Abstract
Numerous microorganisms, including bacteria, yeasts, and molds, constitute the complex ecosystem present in milk and fermented dairy products. Our aim was to describe the bacterial ecosystem of various cheeses that differ by production technology and therefore by their bacterial content. For this purpose, we developed a rapid, semisystematic approach based on genetic profiling by temporal temperature gradient electrophoresis (TTGE) for bacteria with low-G+C-content genomes and denaturing gradient gel electrophoresis (DGGE) for those with medium- and high-G+C-content genomes. Bacteria in the unknown ecosystems were assigned an identity by comparison with a comprehensive bacterial reference database of ∼150 species that included useful dairy microorganisms (lactic acid bacteria), spoilage bacteria (e.g., Pseudomonas and Enterobacteriaceae), and pathogenic bacteria (e.g., Listeria monocytogenes and Staphylococcus aureus). Our analyses provide a high resolution of bacteria comprising the ecosystems of different commercial cheeses and identify species that could not be discerned by conventional methods; at least two species, belonging to the Halomonas and Pseudoalteromonas genera, are identified for the first time in a dairy ecosystem. Our analyses also reveal a surprising difference in ecosystems of the cheese surface versus those of the interior; the aerobic surface bacteria are generally G+C rich and represent diverse species, while the cheese interior comprises fewer species that are generally low in G+C content. TTGE and DGGE have proven here to be powerful methods to rapidly identify a broad range of bacterial species within dairy products.
Numerous dairy products are home to a complex microbial ecosystem, which is responsible for the broad diversity of tastes, aromas, and textures that are associated with them. Many bacteria make a positive contribution to the organoleptic qualities of cheeses or fermented milk, while others may have adverse effects or may even constitute a health risk. Cheese processing is largely based on fermentation by lactic acid bacteria (LAB), which are both deliberately added as starter cultures or adventitiously present in the biotope and selected during the fermentation process. Furthermore, raw milk bacteria, including nonstarter LAB, reportedly enhance cheese flavor and diversity (30, 32, 42). Ripened cheeses are characterized by a succession of largely undefined microbial communities on their surface (6, 59). These aerobic microorganisms have a strong impact on the appearance, odor, flavor, and texture development of the respective cheese products (6). Nondesirable microorganisms, such as the psychrotrophic Pseudomonas fluorescens (52) or certain proteolytic LAB, may cause flavor defects (e.g., bitterness and putrid flavors) in milks and cheeses (7, 8, 51). The presence of Escherichia coli, Listeria monocytogenes, and Staphylococcus aureus in raw milks and cheeses constitutes a health risk (3, 10, 12, 33, 41, 49). The above descriptions illustrate the present indeterminate state of the relatively complex cheese ecosystem. Enumeration of dairy microorganisms was previously based on bacterial cultivation, followed by identification of the dominating microorganisms by phenotypic methods (11, 18, 53). These approaches are tedious, restricted to cultivatable bacteria, and liable to introduce serious biases to community analyses (4, 50). Recent advances in molecular biology and phylogeny analysis techniques have opened the field of microbial ecology and have replaced the less-accurate bacteriological tests. Molecular approaches based on 16S rRNA genes (rDNAs) have facilitated a culture-independent approach for analysis of complex ecosystems (1, 2, 23, 40). In particular, single-strand conformation polymorphism, temporal temperature gradient gel electrophoresis (TTGE), and related denaturing gradient gel electrophoresis (DGGE) methods of PCR-amplified rDNA fragment separation have been applied to a variety of environmental studies for analyzing microbial communities (13, 17, 19, 20, 35, 36, 39, 45, 60). In both TTGE and DGGE, DNA fragments of the same length but with different sequences are separated, based on decreased electrophoretic mobility of partially melted double-strand DNA molecules. Separation is performed with polylacrylamide gels containing a linear gradient of chemical denaturant gradient (DGGE) or a linear temperature gradient (TTGE). TTGE and DGGE are now frequently applied in microbial ecology to compare the compositions of complex microbial communities and study their dynamics.
We recently applied TTGE to describe the diversity of LAB in commercial dairy products by setting up a bacterial database that allows rapid identification of the unknown bands (39). This database essentially included bacteria with a low-G+C-content genome, i.e., numerous LAB and a few dairy Staphylococcus species. In the present study, we modified our approach to expand the bacterial database to other species of dairy interest, including psychrotrophic and spoilage bacteria, pathogens, and bacteria present on the cheese surface. One limitation of TTGE is the poor resolution of species having high-G+C-content genomes. We therefore made combined use of TTGE and DGGE, which is more suitable for these bacterial species. Using this combined approach, new strains were identified, and the profiles of commercial cheeses were described and compared. Our results confirm the usefulness of these approaches for identifying different cheese ecosystems and have revealed some unexpected features of the flora that distinguish the cheese surface from the interior.
MATERIALS AND METHODS
Choice of bacterial strains.
The bacterial strains used in this study for the construction of the database species are listed in Table 1. They originate from culture collections or correspond to bacteria isolated from raw milk and dairy environments (see below for the species identification). We selected 150 bacterial species of dairy interest, including useful dairy microorganisms (LAB), spoilage bacteria (e.g., Pseudomonas and Enterobacteriaceae) and a few pathogenic bacteria (e.g., Listeria species and Staphylococcus aureus). Several strains from each species group were generally selected, except in cases where only one strain was available. Genomic DNA was prepared as previously described (14).
TABLE 1.
Bacterial strains included in the reference set
| Species or subspecies | Strain(s) (% identification by API)a |
|---|---|
| Hafnia alvei | AF15 (99.8), AF16 (99.9), AF17 (99.7), AF22 (99.7), AF24 (99.9), AF26 (99.8) |
| Bacillus licheniformis | AF5 (99.9), AF7 (99.9), AF8 (99.9), AF10 (99.9), CIP52.71T (99.9) |
| Bacillus pumilus | CIP77.25 (99.9) |
| Bacillus subtilis | AF6 (95.6), CIP52.65T (93.4) |
| Bacillus megaterium | AF4 (99.9) |
| Bacillus sphaericus | AF44 (89.1), AF45 (89.9), AF46 (89.9) |
| Bacillus lentus | AF9 (98.3) |
| Bacillus circulans | 12 (99% homology with V3 of B. circulans; GenBank AY043084) |
| Streptococcus uberis | Clone C4 (99% homology with V3 of S. uberis; GenBank AB002527) |
| Streptococcus agalactiae | CIP103227T (99.1), CIP82.41 (99.9) |
| Streptococcus dysgalactiae subsp. | CIP102914T (99.9), CIP55.119 (99.3) |
| Streptococcus equinus | CIP103232 (99.9) |
| Streptococcus bovis | CIP102302T (99.7) |
| Staphylococcus epidermidis | AF42 (94.5), CNRZ478 |
| Staphylococcus aureus | AF43 (92.7), CNRZ740 |
| Staphylococcus haemolyticus | AF49 (99.9), AF50 (99.9), CIP81.56T (99.9), AF69 |
| Staphylococcus chromogenes | AF41, CIP81.59T (74.2) |
| Staphylococcus simulans | CIP81.64T (99.9) |
| Staphylococcus lentus | CIP81.63T (99.9) |
| Staphylococcus warneri | CIP103960 (99.5) |
| Staphylococcus sciuri | CIP105826 (99.9), URLGA2, URLGA13 |
| Staphylococcus equorum | CIP103502T |
| Staphylococcus cohnii | CIP81.54T (91.9) |
| Staphylococcus capitis | CIP81.53T (99.9) |
| Pseudomonas alcaligenes | AF19 (95), AF25 (95.5) |
| Pseudomonas fluorescens | AF16 (99.9), AF17 (99.9), AF23 (99.9), AF24 (99.9), CIP69.13T (99.9), CIP63.47 (98.7) |
| Pseudomonas fragi | CIP55.4T (99.9) |
| Pseudomonas putida | CIP55.5 (99.3) |
| Pseudomonas stutzeri | AF51 (99.9) |
| Pseudomonas aeruginosa | CIP100720T (99.9), CIP104060 (99.9) |
| Enterobacter cloacae | AF27 (99.9), AF98 (99.8), AF99 (96.7), CIP60.85T (99.9) |
| Enterobacter sakazakii | AF20 (95.1), AF21 (98), CIP5733 (99.9) |
| Enterobacter amnigenus | CIP103169T (88.5) |
| Enterobacter intermedius | AF92 (99.4), AF94 (99.4), AF95 (99) |
| Aeromonas hydrophila | AF28 (99.9), AF29 (99.9) |
| Aeromonas sobria | AF66 (99.2), AF71 (99.2) |
| Alcaligenes tolerans | CIP55.94 (94.2), CIP55.95 |
| Alcaligenes faecalis | CIP62.32 (92.9), CIP60.80T (92.9) |
| Klebsiella pneumoniae subsp. pneumoniae | AF65 (99.9), AF97 (99.9), CIP82.91T (99.9) CIP52.145 (99.9) |
| Klebsiella oxytoca | AF70 (99.7), AF90 (99.9), AF11 (99.9), CIP103434T (99.9), AF82 (99.9) |
| Klebsiella terrigena | AF56 (99.9), AF57 (99.9), AF77 (99.9), AF87 (99.9), AF88 (99.9), AF89 (99.9), AF91 (99.7) |
| Acinetobacter baumannii | AF12 (99), CIP70.34T (99.9) |
| Acinetobacter johnsonii | AF47 (83), AF48 (87.4) |
| Acinetobacterspecies | CIP104272 (84.9) |
| Acinetobacter lwoffii | AF63 (99.5) |
| Listeria innocua | AF1 (99.8), AF2 (99.8), AF3 (99.8) |
| Escherichia coli | AF13 (99.9), AF14 (99.9), AF18 (99.9), AF76 (99.5), AF80 (99.9) |
| Stenotrophomonas maltophilia | AF40 (99.9) |
| Buttiauxella agrestis | AF30 (99.9), AF31 (99.9), AF32 (99.9), AF33 (98.7), AF15 (99.8) |
| Citrobacter freundii | AF53 (99.9), AF37 (99.9), AF38 (99.9), AF39 (99.9), AF54 (99.9), AF55 (99.9), AF58 (99.9) |
| Serratia liquefaciens | AF61 (99.9), AF62 (99.9), AF64 (99.9), AF67 (99.9), AF68 (99.9), AF34 (99.9), AF35 (99.9), AF36 (99.9), AF81, CIP103238T (99.9), CIP60.85 (99.9) |
| Serratia marcescens | AF52 (99.9), AF59 (99.9), AF78 (99.9), AF79 (99.9), AF93 (99.9) |
| Serratia fonticola | CIP78.64T (99.9), CIP52.191 (99.9) |
| Chryseobacterium species | clone 2 (91% homology with V3 of Chryseobacterium; GenBank AF207077); CIP104270 (99) |
| Clostridium butyricum | CIP60.51 (99.3) |
| Clostridium sporogenes | CIP100651 |
| Comamonas species | AF96 (97.3) |
| Raoultella planticola | CIP81.36 (98.6), AF60 (98.6) |
| Pantoea spp. | AF72 (99.9), AF73 (99.9), AF74 (99.9) |
| Kluyvera ascorbata | AF85 (99.9), AF86 (99.9) |
| Kluyvera cryocrescens | AF83 (94.1), AF84 (94.1) |
| Microbacterium lacticum | CIP101097 |
| Geobacillus stearothermophilus | CIP67.5 |
| Moraxella bovis | CIP70.40 |
| Lactobacillus buchneri | CNRZ36R, CNRZ214 |
| Aerococcus viridans | URLGA23ag, URLGA23ap |
| Brevibacterium linens | CNRZ910, CNRZ940G, CIP101125OT, CNRZ915, CNRZ929O, CNRZ931 |
| Brevibacterium casei | CNRZ912, CIP102111T |
| Brevibacterium epidermidis | CIP102110BT |
| Brevibacterium iodenum | LMG2201BT |
| Brevibacterium species | CNRZ971, CNRZ938; CNRZ909 |
| Arthrobacter sulfureus | LMG16694T |
| Arthrobacter citreus | CNRZ928T |
| Arthrobacter nicotianae | LMG16305J2T, LMG16305BT |
| Arthrobacter protophormiae | LMG16324T |
| Arthrobacter uratoxydans | LMG16220-2T |
| Arthrobacter globiformis | CNRZ907, CNRZ908 |
| Arthrobacter spp. RAPD group I | CNRZ2057, CNRZ900, CNRZ2052 |
| Arthrobacter spp. RAPD group II | CNRZ2075, CNRZ983, CNRZ2062 |
| Corynebacterium variabile | CIP102112GT, CNRZ2076, CNRZ923b |
| Corynebacterium vitaeruminis | CIP82.7pT, CNRZ929J2T |
| Corynebacterium ammoniagenes | CIP101283T, CNRZ931T, CNRZ922G |
| Corynebacterium spp. | CNRZ2069, CNRZ921, CNRZ978, CNRZ2065 |
| Brachybacterium alimentarium | CNRZ929J4, CNRZ911J, CNRZ925T |
| Brachybacterium tyrofermentans | CNRZ926T |
| Micrococcus lilae | CNRZ882 |
| Micrococcus luteus | CNRZ881 |
| Kytococcus sedentarius | CNRZ880 |
| Kocuria rosea | AF79 (99) |
| Kocuria varians | CIP81.73 |
| Kocuria kristinae | CNRZ872 |
| Propionibacterium thoenii | ATCC4874, CNRZ924, CNRZ83, CNRZ732, ATCC4872, CNRZ724 |
| Propionibacterium freudenreichii | CNRZ81T, CNRZ82, CNRZ88, CNRZ729, CNRZ288, ATCC 13678, ATCC 9616 |
| Propionibacterium acidipropionici | CNRZ733, ATCC 965, CNRZ287, NCIMB8895, CIP3025T, ATCC 9616 |
| Propionibacterium jensenii | ATCC4870, CNRZ85R, CNRZ8728, NCIMB8904, NCIMB8069 |
Abbreviations: ATCC, American Type Culture Collection (Rockville, Md.); CIP, Collection of the Institute Pasteur (Paris, France); CNRZ, Collection of the Centre National de la Recherche Zootechnique (INRA de Jouy-en-Josas, Jouy-en-Josas, France); LMG, Collection of the Laboratorium voor Microbiologie (University of Gand, Gand, Belgium); NCIMB, National Collection of Industrial and Marine Bacteria (Aberdeen, Scotland); AF, isolated strains from raw milk (AFSSA, Maisons-Alfort, France), URLGA, isolated strains from dairy environment (INRA de Jouy-en-Josas); RAPD, randomly amplified polymorphic DNA. See Ogier et al. (39) for the LAB strains and Staphylococcus strains of dairy interest included in the low-G+C-content species database.
Bacterial species identification from raw milk isolates.
The strains isolated from different medium plates were purified and characterized by biochemical tests (Gram stain coloration, mobility, and catalase and oxidase tests) and the API system (BioMérieux, Marcy-l'Etoile, France) to identify the species level (Table 1).
PCR amplification.
TTGE samples were prepared by performing two successive PCRs with the GenAmp system, model 2400 (Perkin-Elmer, Courtaboeuf, France). First, a 700-bp fragment of the 16S rDNA including the V3 region was amplified with primers W01 and W012 as previously described (39). Second, the 700-bp fragment was used to amplify the V3 region with primers HDA1-GC and HDA2 as previously described (39). PCR mixtures and the amplification program were the same as described by Ogier et al. (39). Sizes and quantities of PCR products were determined by 2% agarose gel electrophoresis (Seakem CTG agarose; TEBU, Le Perray-en-Yvelines, France).
TTGE analysis.
PCR products obtained from V3 region amplification were submitted for TTGE analyses. TTGE was performed with the DCode universal mutation detection system (Bio-Rad, Marnes-la-Coquette, France) on 16 cm × 16 cm × 1 mm gels. Polyacrylamide gels (8%) were prepared and run with 1× TAE buffer diluted from 50× TAE buffer (2 M Tris base, 1 M glacial acetic acid, and 50 mM EDTA). Gels were prepared with 8% (wt/vol) acrylamide stock solutions (37.5:1) and a final urea concentration of 6 M. Five-microliter samples of PCR products (∼100 ng of DNA) were deposited in wells, under previously described running conditions (39). After runs, gels were stained for 15 min with an ethidium bromide solution (0.5 μg/ml of 1× TAE buffer), rinsed for 20 min in 1× TAE buffer, and photographed on a UV transillumination table.
DGGE analysis.
Denaturing gradient gel electrophoresis analysis of the V3 amplicons was performed with the Bio-Rad DCode universal mutation detection system. The denaturing gradient gel contained a 40 to 70% gradient of urea and formamide increasing in the direction of electrophoresis. A 100% denaturing solution consisted of 7 M urea and 40% (vol/vol) deionized formamide. Electrophoresis was conducted with 1× TAE buffer (92 V at 60°C for 16 h for one gel). DNA bands were visualized as for TTGE analysis.
Gel analysis and reference database setup.
TTGE and DGGE gels were standardized by including an identification ladder made up of reference species. The ladder consisted of four bacterial strains for the low-G+C-content (TTGE) conditions (39) and six bacterial strains for the high-G+C-content (DGGE) conditions (Kytococcus sedentarius CNRZ880, Arthrobacter citreus CNRZ928T, Micrococcus kristinae CNRZ872, Bacillus pumilus ATCC7725, Propionibacterium jensenii Z87). An ordered data set was generated with GelCompar software (Applied-Maths, Sint-Martens-Latem, Belgium), a data-processing tool. For this purpose, the photographed gels were converted into a file image, which was then analyzed by GelCompar. The software standardizes TTGE and DGGE profiles to minimize migration differences between gels (39). The molecular fingerprints of each bacterial species were integrated into the GelCompar database. We also used WinMelt software (Bio-Rad), which calculates the melting temperature (Tm) of PCR-amplified fragments (25) to predict their migration positions on TTGE and DGGE gels.
Commercial cheeses. (i) Identification of bacterial species in commercial cheeses by culture-dependent and culture-independent methods.
We chose four commercial cheeses that were made according to different technologies. We analyzed two raw milk cheeses: Morbier (France), a semihard cheese, and Munster (France), a red smear cheese. We also analyzed two pasteurized milk cheeses: Epoisses (France), a red smear cheese, and Leerdamer (The Netherlands), a Swiss-type cheese.
(ii) Comparison of bacterial microflora present in the core versus the cheese surface.
Analyses were performed with six different commercial cheeses (sample cheeses different than those used above): two Swiss-type cheeses, Comté (France) and Beaufort (France); two semihard cheeses, Saint Nectaire (France) and Morbier (France); and two red-smear cheeses, Epoisses (France) and Langres (France). For each type of commercial cheese, we analyzed samples from two different producers. Four samples were tested per cheese; two originated from the cheese core, and two originated from the cheese surface.
Determination of bacterial counts.
Cheese samples (each, 10 g of nonfractionated cheese) were emulsified in 100 ml of sterile 2% (wt/vol) trisodium citrate (Merck Eurolab, Fontenay-sous-Bois, France) and homogenized with an Ultra-Turrax mechanical blender at 19,000 rpm for 45 s (T-25 IKA; Labo Moderne, Paris, France) to disrupt lactococcal chains (29). Serial dilutions were prepared in sterile 1% (wt/vol) peptone (Merck Eurolab) and plated on selective agar medium with a spiral plater (Spiral System, Cincinnati, Ohio). Lactococci and streptococci on M17 agar (Difco, Elancourt, France) were counted after 48 h of incubation at 30 or 42°C (56). Lactobacilli were counted on modified MRS agar plates (Difco) (pH adjusted to 5.2) after incubation for 72 h in anaerobic conditions, either at 30°C for mesophilic lactobacilli or at 42°C for thermophilic lactobacilli (15). The Leuconostoc population was estimated on MSE agar (Difco) after 48 h of incubation at 30°C (31).
Coryneforms were isolated on brain heart infusion (BHI) medium (Difco) supplemented with 5% (wt/vol) NaCl, and plates were incubated at 25°C for 3 days, following an exposure to daylight for 4 days at room temperature to enhance pigment production (J. J. Gratadoux, URLGA, personal communication). Staphylococci were isolated on mannitol salt agar medium (Difco) supplemented with 1% (wt/vol) calcium bicarbonate, and plates were incubated at 37°C for 2 days (47). Enterococci were numerated on bile esculin azide agar medium (Difco) after incubation at 37°C for 3 days under anaerobic conditions (46). Propionibacteria were isolated on yeast extract-sodium lactate medium, and plates were incubated at 25°C for 7 days under anaerobic conditions (58). Gram-negative bacteria were isolated on violet red bile agar medium (Difco), by incubation for 2 days at 30°C (48).
Genomic DNA extraction in cheeses.
Cheese samples for Morbier, Epoisses, Leerdamer, and Munster cheeses (each sample, 5 g of nonfractionated cheese) and for Beaufort, Comté, Saint Nectaire, Morbier, Epoisses, and Langres cheeses (each sample, 3 g of surface cheese or 3 g of core cheese) were dissolved in 40 ml of sterile 2% (wt/vol) trisodium citrate and homogenized (19,000 rpm/min) with an Ultra-Turrax blender until solutions were opaque. Fifty milligrams of pronase (Boerhinger, Mannheim, Germany) and 100 μl of β-mercaptoethanol were added to each sample, followed by 3 h of incubation at 52°C. Bacteria were washed twice by centrifugation at 13,000 × g for 15 min. Pellets were first resuspended in sterile water and then in 10 ml of Tris-EDTA-saccharose (TES) buffer (25 mM Tris HCl, 0.1 M EDTA, 25% [wt/vol] saccharose [pH 8]). Cells were recentrifuged, resuspended in 500 μl of TES buffer, transferred into Eppendorf tubes, and cooled in ice for 10 min.
Cell lysis was performed by glass bead (150 to 200 μm) (Sigma, Saint Quentin Fallavier, France) treatment in the presence of TES buffer with the Fast Prep apparatus (FP120; Bio101 Savant; Ozyme, Saint-Quentin-en-Yvelines, France) (two cycles of 40 s each of shaking and 3 min of storage in ice). After settling, the supernatant (2 × 200 μl) was stored for 10 min in ice. DNA was then extracted by the phenol-chloroform method as previously described (14). The DNA pellet was dissolved in 100 μl of Tris-EDTA buffer plus RNase (Sigma, Saint Quentin Fallavier, France) and then examined by 0.8% agarose gel electrophoresis.
Specific primer tests.
Specific primers based on previously published sequences were synthesized by MWG Biotech AG (Ebersberg, Germany) and the analyses were carried out with DNA extracted directly from the sample cheeses. Primers were prepared at a final concentration of 60 μM in deionized autoclaved water. PCR was performed with the Perkin-Elmer GenAmp system, model 2400, and all reactions were carried out following previously published conditions (see Table 4). Sizes of PCR products were determined by 2% agarose gel electrophoresis (TEBU Seakem CTG agarose).
TABLE 4.
Primers used in this study for the specific PCR tests
| Primer | Sequence (5′-3′) of reverse primer | Specificity of targeta | Source or referenceb | PCR response for cheese
|
|||
|---|---|---|---|---|---|---|---|
| Morbier | Epoisses | Munster | Leerdamer | ||||
| Ec1 | Unpublished data | Ec. casseliflavus | Firmesse | − | − | − | |
| Ec2 | |||||||
| Efm1 | Unpublished data | Ec. faecium | Firmesse | − | − | − | |
| Efm2 | |||||||
| Ed1 | Unpublished data | Ec. durans | Firmesse | − | − | ||
| Ed2 | |||||||
| Eh1 | Unpublished data | Ec. hirae | Firmesse | − | − | ||
| Eh2 | |||||||
| Efs1 | Unpublished data | Ec. faecalis | Firmesse | − | − | ||
| Efs2 | |||||||
| fPs16S | ACT GAC ACT GAG GTG CGA AAG CG | Pseudomonas | 26 | + | + | ||
| rPs16S | ACC GTA TGC GCT TCT TCA CTT GAC C | ||||||
| Laci 01 | GAC CGC ATG ATC AGC TTA TA | Lb. acidophilus | 22 | + | − | − | |
| Laci 02 | AGT CTC TCA ACT CGG CTA TG | ||||||
| SLH19857S | CTA GAC AAT CAA TTG CAC CG | Lb. helveticus | 43 | − | − | − | − |
| LH29860 | TAC CAG TTC TTC TTG AAG CC | ||||||
| Ldel01 | ACA TGC ATC GCA TGA TTC AAG | Lb. delbrueckii | 22 | + | − | − | |
| Ldel02 | AAC TCG GCT ACG CAT CAT TG | ||||||
| fSt sap | TCA AAA AGT TTT CTA AAA AAT TTA C | S. saprophyticus | 28 | − | |||
| rSt sap | ACG GGC GTC CAC AAA ATC AAT AGG A | ||||||
| Lfpr16S-23S | GCCGCCTAAGGTGGGACAGAT | Lb. plantarum | 61 | + | + | ||
| PlanII | TTACCTAACGGTAAATGCGA | ||||||
| IRL | TTTGAGAGTTTGATCCTGG | Lc. raffinolactis | 44 | − | + | − | |
| PipLraR | CGTCACTGAGGGCTGGAT | ||||||
| STAE-EpI | TCTACGAAGATGAGGGATA | S. epidermidis | 21 | − | − | − | |
| STAE-EpII | TTTCCACCATATTTTGAATTGT | ||||||
| STAA-AuI | TCTTCAGAAGATGCGGAATA | S. aureus | 21 | − | − | − | |
| STAA-AuII | TAAGTCAAACGTTAACATACG | ||||||
| Lnm1 | TGTCGCATGACACAAAGTTA | Ln. mesenteroides | 9 | − | + | − | + |
| Lnm2 | ATCATTTCCTATTCTAGCTG | ||||||
| Brevib | Unpublished data | B. linens | Furlan | + | + | − | − |
| Blin | |||||||
| Cvar | Unpublished data | C. variabile | Furlan | + | − | − | − |
| Corb | |||||||
Abbreviations: Lb., Lactobacillus; Lc., Lactococcus; Ln., Leuconostoc; Ec., Enterococcus; S., Staphylococus; St., Streptococcus; B., Brevibacterium; C., Corynebacterium.
Firmesse, O. Firmesse et al., unpublished data; Furlan, S. Furlan et al., unpublished data.
Sequencing of TTGE and DGGE fragments.
Bands obtained by TTGE and DGGE analyses of commercial cheeses were excised from the denaturing gels, purified, cloned, and sequenced as previously described (39). Sequences were compared to sequences in the Ribosomal Database Project (27) to determine the closest known relative species of the V3 16S rDNA fragment.
RESULTS
The bacterial species database.
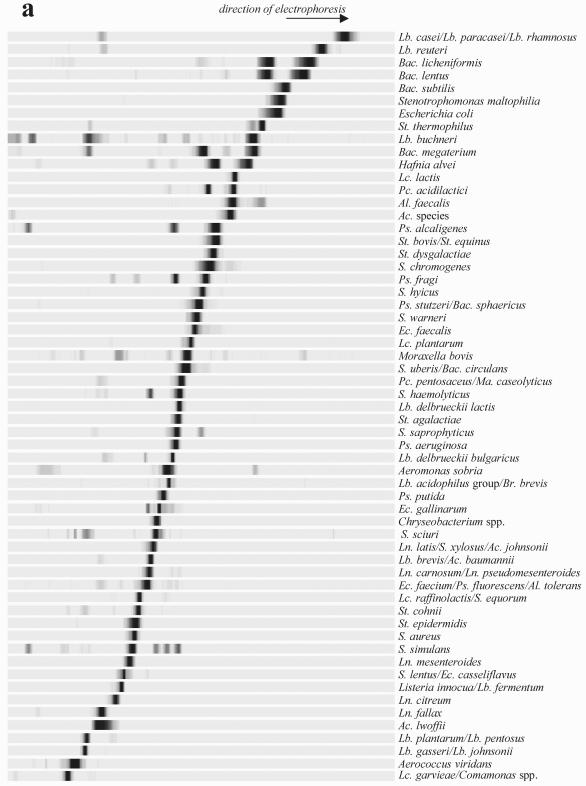
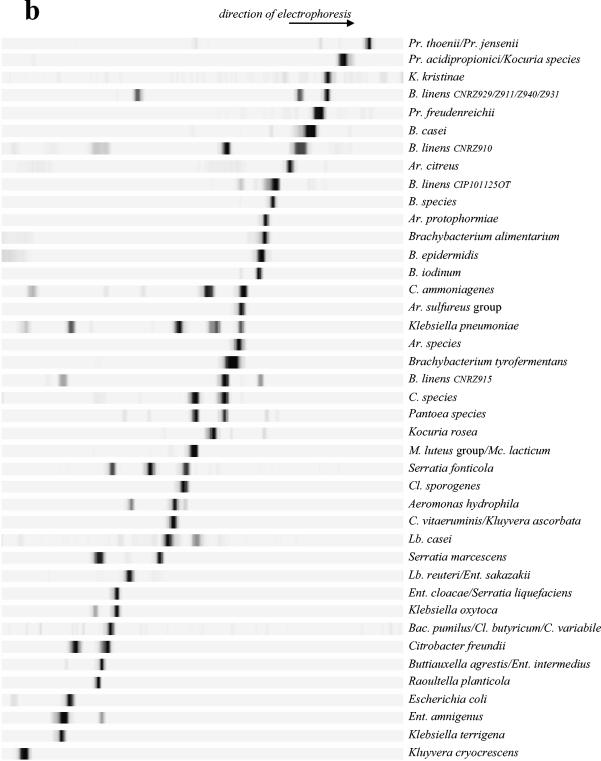
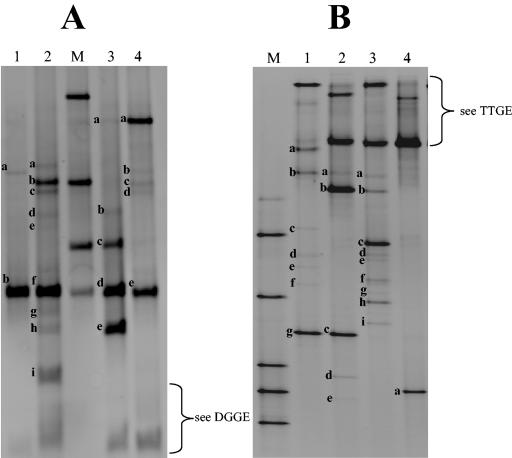
The molecular fingerprints of bacteria that correspond to common dairy species were determined by amplification of the V3 fragment with the HDA1 and HDA2 primers, followed by denaturing gel analysis of the resulting DNA products (Fig. 1). We tested different running conditions in order to determine the optimal parameters for separating the bacteria under study. For the low-G+C-content species bacteria (calculated Tm of the V3 sequence, <75°C with the Win Melt software), optimal resolution was achieved by TTGE; for the medium- and high-G+C-content species bacteria (calculated Tm of the V3 sequence, >75°C), optimal resolution was achieved by DGGE. The gel running parameters were optimized for each method (see Materials and Methods). All the strains described in Table 1 (including culture collection strains and isolates originating from microbial analyses of dairy samples) were run under TTGE or DGGE conditions to determine their position migrations. Note that some species bacteria (with calculated Tm values of the V3 sequence that were ±75°C) were run under both TTGE and DGGE conditions, i.e., Escherichia coli or Lactobacillus reuteri; their molecular fingerprints were integrated into the two-species database (Fig. 1). As a rule, bands generated by strains of the same species migrated identically (data not shown). We observed a few exceptions with P. fluorescens, Staphylococcus haemolyticus, Klebsiella oxytoca, and Brevibacterium linens species (Fig. 2). Strains of Pseudomonas fluorescens isolated from dairy products showed the same migration distance, whereas strain CIP69.13T, which originated from a water tank, and strain CIP6347, isolated from soil, migrated at a different position. Migration anomalies were also observed for species of Staphylococcus haemolyticus and Klebsiella oxytoca, which originated from different biotopes (Fig. 2). The heterogeneity of the V3 sequence within a species could be explained here by differences in ecological origins (as described for Pseudomonas fluorescens) (5, 16, 34). Only the strains originating from dairy environments are conserved in the database species. However, Brevibacterium linens strains exhibited significant variability in V3 sequence migration (Fig. 2), although all strains used were isolated from dairy products. This may be explained by genetic strain variability, as revealed by heterogeneity of the randomly amplified polymorphic DNA profiles of numerous strains of Brevibacterium linens (E. Lepage, personal communication).
FIG. 1.
Low-G+C-content (TTGE) and high-G+C-content (DGGE) species database. The V3 fragments originated from pure bacterial strains are separated by TTGE electrophoresis (a) or DGGE electrophoresis (b) according to their respective Tm values. The gels are then standardized using the GelCompar software (Applied-Maths) and transferred into the PowerPoint software. The profiles are then gathered according to their migration positions. Each species (indicated on the right of the profile) is characterized by a specific TTGE or DGGE fingerprint. The resolution is discussed in the text. Abbreviations: Ac., Acinetobacter; Al., Alcaligenes; Ar., Arthrobacter; B., Brevibacterium; Bac., Bacillus; Br., Brevibacillus; C., Corynebacterium; Cl., Clostridium; Ec., Enterococcus; Ent., Enterobacter; Lb., Lactobacillus; Lc., Lactococcus; Ln., Leuconostoc; M., Micrococcus; Ma., Macrococcus; Mc., Microbacterium; Pc., Pediococcus; Ps., Pseudomonas; Pr., Propionibacterium; S., Staphylococcus; St., Streptococcus.
FIG. 2.
Heterogeneity of the V3 fingerprints between strains belonging to the same species: Pseudomonas fluorescens (A), Staphylococcus haemolyticus (B), Klebsiella oxytoca (C), Brevibacterium linens (D). The V3 fragments originated from pure bacterial strains are separated by TTGE electrophoresis (Pseudomonas fluorescens and Staphylococcus haemolyticus) or DGGE electrophoresis (Brevibacterium linens and Klebsiella oxytoca). The gels are then standardized with GelCompar software (Applied-Maths) and transferred into the PowerPoint software. Strain numbers are indicated on the right of the profile. The numbers in parentheses correspond to biotope origins of the strains: 1, water tank; 2, soil; 3, dairy origin; 4, pharynx; 5, human skin. Strains differing by their ecologic origin generally showed a different migration behavior, as discussed in the text.
In some cases, the close phylogenetic relationships between species belonging to the same genus made it impossible to differentiate them, as described for the Lactobacillus acidophilus group or Enterococcus faecium group (39). For example, the members of the Listeria genus could not be differentiated using primers HDA1 and HDA2; the V3 sequences of Listeria monocytogenes, Listeria ivanovii, and Listeria innocua were similar. The different species of the Arthrobacter sulfureus group (Arthrobacter sulfureus, Arthrobacter nicotianae, Arthrobacter urotoxydans, and Arthrobacter globiformis) or the Micrococcus luteus group (Micrococcus lylae, Kytococcus sedentarius, Micrococcus luteus, and Microbacterium lacticum) could not be separated by DGGE electrophoresis (see Fig. 1b).
In a few cases, species belonging to different genera could not be differentiated, i.e., “Alcaligenes tolerans,” Pseudomonas fluorescens, and Enterococcus faecium (Fig. 1a) or Buttiauxella agrestis and Enterobacter intermedius (Fig. 1b). Despite sequence differences, the Tm values of comigrated fragments from these bacteria were similar (data not shown). Some strains were characterized by two bands of similar intensities (e.g., Hafnia alvei and Bacillus licheniformis) (Fig. 1a). The presence of two amplified V3 segments is probably due to the heterogeneity of the 16S rDNA operon (38).
The band position of each bacterial species was stored in a GelCompar database (Applied-Maths). Our reference database was then used to identify the bacterial species present in commercial cheeses. The potential of TTGE and DGGE for ecosystem analyses was compared to the culture-dependent analysis approach.
Identification of bacterial species.
Analyses of commercial cheeses were carried out by (i) culture-dependent and (ii) culture-independent methods.
(i) Culture-dependent analysis.
Microbial analyses of cheeses were performed by plating samples on different media to select different groups of bacteria (Table 2). For Munster cheese, plate counts revealed a high level of presumed LAB (lactococci, streptococci, and mesophilic and thermophilic lactobacilli) and salt-tolerant bacteria (as determined by growth on BHI medium). Epoisses cheese was characterized by low numbers of bacteria that could grow on MRS medium. We could not enumerate colonies on M17 plates, because they were overgrown by gram-negative bacteria that originated from the cheese.
TABLE 2.
Microbial enumeration of commercial cheese samples on different media
| Sample | Bacterial count (log10 of sample)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VRBA, 30°C (total coliforms) | M17, 30°C (Lactococcus spp.) | M17, 42°C (Streptococcus spp.) | MRS, 30°C (mesophilic Lactobacillus) | MRS, 42°C (thermophilic Lactobacillus) | BHI + 5% NaCl, 25°C (salt-tolerant flora) | MSA, 37°C (Staphylococcus spp.) | MSE, 30°C (Leuconostoc spp.) | YEL, 25°C (Propionibacterium spp.) | BEA, 37°C (Enterococcus spp.) | |
| Leerdamer | 3.6 | 7.62 | 6 | 5.5 | 4.9 | 8.9 | 7.4 | <2.2 | 7.4 | 6.1 |
| Munster | 6.5 | 9.1 | 9 | 7.2 | 7.3 | 8.4 (7.1)c | 5.8 | <2.2 | 6.1 | |
| Morbier | 6.1 | 9 | 8.9 | 8 | 7.6 | 8.9 (8)c | 7.4 | <2.2 | 6.1 | |
| Epoisses | 6.3 | —b | —b | 6 | 5 | 9.3 (6.5)c | 6.2 | <2.2 | 4.7 | |
Bacterial counts under different culture conditions (medium and temperature). VRBA, violet red bile agar; YEl, yeast extract-sodium lactate; BEA, bile esculin azide agar.
—, overgown by gram-negative bacteria.
Numbers in parentheses indicate bacterial counts of orange colonies.
Morbier cheese was characterized as having a diverse bacterial content, including presumed lactococci, streptococci, mesophilic and thermophilic lactobacilli, and salt-tolerant bacteria. The dominant bacteria of Leerdamer cheese consisted of salt-tolerant bacteria and presumed lactococci and propionibacteria. With all four cheeses, we enumerated a high-salt-tolerant bacterial population, which represents a major group of bacteria inhabiting the cheese surface. A wide variety of bacterial colonies, differing in their morphology and color, grew on BHI medium supplemented with 5% NaCl, indicative of the great biodiversity of bacteria species among the salt-tolerant microorganisms. However, we were unable to differentiate the salt-tolerant species on plates. After a few days at room temperature, orange colonies appeared and may have corresponded to Brevibacterium linens. Gram-negative bacteria were present at high levels in the different cheeses (except for the Leerdamer cheese); enumeration of colonies growing on violet red bile agar medium revealed bacterial counts higher than 106 UFC/g.
(ii) Culture-independent analysis.
To investigate microbial diversity of cheeses with molecular tools, genomic bacterial DNA was directly extracted from the cheeses, followed by amplification of the V3 regions of 16S rDNA. Separation of resulting amplicons was performed by TTGE and DGGE electrophoresis (Fig. 3). The molecular fingerprint was identified by comparing the migration position of a band to that of a species in the database or by DNA sequencing of the unknown band. Confirmation was achieved by specific PCR analysis of DNA extracted directly from the sample.
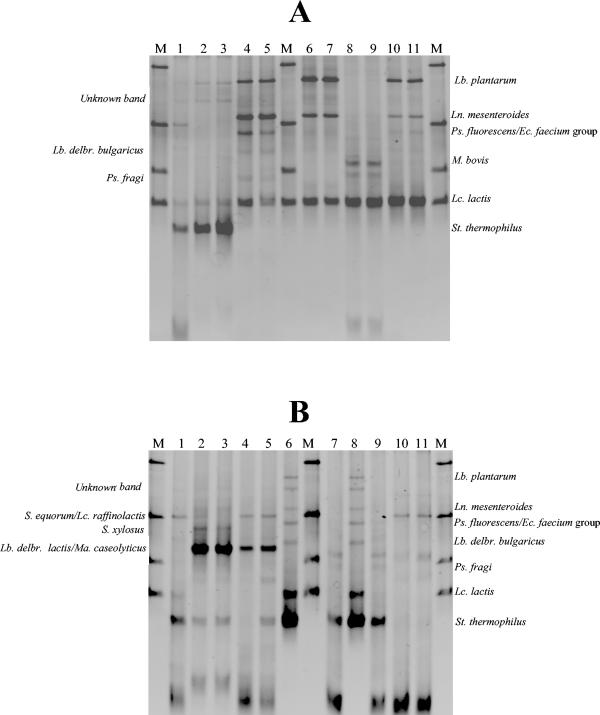
FIG. 3.
Photographed gel (ethidium bromide revelation) after TTGE (A) and DGGE (B) electrophoresis of V3 16S rDNA fragments from various commercial dairy products differing by their technology processes. Genomic DNA was extracted from nonfractionated cheese sample, followed by amplification of the V3 16S rDNA region and separation of the amplicons by gel electrophoresis. After standardization of the gel with GelCompar software, the bands were identified by comparison with the species database. We have indicated the position of the respective assigned bands by lowercase letters. See Table 5 for a list of band identities corresponding to each labeled band. (A) Lane M, standardization ladder; lane 1, Leerdamer cheese; lane 2, Morbier cheese; lane 3, Munster cheese; lane 4, Epoisses cheese. (B) Lane M, standardization ladder; lane 1, Epoisses cheese; lane 2, Munster cheese; lane 3, Morbier cheese; lane 4, Leerdamer cheese.
(a) Identification of the low-G+C-content bacteria species by TTGE (Fig. 3A).
The Morbier cheese profile includes nine bands, seven of which were assigned by using the species database (Fig. 3A, lane 2). Two bands were directly (unambiguously) assigned to Lactococcus lactis (band f) and Streptococcus thermophilus (band h). The presence of these two species was confirmed by cloning and sequencing of the two corresponding bands (Table 3). The other bands were assigned to several possible comigrating species, and we therefore performed further analyses using specific species primers to identify them (Table 4). For example, the presence of Lactobacillus delbrueckii subsp. lactis in the Morbier cheese (Fig. 3A, lane 2, band e) was confirmed by specific PCR, whereas the comigrating species Staphylococcus saprophyticus was not found (Table 4). Using combination of different specific PCR tests (Table 4), the bands a, b, and d (Fig. 3A, lane 2) were presumably identified as Staphylococcus lentus, Staphylococcus equorum, and Lactobacillus acidophilus. The unassigned band g (Fig. 3A, lane 2) was excised from the gel, cloned, sequenced, and identified as Lactobacillus buchneri (Table 3); its migration profile was confirmed by running two pure strains on TTGE, and then it was added to the reference database. We were unable to identify band c in the data shown in Fig. 3A. The strong intensity of the band corresponding to Lactococcus lactis indicates that it is the dominant species in Morbier cheese. Note that band intensities seem to reflect the relative proportion of each species in the total bacterial population (35).
TABLE 3.
Identification by cloning sequencing of V3 fragments excised from TTGE and DGGE patterns of total microbial community cheeses
| Sample cheese | Fragments (method[s]) | Clone(s) no. | Closest sequence relative (species)a | % Identity | GenBank accession no. |
|---|---|---|---|---|---|
| Morbier | f (TTGE) | 45 | Lc. lactis sup sp. lactis | 99 | AJ419572 |
| Munster | d (TTGE) | 41, 42 | Lc. lactis lactis | 99 | AF530330.1 |
| Morbier | g (TTGE) | 9, 21 | Lb. buchneri | 99 | AY026751.1 |
| Epoisses | a (DGGE) | 9, 18, 19 | Pse. agarovorans | 100 | AY100680.1 |
| Morbier | b (DGGE) and h (TTGE) | 2, 10, 19 | Uncultured St. | 100 | AF408263.1 |
| Munster | b (DGGE) and e (TTGE) | 47 | St. thermophilus | 99 | AY188354 |
| Morbier | c (DGGE) | 24 | C. variabile | 100 | AJ222816.1 |
| Morbier | e (DGGE) | 19 | Ar. species | 91 | AB017540.1 |
| Epoisses | f (DGGE) | 7 | H. variabilis | 96 | AF173968 |
| Morbier | g (DGGE) | 22 | C. casei | 94 | AF267152.1 |
| Morbier | h (DGGE) | 29 | C. mastitidis | 97 | Y09806.1 |
| Epoisses | g (DGGE) | 22, 25 | Ar. species | 100 | AF487785.1 |
| Ar. woluwensis | 100 | AY112986.1 | |||
| Ar. sulfonivorans | 100 | AF235091 | |||
| Ar. nicotiane | 100 | AJ315492.1 | |||
| Ar. globiformis | 100 | M23411.1 | |||
| Ar. sulfureus | 100 | AB046358.1 |
Abbreviations: Lc., Lactococcus; Lb., Lactobacillus; Pse., Pseudoalteromonas; C., Corynebacterium; Ar., Arthrobacter; H., Halomonas.
The Munster cheese (Fig. 3A, lane 3) sample was characterized by three intense bands, identified as Enterococcus faecalis (band c), Lactococcus lactis (band d), and Streptococcus thermophilus (band e), and two bands of lower intensity, respectively, assigned to Lactobacillus plantarum (band a) and Lactobacillus acidophilus group (band b). We confirmed the assignations to Lactococcus lactis and Streptococcus thermophilus by cloning sequences of the corresponding bands (Table 3). With specific primer tests, Lactobacillus plantarum was found, but the assignation of band c shown in Fig. 3A to Enterococcus faecalis was not confirmed (Table 4).
The Epoisses cheese profile (Fig. 3A, lane 4) includes two intense bands, a and e (Lactobacillus plantarum and Lactococcus lactis), and three bands, b, c, and d, of lower intensity (Leuconostoc mesenteroides, Lactococcus raffinolactis-Staphylococcus equorum, and Pseudomonas fluorescens-Enterococcus faecium group-“Alcaligenes tolerans”). The presence of Lactobacillus plantarum, Leuconostoc mesenteroides, Lactococcus raffinolactis, and Pseudomonas species in the Epoisses cheese was confirmed with specific primers; members of the Enterococcus faecium group were not found (Table 4).
In pasteurized Leerdamer cheese (Fig. 3A, lane 1), the TTGE analysis revealed the presence of only two species: Leuconostoc mesenteroides (band a), confirmed using specific primer tests (Table 4), and Lactococcus lactis (band b).
TTGE band identities for each sample cheese are listed in Table 5.
TABLE 5.
Approaches used to identify bands of TTGE and DGGE gelsa
| Cheese | Band and presumed species in gel
|
Band and identification by sequencing
|
Band and species revealed by specific PCR
|
|||
|---|---|---|---|---|---|---|
| TGGE | DGGE | TGGE | DGGE | TGGE | DGGE | |
| Morbier | a, Ec. casseliflavus-S. lentus; b, Lc. raffinolactis-S. equorum; c, unknown band; d, Lb. acidophilus group; e, Lb. delb. subsp. lactis-S. saprophyticus; f, Lc. lactis; g, Lb. buchneri; h, St. thermophilus; i, Escherichia coli | a, Lb. buchneri; b, St. thermophilus; c, C. variabile; d, Lb. reuteri-Enterobacter sakazakii; e, Ar. species; f, Lb. casei; g, C. casei; h, M. luteus group-Corynebacterium sp.; i, B. linens | f, Lc. lactis; g, Lb. buchneri; h, St. thermophilus | b, Uncultured, St.; c, C. variabile; e, Ar. species; g, C. casei; h, Corynebacterium mastidis | d, Lb. acidophilus; e, Lb. delb. subsp. lactis | c, C. variabile; i, B. linens |
| Munster | a, Lb. plantarum-Lb. pentosus; b, Lb. acidophilus group; c, Ec. faecalis; d, Lc. lactis; e, St. thermophilus | a, Kluyvera cryocrescens; b, St. thermophilus; c, Ar. sulfureus group; d, B. linens; e, B. linens | d, Lc. Lactis; e, St. thermophilus | b, St. thermophilus; c, Ar. sulfureus | a, Lb. plantarum; b, Lb. acidophilus | |
| Epoisses | a, Lb. plantarum-Lb. pentosus; b, Ln. mesenteroides; c, Lc. raffinolactis-S. equorum; d, Ec. faecium group-Ps. fluorescens-Al. tolerans; e, Lc. lactis | a, Pseudoalteromonas agarivorans; b, Kluyvera cryocrescens; c, Buttiauxella agrestis-Enterobacter intermedius; d, B. linens; e, unknown band; f, Halomonas variabilis; g, Ar. sulfureus group | a, Pseudoalteromonas agarivorans; f, Halomonas variabilis; g, Ar. sulfureus | a, Lb. plantarum; b, Ln. mesenteroides; c, Lc. raffinolactis; d, Ps. genus | d, B. linens | |
| Leerdamer | a, Ln. mesenteroides; b, Lc. lactis | a, Propionibacterium freudenreichii | a, Ln. mesenteroides | |||
Presumed bacterial species were assigned with the reference database. Bands are shown in Fig. 3; see the legend to Fig. 3 for details of analyses and lanes. Bands are identified by lowercase letters. Abbreviations: Al., Alcaligenes; Ar., Arthrobacter; B., Brevibacterium; C., Corynebacterium; Ec., Enterococcus; Lb., Lactobacillus; Lc., Lactococcus; Ln., Leuconostoc; M., Micrococcus; Ps., Pseudomonas; S., Staphylococcus; St., Streptococcus; delb., delbrueckii.
(b) Identification of the medium- and high-G+C-content % bacteria by DGGE (Fig. 3B).
The DGGE pattern of Leerdamer (Fig. 3B, lane 4) cheese was very simple and was confined to one main band (Propionibacterium freudenreichii). The presence of Propionibacterium freudenreichii in the Leerdamer cheese is consistent with its process technology, as propionibacteria are added to pasteurized milk.
With the Munster cheese (Fig. 3B, lane 2), we detected five bands; three bands, a, b, and c, were respectively assigned to Kluyvera cryocrescens, Streptococcus thermophilus, and the Arthrobacter sulfureus group; two bands, d and e, were assigned to Brevibacterium linens. However, we did not confirm the presence of Brevibacterium linens in the Munster cheese with specific primers designed by S. Furlan et al. (unpublished data) (Table 4).
With Morbier cheese, the DGGE method revealed 9 bands. The species database allowed us to identify six of them (Fig. 3B, lane 3) as corresponding to Lactobacillus buchneri (band a), Streptococcus thermophilus (band b), Lactobacillus reuteri-Enterobacter sakazakii (band d), Lactobacillus casei (band f), the Micrococcus luteus group (band h), and Brevibacterium linens (band i). The presence of Brevibacterium linens in the Morbier cheese was confirmed by specific PCR testing (Table 4). However, a very faint band (not visible on the gel photograph) was assigned to E. coli. Three of the unassigned bands were cloned and sequenced; sequence comparison against the GenBank database allowed us to identify them as Corynebacterium variabile (band c), Arthrobacter species (band e), and Corynebacterium casei (band g). We also identified the Corynebacterium mastitidis species bacteria by sequencing a clone originating from band h (Fig. 3B, lane 3). Each new species fingerprint was added to the database after validation with pure strains.
We detected seven bands in the DGGE pattern of the Epoisses cheese (Fig. 3B, lane 1); four bands were assigned to Kluyvera cryocrescens (band b), Buttiauxella agrestis-Enterobacter intermedius (band c), Brevibacterium linens (band d), and the Arthrobacter sulfureus group (band g). The presence of Brevibacterium linens was confirmed by a specific PCR test (Table 4). Sequencing of the unassigned bands a and f (Fig. 3B, lane 1) revealed the presence of Pseudoalteromonas and Halomonas, although the habitat of these two genera is generally salt water (57). We did not succeed in sequencing the unknown band e (Fig. 3B, lane 1). We were also unable to determine the precise identity of band c (Fig. 3B, lane 1) corresponding to comigrating bacterial species Buttiauxella agrestis and Enterobacter intermedius. Identification of some bands corresponding to comigrating species awaits the use of specific primers. DGGE band identities for sample cheeses are listed in Table 5.
We noted that the intensity of bands corresponding to Brevibacterium linens was very low in Epoisses, and its presence was not confirmed in Munster cheeses. This was unexpected, as these cheeses belong to the red smear cheese type, in which Brevibacterium linens is a common colonizer of the cheese surface and produces red pigment.
Comparison of the bacterial microflora between surface and core cheese samples.
We used (i) TTGE and (ii) DGGE to separately analyze core or surface fractions of six commercial cheeses, some of which originate from two different commercial producers. Two samplings of each cheese fraction were analyzed.
(i) TTGE analysis.
We performed TTGE analysis of core and surface samples of Langres, Epoisses, Morbier, Beaufort, Comté, and Saint Nectaire cheeses, each coming from one producer (Fig. 4). Two samples of each cheese fraction were analyzed (Fig. 4). TTGE profiles of core Beaufort cheeses (data not shown) are nearly identical to those of core Comté cheeses. No differences were seen in TTGE patterns between samples originating from the same part of the cheese. However, TTGE patterns revealed differences in bacterial composition between the cheese core and cheese surface. The cheese core was generally inhabited by various LAB (Lactococcus lactis, Lactobacillus plantarum, Leuconostoc mesenteroides, and Streptococcus thermophilus), whereas the cheese surface was generally inhabited by staphylococci and gram-negative bacteria (Moraxella bovis and Pseudomonas sp.).
FIG. 4.
Comparison of bacterial microflora present in surface versus core cheeses samples by TTGE electrophoresis of V3 amplified fragments. For each sample cheese, genomic DNA extraction was performed either with two surface cheese samples (indicated by S1 or S2) or with two core cheese samples (indicated by C1 or C2), followed by amplification of the V3 16S rDNA region and separation of the amplicons by TTGE. The bands are compared with the reference strain fingerprints of the data bank; assignments of major bands are indicated on the right. Analysis of the profile is discussed in the text. (A) Lanes: M, identification ladder; 1, Morbier cheese S1; 2, Morbier cheese C1; 3, Morbier cheese C2; 4, Langres cheese S1; 5, Langres cheese S2; 6, Langres cheese C1; 7, Langres cheese C2; 8, Epoisses cheese S1; 9, Epoisses cheese S2; 10, Epoisses cheese C1; 11, Epoisses cheese C2. (B) Lanes: M, identification ladder; 1, Morbier cheese S2; 2, Comté cheese C1; 3, Comté cheese C2; 4, Comté cheese S1; 5, Comté cheese S2; 6, Saint Nectaire cheese C1; 7, Saint Nectaire cheese S1; 8, Saint Nectaire cheese C2; 9, Saint Nectaire cheese S2; 10, Beaufort cheese S1; 11, Beaufort cheese S2. Abbreviations: Ec., Enterococcus; Lb., Lactobacillus; Lc., Lactococcus; Ln., Leuconostoc; Ma., Macrococcus; Ps., Pseudomonas; S., Staphylococcus; St., Streptococcus.
We observed homogeneity in bacterial composition and cheese technology. For example, we identified Lactococcus lactis, Leuconostoc mesenteroides, and Lactobacillus plantarum as major components of the red smear cheeses (Langres and Epoisses cheese cores) or Streptococcus thermophilus and Lactobacillus delbrueckii subsp. lactis in Swiss-type cheeses (Comté and Beaufort cheese cores). The LAB species found in the cheese cores probably originated from the starters added by manufacturers.
The differences between bacterial compositions of the cheese core and cheese surface coincide with difference of pH between these fractions (Table 6). The increase in pH of the cheese surface allowed the growth of nonstarter bacterial species, such as staphylococci or gram-negative bacteria.
TABLE 6.
pH of core and surface commercial cheeses samples
| Sample site | pH
|
|||||
|---|---|---|---|---|---|---|
| Epoisses | Langres | Morbier | Saint Nectaire | Comté | Beaufort | |
| Core | 6.21 | 4.96 | 5.28 | 6.21 | 5.88 | 5.90 |
| Surface | 6.61 | 6.18 | 6.83 | 7.49 | 7.06 | 6.91 |
(ii) DGGE analysis.
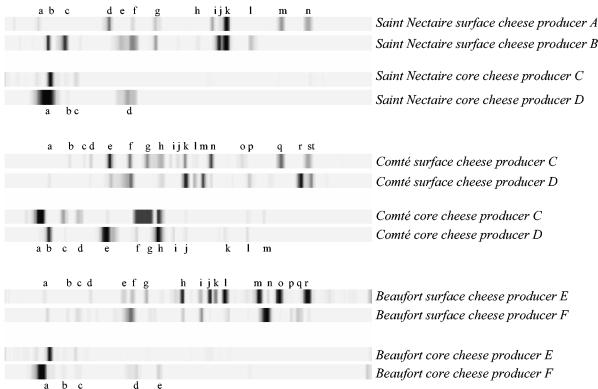
We performed DGGE analysis of core and surface samples of Beaufort, Comté and Saint Nectaire, each coming from two different producers (Fig. 5). As with TTGE, the DGGE patterns of two samples originating from the same part of the cheese were identical (data not shown). Cheeses of the same type but from different producers displayed similarities in bacterial composition, as seen by numerous common bands in DGGE profiles. In view of the complexity of the surface profiles, it appears that bacteria of the cheese surface are generally far more diversified than those in the core. Indeed, the total number of bands is significantly higher in the surface than in core samples. As described above, more-permissive surface conditions (due to higher pH levels) may favor the outgrowth of a greater variety of bacteria. Presumed identification of some of the bands (Table 7) suggests that many of the surface bacteria belong to the coryneform group. Several DGGE bands were unassigned and may correspond to undescribed bacterial species (numerous coryneform species remain poorly described in the literature). Thus, full characterization of the cheese surface awaits completion of the reference database.
FIG. 5.
Comparison of bacterial microflora present in surface cheese samples versus core cheeses samples by DGGE of V3 amplified fragments. For each sample cheese, genomic DNA extraction was performed either from surface cheese samples or from core cheese samples, followed by amplification of the V3 16S rDNA region and separation of the amplicons by DGGE. Analyses were performed with sample cheeses originating from different producers (A to F). After standardization of band migrations was carried out with GelCompar software, bands (indicated by lowercase letters) were identified by comparison with known species in the reference database. Profile analyses are discussed in the text; Table 7 recapitulates the presumed species bacteria corresponding to each labeled band.
TABLE 7.
Presumed species bacteria present in core and surface commercial cheeses by assignations of the DGGE bands to the database species
| Cheese | Sample site | Band and presumed bacterial speciesa |
|---|---|---|
| Saint Nectaire | Core | a, St. thermophilus; b, E. coli; c, unknown band; d, B. linens |
| Surface | a and b, St. thermophilus; c, E. coli; d, Ent. cloacae-Kl. oxytoca; e, Ent. sakazakii; f, unknown band; g, Lb. casei; h, K. rosea; i, B. linens; j, Ar. species; k, Ar. sulfureus group; l, B. species; m, B. linens; n, unknown band | |
| Comté | Core | a and b, St. thermophilus; c, E. coli; d, unknown band; e, Ent. cloacae-Kl. oxytoca; f, S. fonticola; g, S. marscesens; h, Lb. casei; i, Cl. sporogenes; j, Br. tyrofermentans; k, Ar. sulfureus group; l, Ar. protophormiae; m, unknown band |
| Surface | a, St. thermophilus; b, E. coli; c, unknown band; d, Kl. oxytoca; e, Ent. cloacae-Kl. oxytoca; f, B. linens; g, S. marscesens; h, C. vitaerumints; i, unknown band; j, M. luteus group-Mc. lacticum; k, Br. tyrofermentans; l, unknown band; m, unknown band; n, B. linens; o, Br. alimentarium-B. species; p, unknown band; q, B. linens; r, K. kristinae; s, unknown band; t, unknown band | |
| Beaufort | Core | a, St. thermophilus; b, E. coli; c, unknown band; d, S. fonticola; e, Lb. casei |
| Surface | a, St. thermophilus; b, E. coli; c, unknown band; d, Kl. oxytoca; e, Ent. sakazakii; f, Ar. species-B. linens; g, S. marcescens; h, M. luteus group-Mc. lacticum; i, K. rosea; j, C. species; k, unknown band; l, Ar. sulfureus group; m, B. linens; n, Ar. citreus; o, B. linens; p, B. casei; q, K. kristinae; r, unknown band |
Abbreviations: Ar., Arthrobacter; B., Brevibacterium; Br., Brachybacterium; C., Corynebacterium; Cl., Clostridium; E., Escherichia, Ent., Enterobacter; K., Kocuria; Kl., Klebsiella; Lb., Lactobacillus; M., Micrococcus; Mc., Microbacterium; S., Serratia, St.,Streptococcus.
These studies revealed an interesting difference between surface and core profiles: the essentially anaerobic core samples are dominated by low-G+C-content bacterial species but are devoid of high-G+C-content bacterial species (Fig. 5). In contrast, aerobic surface cheese samples produced complex DGGE patterns, revealing the presence of a great diversity of bacteria with a high-G+C-content genome (more than 10 bands per sample). These results indicate a strong bias in G+C content between the bacterial flora present at the surface versus that present in the core. These observations are in agreement with the study of Naya et al. (37), suggesting that aerobiosis selects for bacteria having high G+C content in their genomes.
DISCUSSION
We combined TTGE and DGGE methods to provide a thorough description of bacterial diversity in dairy products. This project was realized by first setting up a comprehensive bacterial reference database, which now comprises more than 150 species. Identification of species in dairy samples is identified by alignment with one of the species in the database. When a new band cannot be assigned through the reference set, it is directly excised and sequenced. The new species assignment is then used to enrich the reference set. By setting up this system, we have a rapid means of describing the flora in a variety of dairy ecosystems. This strategy is much less time consuming and more specific than conventional analysis by cloning and sequencing. In addition, ambiguities in species assignments are readily resolved by either (i) cloning and sequencing the V3 bands or (ii) modifying the PCR primers to narrow down the species assignments. The latter approach was also used to validate some of the species assignments made using the reference database.
The potential of the molecular approach to describe bacterial composition of different cheeses was compared to the culture-dependent method. Results obtained by the molecular approach reflected, to a large extent, those obtained by microbial enumerations on different media, but the molecular approach was advantageous in its rapidity and specificity. Indeed, some media are not very selective (2, 20, 45). For example, Ercolini et al. (20) analyzed cells harvested from a variety of viable count media and identified staphylococci on M17 and MRS agar, enterococci on mannitol salt agar, and Leuconostoc on M17 agar plates. The molecular approach was thus more informative and generally allowed bacterial identification at the species level within 2 days.
In molecular studies of microbial communities, DNA extraction is the most critical step. Isolated DNA should reflect the existing genetic diversity, but microorganisms, particularly the gram-positive bacteria, may not all lyse equally well. An effective and reproducible method for DNA isolation from dairy products was developed in this study (Materials and Methods) to assure representative extraction of microbial members of cheese ecosystems. Using TTGE and DGGE, we identified about 17 different bacterial species in Morbier cheeses manufactured with raw milk but only 3 bacterial species in Leerdamer cheese manufactured with pasteurized milk. This microbial diversity reflected the dominant bacterial species of the ecosystem (39).
The molecular approach allowed us to identify some species that are of particular interest: Lactobacillus buchneri, present in Morbier cheese, is a heterofermentative lactobacillus responsible for off-flavor defects caused by biogenic amine production (24, 54, 55). Specific detection of this microorganism could be of interest to the dairy industry. We also note that Propionibacterium freudenreichii was rapidly identified in Leerdamer cheese by DGGE; this microorganism is particularly difficult to cultivate on artificial medium and requires an incubation time of at least 7 days. Unexpectedly, we identified Pseudoalteromonas species and Halomonas species by sequencing two unassigned bands found in the Epoisses cheese profiles. This is the first time that these two salt-tolerant bacterial species are found in dairy products; their identification was enabled by molecular methods. The presence of these bacteria may be explained by surface cheese treatments performed during the cheese ripening process that includes washes in saline water. It will be of interest to know whether these unexpected bacterial species play a significant role in the cheese ripening process, or in aroma formation.
The combination approach using TTGE and DGGE has proved valuable in describing bacterial diversity at the cheese surface. The microbial communities present on Morbier, Epoisses, and Munster cheese surfaces were largely undefined (59). Most of the bacteria were described as coryneform, but species level classification to date has proved unsuccessful. Identification of coryneforms has been mainly based on determination of the types of peptidoglycan, menaquinones, and fatty acids in the cell wall (6). Some of these analyses are labor intensive, which means that only small numbers of isolates can be analyzed. Using the DGGE method, we identified numerous coryneform species in cheese samples, including Corynebacterium variabile, Corynebacterium mastitidis, Corynebacterium casei, Arthrobacter species, and Brevibacterium linens. Unexpectedly, Brevibacterium linens was not the major bacterial species in Epoisses and Munster cheeses, although it is often inoculated onto the surface of these cheeses during the early ripening process. Our results are in agreement with those of Brennan et al. (6), who also reported that Brevibacterium linens is not recovered from the surface of cheeses that were deliberately smeared with Brevibacterium linens BL2. Moreover, the taxonomy of the Brevibacterium linens group is still unclear; using genotypic methods, E. Lepage et al. (personal communication) reclassified numerous strains of Brevibacterium linens as members of the Arthrobacter genus. In fact, coryneform strains were often classified as part of the Brevibacterium linens group because they formed orange colonies on plates.
Information concerning the natural bacterial species composition of cheese ecosystems, as described in this work, should enable industrial dairy producers to design ripening cultures by selecting appropriate species. For example, this approach would provide a positive alternative to the use of traditional methods of “old-young smearing” which is considered the source of undesirable microorganisms like Listeria monocytogenes or Staphylococcus aureus (59). Until now, attempts to design a well-defined red smear culture failed; the resulting cheeses were of low quality, due to the lack of knowledge concerning the natural composition of red smear (59).
Analyses of the cheese core and surface as separate fractions revealed two more fundamental characteristics of the cheese ecosystems. First, microbial composition of a cheese is nonuniform, and the diversity of surface microflora appears to be richer than that of the core microflora. This is in agreement with the more-selective conditions occurring in the core of the cheese where the pH is lower than on the surface. Ercolini et al. (20) also observed a spatial distribution of bacterial species in Stilton cheese, based on metabolite availability and competition between bacteria. Second, we observed that bacteria of the cheese surface generally correspond to high-G+C-content genomes, whereas bacteria present in the core generally have low-G+C-content genomes. These results are consistent with a recent study (37), in which a correlation was made between bacteria having a high-G+C-content genome and the capacity to live under aerobic conditions.
In this study, TTGE and DGGE have proven to be powerful methods to describe the bacterial diversity of various cheeses differing in their technology and microbial composition. Improved knowledge about the composition and location of different bacterial species present in cheese may prove valuable in controlling development of specific microflora during the ripening process.
Acknowledgments
We are very grateful to Alexandra Gruss for reading the manuscript and for helpful discussions. We especially thank E. Lepage for technical advice.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ampe, F., N. ben Omar, C. Moizan, C. Wacher, and J. P. Guyot. 1999. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65:5464-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araujo, V. S., V. A. Pagliares, M. L. Queiroz, and A. C. Freitas-Almeida. 2002. Occurrence of Staphylococcus and enteropathogens in soft cheese commercialized in the city of Rio de Janeiro, Brazil. J. Appl. Microbiol. 92:1172-1177. [DOI] [PubMed] [Google Scholar]
- 4.Boivin-Jahns, V., A. Bianchi, R. Ruimy, J. Garcin, S. Daumas, and R. Christen. 1995. Comparison of phenotypical and molecular methods for the identification of bacterial strains isolated from a deep subsurface environment. Appl. Environ. Microbiol. 61:3400-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bossis, E., P. Lemanceau, X. Latour, and L. Gardan. 2000. The taxonomy of Pseudomonas fluorescens and Pseudomonas putida: current status and need for revision. Agronomie 20:51-63. [Google Scholar]
- 6.Brennan, N. M., A. C. Ward, T. P. Beresford, P. F. Fox, M. Goodfellow, and T. M. Cogan. 2002. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl. Environ. Microbiol. 68:820-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celestino, E. L., M. Iyer, and H. Roginski. 1996. The effects of refrigerated storage on the quality of raw milk. Aust. J. Dairy Technol. 51:59-63. [Google Scholar]
- 8.Champagne, C. P., R. R. Laing, D. Roy, A. A. Mafu, and M. W. Griffiths. 1994. Psychrotrophs in dairy products: their effects and their control. Crit. Rev. Food Sci. Nutr. 34:21-30. [DOI] [PubMed] [Google Scholar]
- 9.Cibik, R., E. Lepage, and P. Talliez. 2000. Molecular diversity of Leuconostoc mesenteroides and Leuconostoc citreum isolated from traditional French cheeses as revealed by RAPD fingerprinting, 16S rDNA sequencing and 16S rDNA fragment amplification. Syst. Appl. Microbiol. 23:267-278. [DOI] [PubMed] [Google Scholar]
- 10.Coia, J. E., Y. Johnston, N. J. Steers, and M. F. Hanson. 2001. A survey of the prevalence of Escherichia coli O157 in raw meats, raw cow's milk and raw-milk cheeses in south-east Scotland. Int. J. Food Microbiol. 66:63-69. [DOI] [PubMed] [Google Scholar]
- 11.Corroler, D., I. Mangin, N. Desmasures, and M. Gueguen. 1998. An ecological study of lactococci isolated from raw milk in the Camembert cheese Registered Designation of Origin area. Appl. Environ. Microbiol. 64:4729-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Buyser, M. L., B. Dufour, M. Maire, and V. Lafarge. 2001. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol. 67:1-17. [DOI] [PubMed] [Google Scholar]
- 13.Delbès, C., M. Leclerc, E. Zumstein, J. J. Godon, and R. Moletta. 2001. A molecular method to study population and activity dynamics in anaerobic digestors. Water Sci. Technol. 43:51-57. [PubMed] [Google Scholar]
- 14.de los Reyes-Gavilan, C., G. K. Y. Limsowtin, P. Tailliez, L. Séchaud, and J. P. Accolas. 1992. A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl. Environ. Microbiol. 58:3429-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Man, J. D., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 16.Dogan, B., and K. J. Boor. 2003. Genetic diversity and spoilage potentials among Pseudomonas spp. isolated from fluid milk products and dairy processing plants. Appl. Environ. Microbiol. 69:130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duthoit, F., J. J. Godon, and M. C. Montel. 2003. Bacterial community dynamics during production of registered designation of origin Salers cheese as evaluated by 16S rRNA gene single-strand conformation polymorphism analysis. Appl. Environ. Microbiol. 69:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eliskases-Lechner, F., and W. Guizinger. 1995. The bacterial flora of surface-ripened cheeses with special regard to coryneforms. Lait 75:571-584. [Google Scholar]
- 19.Ercolini, D., G. Moschetti, G. Blaiotta, and S. Coppola. 2001. Behavior of variable V3 region from 16s rDNA of lactic acid bacteria in denaturing gradient gel electrophoresis. Curr. Microbiol. 42:199-202. [DOI] [PubMed] [Google Scholar]
- 20.Ercolini, D., P. J. Hill, and C. E. R. Dodd. 2003. Bacterial community structure and location in Stilton cheese. Appl. Environ. Microbiol. 69:3540-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsman, P., A. Tilsala-Timisjarvi, and T. Alatossava. 1997. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S-23S rRNA spacer regions. Microbiology 143:3491-3500. [DOI] [PubMed] [Google Scholar]
- 22.Furet, J. P., P. Quénée, and P. Talliez. 2002. Identification des bactéries lactiques par PCR quantitative. Sci. Aliments 2233-2244.
- 23.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joosten, H. M. L. J., and M. D. Northolt. 1989. Detection, growth, and amine-producing capacity of lactobacilli in cheese. Appl. Environ. Microbiol. 55:2356-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lerman, L. S., and K. Silverstein. 1987. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 155:482-501. [DOI] [PubMed] [Google Scholar]
- 26.Locatelli, L., S. Tarnawski, J. Hamelin, P. Rossi, M. Aragno, and N. Fromin. 2002. Specific PCR amplification for the genus Pseudomonas targeting the 3′ half of 16S rDNA and the whole 16S-23S rDNA spacer. Syst. Appl. Microbiol. 25:220-227. [DOI] [PubMed] [Google Scholar]
- 27.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martineau, F., F. J. Picard, L. Grenier, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2000. Multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci isolated from patients infected after cardiac surgery. The ESPRIT Trial. J. Antimicrob. Chemother. 46:527-534. [DOI] [PubMed] [Google Scholar]
- 29.Martley, F. G., and R. C. Lawrence. 1972. Cheddar cheese flavour. II. Characteristics of single strain starters associated with good or poor flavour development. N. Z. J. Dairy Sci. Technol. 7:38-44. [Google Scholar]
- 30.Martley, F. G., and V. L. Crow. 1993. Interactions between non-starter microorganisms during cheese manufacture and ripening. Int. Dairy J. 3:461-483. [Google Scholar]
- 31.Mayeux, J. V., W. E. Sandine, and P. R. Elliker. 1962. A selective medium for detecting Leuconostoc in mixed-strain starter cultures. J. Dairy Sci. 45:655. [Google Scholar]
- 32.McSweeney, P. L. H., P. F. Fox, J. A. Lucey, K. N. Jordan, and T. M. Cogan. 1993. Contribution of the indigenous microflora to the maturation of Cheddar cheese. Int. Dairy J. 3:613-634. [Google Scholar]
- 33.Meyer-Broseta, S., A. Diot, S. Bastian, J. Riviere, and O. Cerf. 2003. Impact of preheating on the behavior of Listeria monocytogenes in a broth that mimics Camembert cheese composition. Int. J. Food Microbiol. 80:1-15.12430767 [Google Scholar]
- 34.Moore, E. R. B., M. Mau, A. Arnscheidt, E. C. Böttger, R. A. Hutson, M. D. Collins, Y. Van de Peer, R. De Wachter, and K. N. Timmis. 1996. The determination and comparison of the 16S rRNA gene sequence of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst. Appl. Microbiol. 19:478-492. [Google Scholar]
- 35.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 37.Naya, H., H. Romero, A. Zavala, B. Alvarez, and H. Musto. 2002. Aerobiosis increases the genomic guanine plus cytosine content (GC%) in prokaryotes. J. Mol. Evol. 55:260-264. [DOI] [PubMed] [Google Scholar]
- 38.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogier, J. C., O. Son, A. Gruss, P. Tailliez, and A. Delacroix-Buchet. 2002. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 68:3691-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pace, N. R., E. A. Angert, E. F. DeLong, T. M. Schmidt, and G. S. Wickham. 1993. New perspective on the natural microbial world, p. 77-83. In R. H. Baltz, G. D. Hegeman, and P. L. Skatrud (ed.), Industrial microorganisms: basic and applied molecular genetics. American Society for Microbiology, Washington, D.C.
- 41.Pak, S. I., U. Spahr, T. Jemmi, and M. D. Salman. 2002. Risk factors for L. monocytogenes contamination of dairy products in Switzerland, 1990-1999. Prev. Vet. Med. 53:55-65. [DOI] [PubMed] [Google Scholar]
- 42.Peterson, S. D., and R. T. Marshall. 1990. Non-starter lactobacilli in Cheddar cheese. A review. J. Dairy Sci. 73:1395-1410. [Google Scholar]
- 43.Pilloud, N., and B. Mollet. 1990. DNA probes for the detection of Lactobacillus helveticus. Syst. Appl. Microbiol. 13:345-349. [Google Scholar]
- 44.Pu, Z. Y., M. Dobos, G. K. Limsowtin, and I. B. Powell. 2002. Integrated polymerase chain reaction-based procedures for the detection and identification of species and subspecies of the gram-positive bacterial genus Lactococcus. J. Appl. Microbiol. 93:353-361. [DOI] [PubMed] [Google Scholar]
- 45.Randazzo, C. L., S. Torriani, A. D. Akkermans, W. M. de Vos, and E. E. Vaughan. 2002. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reuter, G. 1992. Culture media for enterococci and group D-streptococci. Int. J. Food Microbiol. 17:101-111. [DOI] [PubMed] [Google Scholar]
- 47.Richard, J. 1984. Evolution de la flore microbienne à la surface des camemberts fabriqués avec du lait cru. Lait 64:496-520. [Google Scholar]
- 48.Richard, J., and H. Zadi. 1983. Inventaire de la flore dominante des camemberts fabriqués avec du lait cru. Lait 63:25-42. [Google Scholar]
- 49.Rudol, M., and S. Scherer. 2001. High incidence of Listeria monocytogenes in European red smear cheese. Int. J. Food Microbiol. 63:91-98. [DOI] [PubMed] [Google Scholar]
- 50.Sørheim, R., V. L. Torsvik, and J. Goksøyr. 1989. Phenotypical divergences between populations of soil bacteria isolated on different media. Microb. Ecol. 17:181-192. [DOI] [PubMed] [Google Scholar]
- 51.Stepaniak, L., L. Jedrychowski, J. Grabska, B. Wroblewska, and F. Sorhang. 1998. Application of immunoassays for protein and peptides in milk and dairy products. Recent Res. Dev. Agri. Food Chem. 2:673-687. [Google Scholar]
- 52.Stevenson, R. G., M. T. Rowe, G. B. Wisdom, and D. Kilpatrick. 2003. Growth kinetics and hydrolytic enzyme production of Pseudomonas spp. isolated from pasteurized milk. J. Dairy Res. 70:293-296. [DOI] [PubMed] [Google Scholar]
- 53.Stiles, M. E., and W. H. Holzapfel. 1997. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 36:1-29. [DOI] [PubMed] [Google Scholar]
- 54.Sumner, S. S., F. Roche, and S. L. Taylor. 1990. Factors controlling histamine production in Swiss cheese inoculated with Lactobacillus buchneri. J. Dairy Sci. 73:3050-3058. [DOI] [PubMed] [Google Scholar]
- 55.Sumner, S. S., H. W. Speckhard, E. B. Somers, and S. L. Taylor. 1985. Isolation of histamine-producing Lactobacillus buchneri from Swiss cheese implicated in a food poisoning outbreak. Appl. Environ. Microbiol. 50:1094-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teske, A., T. Brinkhoff, G. Muyzer, D. P. Moser, J. Rethmeier, and H. W. Jannasch. 2000. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thierry, A., M. B. Maillard, and M. Yvon. 2002. Conversion of l-leucine to isovaleric acid by Propionibacterium freudenreichii TL 34 and ITGP23. Appl. Environ. Microbiol. 68:608-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valdes-Stauber, N., S. Scherer, and H. Seiler. 1997. Identification of yeasts and coryneform bacteria from the surface microflora of brick cheeses. Int. J. Food Microbiol. 34:115-129. [DOI] [PubMed] [Google Scholar]
- 60.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]