of bacterial decline over the 16 days immediately after waste spreading. A dash (-) was used to denote declines that were so variable that no meaningful trend line (R2 < 0.65) could be fitted to the results.
Winter decline experiments compared rates of pathogen decline between unincorporated and immediately incorporated wastes only (Table 4). As observed in the summer decline experiments, initial rates of decline were significantly slower (P < 0.05 [ANOVA]) when wastes were immediately incorporated into the soil in winter. Although there were no apparent differences in the setup of the winter and summer decline experiments, we noted there were larger SDs in the winter treatment. At times, the variation made calculation of D values impossible because straight-line fitting with an R2 of >0.65 was not possible for some data sets (Table 4). We believe excessive variation is also a likely reason why the levels of E. coli O157 in dairy cattle FYM and pig slurry and C. jejuni in pig slurry appear to decline more rapidly when these wastes were incorporated into the soil.
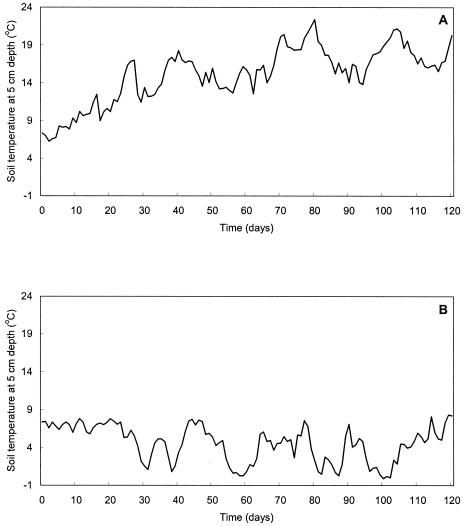
In the present study, pathogens declined at similar rates during summer and winter (P > 0.05). This was surprising because in vitro and on-farm studies have shown temperature-dependent differences in decline (28) with bacteria surviving longer at lower temperatures (6). These previous studies, some of which were laboratory-based (38), showed differences in declines at different temperatures and used constant temperature incubators, as well as single batches of wastes. In the present study, electronic temperature recording at a soil depth of 5 cm was undertaken every 4 h over the course of the experiment; mean results are shown in Fig. 2. Since we had no control over ambient temperatures, our experiments were subject to a constantly changing range of temperatures. Furthermore, soil temperatures for winter and summer experiments were within 5°C of each other over the first 3 weeks that the experiments were running. When combined with sampling variation, this difference may have been too small to show any difference. In addition, there was low-level almost-continuous rainfall during over the first 2 weeks of the winter decline experiments. The summer rainfall, although higher in volume, was more sporadic. A previous study has shown that leaching and movement of E. coli O157 occurs as a response to simulated rainfall in sandy loam soils (14). Thus, it is possible the decline we observed was the result of bacterial death and rain-assisted movement of bacteria. A further potential explanation for this finding may be that it is appropriate only to compare bacterial decline in wastes and soils for a single seasonal experiment with subsamples from a single batch of manure. Wastes do have variable physical properties and chemical compositions (22), and interbatch differences may be significant enough to mask correlations with factors, such as temperature, thereby confounding seasonal comparisons (22). The pH, dry-matter content, conductivity, and ammonium concentration were determined for each sample during the summer experiment only. Although the original manures had different pH values, dry matter, and conductivities, spreading the waste onto or into a large mass of soil normalized these differences. After waste incorporation into soil, dry matter (mean, 862 g kg−1), pH (mean, 7.45), and conductivity (mean, 22,650 μSi cm−2) did not change appreciably over the course of the experiments.
FIG. 2.
Temperature of soil at 5 cm depth during summer (A) and winter (B) decline experiments.
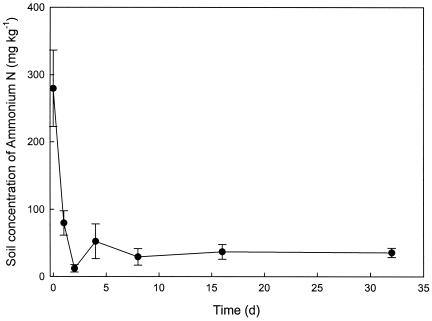
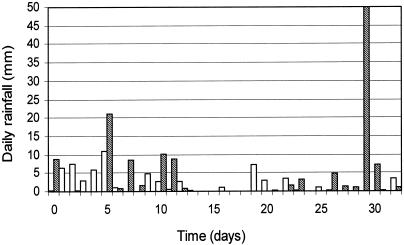
Figure 3 shows the changes in soil ammonium-N levels (calculated as the mean of all 27 experimental plots during the summer decline) over the first 32 days that the experiment ran. Although manures were applied and incorporated into the soil at almost the maximum DEFRA-specified loadings (26), there was a rapid decrease in soil levels of ammonium-N. Thus, it seems likely that antimicrobial ammonium-N was not involved in the observed decline of zoonotic bacteria. This finding is interesting because it helps to explain the counterintuitive, but reproducible, findings of previous studies which have shown enhanced survival or growth of bacteria in manure amended soils (14, 21). Similar sharp decreases in ammonium have been reported previously to be the result of conversion of ammonium-N to nitrate by indigenous soil bacteria (9). Furthermore, sandy soils drain readily, and excess rainfall is likely to result in nitrate and, to a lesser extent, ammonium-N leaching. Over the first 5 days of the summer experiment 30 mm of rain was recorded as two distinct events (Fig. 4) that likely contributed to the ammonium-N decline.
FIG. 3.
Changes in soil concentration of ammonium-N over time. Livestock wastes were applied to sandy loam soil and incorporated immediately during summer. Samples were collected and analyzed for ammonium-N as described in Materials and Methods. d, days.
FIG. 4.
Rainfall over the 32 days after the experimental plots were spread with livestock manure inoculated with zoonotic agents in summer (▴) and winter (□).
Table 5 summarizes the periods that zoonotic agents were isolated from the experimental plots. These intervals are the worst case since they are the longest survival times from either the winter or summer experiments. In the present study, pathogens were inoculated into wastes at the maximum levels measured during a national survey of zoonotic agents in fresh (unstored) livestock wastes (17). Since Table 5 takes no account of the bacterial decline that occurs during storage, it is an indication of the longest periods of time that should be left to ensure zoonotic agents have declined completely. For the batches of waste used in the present study, no zoonotic bacteria survived longer than 120 days.
TABLE 5.
Maximum isolation times for zoonotic agentsa
| Waste typeb | Isolation time maximum (days) for:
|
|||
|---|---|---|---|---|
| Salmonella sp. | E. coli O157 | Listeria sp. | Campylobacter sp. | |
| Dairy cattle FYM | 120 | 34 | 120 | 64 |
| Dairy cattle slurry | 120 | 64 | 120 | 34 |
| Beef cattle FYM* | 34 | 64 | 120 | 120 |
| Beef cattle slurry* | 120 | 16 | 120 | 64 |
| Pig FYM | 120 | 16 | 120 | 34 |
| Pig Slurry | 56 | 16 | 120 | 36 |
| Poultry FYM | 56 | 16 | 56 | 64 |
| Sheep FYM* | 120 | 16 | 120 | 34 |
| Dirty water | 120 | 34 | 120 | 120 |
The maximum isolation time was the longest recorded length of time (in days) until zoonotic agents could no longer be isolated. Bacteria were introduced into livestock wastes that were spread onto a sand loam and immediately incorporated. The majority of data shown are the longest times observed from either of two decline experiments that ran separately during a summer or a winter season.
Wastes marked with an asterisk were studied during a summer season only.
A number of previous studies have published summaries from both naturally contaminated and inoculated wastes from both field- and laboratory-based studies (3, 14, 15, 29). Previous studies have used a variety of designs and different analysis methods. Although this makes direct comparisons difficult, it provides a likely explanation for why the findings of previous studies both agree and conflict with the findings reported here. Baloda et al. (3) determined that after 21 days Salmonella could no longer be isolated from soil spread with naturally contaminated (unreported levels) slurry. Gessel et al. (15) were able to isolate S. enterica serotype Anatum for only 7 days with a starting inoculum of similar concentration (2 × 106 CFU g−1) to the present study. A range of survival times have also been reported for E. coli O157. Jiang et al. (22) were able to isolate E. coli O157:H7 derived from a five-strain inoculum for over 200 days in soil-manure mixtures, although the initial inoculum was higher than we could obtain for field-based studies. Exceptionally high E. coli O157 inoculum levels of 108 declined to only 5 × 106 after 130 days in wastes applied to soil sown with grass under laboratory conditions (28). A number of previous studies have reported that survival of pathogens is at least partly dependent on levels and types of soil microbiota (12, 21). Since this is an additional factor influencing bacterial decline in soils, it may be helpful for future studies to make attempts to characterize indigenous populations since this will make it easier to make interstudy comparisons.
Disposal of livestock waste is a complex problem, and there are numerous factors that should be considered when deciding the best approach. Livestock wastes contain a variety of nitrogenous compounds and may contain toxic elements such as copper and selenium. The environmental impacts of high levels of these chemicals must be considered when determining how best to dispose of livestock wastes. Although the overall contribution of waste spreading to levels of food-borne illness is currently uncertain, a number of cases of food-borne illness have been linked to animal manures (10, 16). A recent British survey has shown that there is a one-in-three chance that a sample of livestock waste will contain either Campylobacter, Listeria, Salmonella, Giardia, E. coli O157, or Cryptosporidium parvum at mean levels of up to 106 g of waste−1 (17). Thus, the microbiological risks associated with waste spreading should also be considered in determining how best to dispose of livestock wastes.
We have shown here clearly that the amount of time contaminated wastes remain on the soil surface influences the rate at which pathogens decline. Hence, incorporating livestock wastes will increase the total time that manure-borne pathogens remain viable in the soil after waste spreading. However, leaving wastes on the soil surface may increase the likelihood of pathogen spread. Previously, it has been reported that zoonotic agents such as E. coli O157 can be isolated from flies (24, 29). Leaving waste on the soil surface increases the likelihood of insect infection and the spread of zoonotic agents to the wider environment. A similar scenario is equally relevant for vermin, birds, and other wildlife likely to scavenge fields spread with wastes (1, 11, 23). Furthermore, leaving wastes on the surface increases the possibility that rainfall heavy enough to cause surface runoff could wash pathogens and manures directly into watercourses where they are likely to last longer than those in terrestrial environments (8, 25, 28, 31, 36).
Smith et al. (34) determined that current agricultural practice in the UK is to leave manures on the soil surface for at least a week after spreading. Such a practice would be conducive to an initial rapid decline in pathogen levels, although according to Smith et al., manures can be incorporated as soon as practicable after spreading in an effort to control odor and ammonia pollution. For similar reasons, spreading methods such as soil injection and band spreading, which restrict the amount of time waste is exposed to UV irradiation from sunlight (4) and the drying effect of the atmosphere can also be used. Livestock waste disposal should be considered holistically using a multidisciplinary approach that takes into account population, food, and the environment. Emissions of greenhouse gases such as ammonia do have long-term importance because their continued release has implications for climate change and global warming. However, the short-term implications of inappropriate waste disposal for the microbiological aspects of food safety should not be overlooked.
Acknowledgments
This study was funded by the B17 Organic Wastes Programme of the United Kingdom Food Standards Agency.
We gratefully acknowledge Barry Petrie and Gemma Simpson for laboratory analyses, Tracey Woodfield for sample collections, and Rob Davies of the Veterinary Laboratories Agency (United Kingdom) for kindly providing recent livestock isolates of Salmonella.
REFERENCES
- 1.Adesiyun, A. A., N. Seepersadsingh, L. Inder, and K. Caesar. 1998. Some bacterial enteropathogens in wildlife and racing pigeons from Trinidad. J. Wildl. Dis. 34:73-80. [DOI] [PubMed] [Google Scholar]
- 2.Bach, S. J., T. A. McAllister, D. M. Veira, V. P. J. Gannon, and R. A. Holley. 2002. Transmission and control of Escherichia coli O157:H7: a review. Can. J. Anim. Sci. 82:475-490. [Google Scholar]
- 3.Baloda, S. B., L. Christensen, and S. Trajcevska. 2001. Persistence of a Salmonella enterica serovar Typhimurium DT12 clone in a piggery and in agricultural soil amended with Salmonella-contaminated slurry. Appl. Environ. Microbiol. 67:2859-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, E. K., S. A. Husseini, M. T. Farran, D. A. Itani, R. H. Houalla, and S. K. Hamadeh. 2002. Soil solarization: a sustainable agriculture approach to reduce microorganisms in chicken manure-treated soil. J. Sustainable Agric. 19:95-104. [Google Scholar]
- 5.Bicudo, J. R., and S. M. Goyal. 2003. Pathogens and manure management systems: a review. Environ. Technol. 24:115-130. [DOI] [PubMed] [Google Scholar]
- 6.Bolton, D. J., C. M. Byrne, J. J. Sheridan, D. A. McDowell, and I. S. Blair. 1999. The survival characteristics of a non-toxigenic strain of Escherichia coli O157:H7. J. Appl. Microbiol. 86:407-411. [DOI] [PubMed] [Google Scholar]
- 7.Brackett, R. E. 1999. Incidence, contributing factors, and control of bacterial pathogens in produce. Postharvest Biol. Technol. 15:305-311. [Google Scholar]
- 8.Chalmers, R. M., H. Aird, and F. J. Bolton. 2000. Waterborne Escherichia coli O157. J. Appl. Microbiol. 88:124S-132S. [DOI] [PubMed] [Google Scholar]
- 9.Chantigny, M. H., D. A. Angers, T. Morvan, and C. Pomar. 2004. Dynamics of pig slurry nitrogen in soil and plant as determined with N-15. Soil Sci. Soc. Am. J. 68:637-643. [Google Scholar]
- 10.Cieslak, P. R., T. J. Barrett, P. M. Griffin, K. F. Gensheimer, G. Beckett, J. Buffington, and M. G. Smith. 1993. Escherichia coli O157:H7 infection from a manured garden. Lancet 342:367.. [DOI] [PubMed] [Google Scholar]
- 11.Clough, H. E., D. Clancy, P. D. O'Neill, and N. P. French. 2003. Bayesian methods for estimating pathogen prevalence within groups of animals from faecal-pat sampling. Prevent. Vet. Med. 58:145-169. [DOI] [PubMed] [Google Scholar]
- 12.Cooley, M. B., W. G. Miller, and R. E. Mandrell. 2003. Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl. Environ. Microbiol. 69:4915-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.den Aantrekker, E. D., R. M. Boom, M. H. Zwietering, and M. van Schothorst. 2002. Quantifying recontamination through factory environments: a review. Int. J. Food Microbiol. 80:117-130. [DOI] [PubMed] [Google Scholar]
- 14.Gagliardi, J. V., and J. S. Karns. 2000. Leaching of Escherichia coli O157 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gessel, P. D., N. C. Hansen, S. M. Goyal, L. J. Johnston, and J. Webb. 2004. Persistence of zoonotic pathogens in surface soil treated with different rates of liquid pig manure. Appl. Soil Ecol. 25:237-243. [Google Scholar]
- 16.Hilborn, E. D., J. H. Mermin, P. A. Mshar, J. L. Hadler, A. Voetsch, M. Swartz, R. Mshar, M. A. Lambert-Fair, J. A. Farrar, M. K. Glynn, and L. Slutsker. 1999. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch. Intern. Med. 159:1758-1764. [DOI] [PubMed] [Google Scholar]
- 17.Hutchison, M. L., A. K. Ashmore, K. M. Crookes, D. W. Wilson, S. J. Grives, B. T. Chambers, C. W. Keevil, and A. Moore. 2002. Enumeration of pathogens in livestock wastes and factors affecting their survival. Proc. Joint CIWEM Aqua-Enviro Technol. Transfer 7th Eur. Biosolids Organic Residuals Conf. 1:S3.15.1-S3.15.7.
- 18.Hutchison, M. L., F. A. Nicholson, K. Smith, W. C. Keevil, and T. A. Moore. 2000. Study of on-farm manure applications to agricultural land and an assessment of the risks of pathogen transfer into the food chain. Ministry of Agriculture Fisheries and Foods, London, United Kingdom.
- 18a.Hutchinson, M. L., L. D. Walters, S. M. Avery, B. A. Synge, and A. Moore. Levels of zoonotic agents in British livestock manures. Lett. Appl. Microbiol., in press. [DOI] [PubMed]
- 19.International Organization for Standardization. 1991. Methods for microbiological examination of food and animal feeding stuffs. Enumeration of micro-organisms: colony count technique at 30°C. Document ISO 4833. International Organization for Standardization, Geneva, Switzerland.
- 20.International Organization for Standardization. 1999. Microbiology of food and animal feeding stuffs. Preparation of test samples, initial suspension, and decimal dilutions for microbiological examination: general rules for the preparation of the initial suspension and decimal dilutions. Document ISO 6887. International Organization for Standardization, Geneva, Switzerland.
- 21.Jiang, X. P., J. Morgan, and M. P. Doyle. 2002. Fate of Escherichia coli O157:H7 in manure-amended soils. Appl. Environ. Microbiol. 68:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, P. W. 1976. The effect of temperature, solids content and pH on the survival of salmonellas in cattle slurry. Br. Vet. J. 132:284-293. [DOI] [PubMed] [Google Scholar]
- 23.Kapperud, G., H. Stenwig, and J. Lassen. 1998. Epidemiology of Salmonella typhimurium O:4-12 infection in Norway: evidence of transmission from an avian wildlife reservoir. Am. J. Epidemiol. 147:774-782. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, M., T. Sasaki, N. Saito, K. Tamura, K. Suzuki, H. Watanabe, and N. Agui. 1999. Houseflies: not simple mechanical vectors of enterohemorrhagic Escherichia coli O157:H7. Am. J. Trop. Med. Hyg. 61:625-629. [DOI] [PubMed] [Google Scholar]
- 25.Lim, C. H., and K. P. Flint. 1989. The effects of nutrients on the survival of Escherichia coli in lake waters. J. Appl. Bacteriol. 66:559-569. [DOI] [PubMed] [Google Scholar]
- 26.Ministry of Agriculture, Fisheries, and Foods. 1999. Managing livestock manures: making better use of livestock manures on arable land. Ministry of Agriculture Fisheries and Foods, London, United Kingdom.
- 27.Ministry of Agriculture, Fisheries, and Foods. 1999. Managing livestock manures: making better use of livestock manures on grassland. Ministry of Agriculture Fisheries and Foods, London, United Kingdom.
- 28.Maule, A. 2000. Survival of verocytotoxigenic Escherichia coli O157 in soil, water and on surfaces. J. Appl. Microbiol. 88:71S-78S. [DOI] [PubMed] [Google Scholar]
- 29.Moriya, K., T. Fujibayashi, T. Yoshihara, A. Matsuda, N. Sumi, N. Umezaki, H. Kurahashi, N. Agui, A. Wada, and H. Watanabe. 1999. Verotoxin-producing Escherichia coli O157:H7 carried by the housefly in Japan. Med. Vet. Entomol. 13:214-216. [DOI] [PubMed] [Google Scholar]
- 30.Ndegwa, P. M., J. Zhu, and A. Luo. 2003. Effects of bioreactor temperature and time on odor-related parameters in aerated swine manure slurries. Environ. Technol. 24:1007-1016. [DOI] [PubMed] [Google Scholar]
- 31.Pelczar, M. J., E. C. S. Chan, and N. R. Krieg. 2001. Foodborne and waterborne diseases, p. 680-714. In M. J. Pelczar, E. C. S. Chan, and N. R. Krieg (ed.), Microbiology concepts and applications. McGraw-Hill Book Co., Inc., New York, N.Y.
- 32.Pell, A. N. 1997. Manure and microbes: public and animal health problem. J. Dairy Sci. 80:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, K. A., A. J. Brewer, J. Crabb, and A. Dauven. 2001. A survey of the production and use of animal manures in England and Wales. II. Poultry manure. Soil Use Manage. 17:48-56. [Google Scholar]
- 34.Solano, M. L., F. Iriarte, P. Ciria, and M. J. Negro. 2001. Performance characteristics of three aeration systems in the composting of sheep manure and straw. J. Agric. Eng. Res. 79:317-329. [Google Scholar]
- 35.Strauch, D., and G. Ballarini. 1994. Hygienic aspects of the production and agricultural use of animal wastes. Zentralbl. Veterinarmed. B 41:176-228. [DOI] [PubMed] [Google Scholar]
- 36.Sturdee, A. P., A. T. Bodley-Tickell, and S. E. Kitchen. 1998. Cryptosporidium in farmed and wild animals and the implications for water contamination, p. 1-66. MAFF CSA2783. Coventry University, Coventry, United Kingdom.
- 37.Vinten, A. J. A., D. R. Lewis, D. R. Fenlon, K. A. Leach, R. Howard, I. Svoboda, and I. Ogden. 2002. Fate of Escherichia coli and Escherichia coli O157 in soils and drainage water following cattle slurry application at three sites in southern Scotland. Soil Use Manage. 18:223-231. [Google Scholar]
- 38.Wang, G., T. Zhao, and M. P. Doyle. 1996. Fate of enterohemorrhagic Escherichia coli O157:H7 in bovine feces. Appl. Environ. Microbiol. 62:2567-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]