Abstract
The study of several Escherichia coli intestinal commensal isolates per individual in 265 healthy human subjects belonging to seven populations distributed worldwide showed that the E. coli population is highly structured, with major differences between the tropical and temperate populations.
Escherichia coli is a commensal inhabitant of the intestinal tracts of healthy humans and many animal species, but it can also cause a wide range of diseases, ranging from diarrhea to extraintestinal infections (8). As it has been proposed that pathogenic E. coli strains are derived from commensal strains by the acquisition of chromosomal or extrachromosomal virulence operons (19), identifying the factors that shape the genetic structure of commensal strains might help us understand the emergence of virulence. E. coli can be considered to have a clonal genetic structure with a low level of recombination (7, 10). Four main phylogenetic groups, A, B1, B2, and D, constitute the bulk of the species (15). A few authors have examined commensal strains from humans (3-5, 13) using population genetics molecular tools (for a pioneer review, see reference 14). A significant locale-specific distribution among groups A, B1, B2, and D has been observed in human commensal strains in three geographically distinct human populations (French, Croatian, and Malian) (9).
To gain insight into the composition of the E. coli human commensal microbiota, we characterized the relative abundance of E. coli phylogenetic groups in a large collection of 1,740 isolates from 265 subjects belonging to seven populations spread over three continents and compared the results to those of previous studies using the same approach.
Bacterial isolates.
Isolates were collected between 1999 and 2001 from seven human populations composed of healthy adult subjects of both sexes from 15 to 65 years of age, except when otherwise stated. The populations were the following: (i) 27 subjects living in the Paris area (mainland France, Europe), (ii) 21 university students living in Brest (Brittany, mainland France), (iii) 25 bank and insurance workers (BIW) living in seven distinct areas of Brittany (mainland France), (iv) 25 pig farmers (PF) living in the same seven areas as the BIW, (v) 93 ethnically homogeneous Wayampi Amerindians living in three villages of southern French Guyana (South America) with no modern sanitary or hygienic facilities, (vi) 46 women living in Cotonou (Benin, Africa), and (vii) 28 subjects living in Bogotá (Colombia, South America). The individuals in the BIW and PF populations in Brittany were matched for county of residence, age (20 to 60 years), and sex (13 men and 12 women). A subset of 25 Amerindians was also matched for age and sex with the BIW and PF populations. These matched populations had neither been hospitalized nor had taken antibiotics for at least 1 month before stool sampling. In all, 1,740 E. coli isolates were obtained after plating fresh fecal samples on Drigalski agar, with 5 or 10 randomly chosen E. coli isolates per individual.
The method of Clermont et al. (6) was first used for the assignment of E. coli isolates to one of the four major phylogenetic groups A, B1, D, or B2. To increase the discriminative power of our analyses, all the combinations of genetic markers described in this method were also used as follows: subgroup A0 (group A), lacking chuA, yjaA, and Tspe4.C2; subgroup A1 (group A), lacking chuA, having yjaA, and lacking Tspe4.C2; subgroup B22 (group B2), having chuA and yjaA and lacking Tspe4.C2; subgroup B23 (group B2), having chuA, yjaA, and Tspe4.C2; subgroup D1 (group D), having chuA and lacking yjaA and Tspe4.C2−; and subgroup D2 (group D), having chuA, lacking yjaA, and having Tspe4.C2. Thus, seven groups and subgroups (A0, A1, B1, B22, B23, D1, and D2) were defined.
In addition, data for three other healthy adult populations comprising (i) 24 women living in Tours (mainland France) (21), (ii) 61 subjects of both sexes living in Tokyo (Japan) (18), and (iii) 88 women living in the Michigan (22) were taken from previously published studies in which one fecal commensal isolate per individual was assigned to one of the four major phylogenetic groups A, B1, D, or B2, as in reference 6.
Population structure analysis.
Analysis of one randomly chosen isolate from each individual in the 10 populations showed that the phylogenetic composition of the human E. coli commensal microbiota varied in a population-specific manner (Table 1; Fisher-Freeman-Halton test, P < 0.001). The number of isolates sampled per individual (1, 5, or 10) did not affect the general pattern of phylogenetic distribution among the seven populations in which it was tested (data not shown).
TABLE 1.
Prevalence of the four major E. coli phylogenetic groups in fecal samples from 10 human populationsa
| Population | nb | Prevalencec (%) in phylogenetic group:
|
|||
|---|---|---|---|---|---|
| A | B1 | D | B2 | ||
| Brittany (PF) | 25 | 32.0 | 28.0 | 24.0 | 16.0 |
| Brest University students | 21 | 14.3 | 23.8 | 28.6 | 33.3 |
| Paris area residents | 27 | 29.6 | 11.1 | 22.2 | 37.1 |
| Brittany (BIW) | 25 | 24.0 | 24.0 | 20.0 | 32.0 |
| Michigan residents | 88 | 20.5 | 12.5 | 19.3 | 47.7 |
| Tokyo residents | 61 | 28.0 | 0.0 | 28.0 | 44.0 |
| Tours residents | 24 | 25.0 | 21.0 | 25.0 | 29.0 |
| Bògotá residents | 28 | 57.1 | 3.6 | 14.3 | 25.0 |
| Cotonou residents | 46 | 50.0 | 32.6 | 0.0 | 17.4 |
| French Guyana Amerindians | 93 | 63.4 | 20.4 | 12.9 | 3.2 |
One isolate per individual was studied.
n, number of individuals.
Data for Michigan, Tokyo, and Tours residents were taken from previously published works (see the text).
The geographic locations of the human populations seem to play an important role in structuring the E. coli populations. In fact, when the 10 populations considered in the study were divided into two major groups according to geographic location and climate (mainland France, Tokyo, and Michigan populations in the temperate zone and French Guyana, Cotonou, and Bogotá populations in the tropical zone), significant differences between the two zones in the percentages of isolates carried were observed for three of the four phylogenetic groups. The prevalence of group A isolates in the temperate zone was half that in the tropical zone (14.3 to 32% versus 50 to 63.4%; χ2 test, P < 0.0001). A high prevalence of D isolates was observed among temperate zone populations, but these isolates were rare in the French Guyana and Bogotá populations (20 to 28.6% versus 12.9 to 14.3%), and even absent in the Cotonou population (χ2 test, P = 0.003). Last, the frequency of B2 group isolates ranged from 3.2% for the French Guyana population to 47.7% for the Michigan population, with a significantly higher prevalence in the temperate populations than in the tropical populations (χ2 test, P < 0.0001) (Table 1).
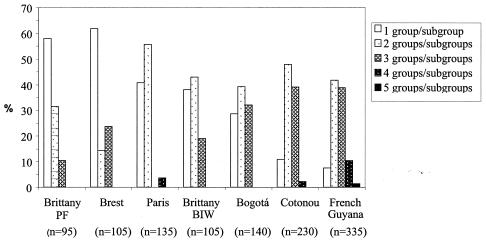
Our data also show that subjects in tropical areas exhibit more-diverse E. coli microbiota than those in temperate areas. The analysis of the numbers of groups and subgroups, defined by the markers of Clermont et al. (6), present in a sample of five isolates per individual indicated that subjects in the French Guyana, Cotonou, and Bogotá populations clearly harbored a more-diverse E. coli community than individuals in mainland France (Fig. 1; Kruskal-Wallis test, P < 0.0001), with averages of 2.4 and 1.7 groups and subgroups, respectively (Wilcoxon test, P < 0.0001). This significant difference was still observed when 10 isolates per subject were compared (data not shown).
FIG. 1.
E. coli intrahost diversity. Diversity of the percentages of the groups and subgroups (A0, A1, B1, D1, D2, B22, and B23) defined by all the combinations of genetic markers described in reference 6 per individual among the seven studied human populations when five isolates were considered per individual. n, number of isolates.
Consequently, geographic and climatic factors seem to play an important role in structuring the E. coli population worldwide. Although the two types (tropical and temperate) of regions chiefly differ in their climates, which in turn determine many biologic processes, the populations sampled in these regions also differ in numerous socioeconomic factors, such as diet and hygienic level. In fact, the temperate zone populations studied here are residents of cities which are highly developed technologically, and diet and/or the way food is processed by refrigeration chains may affect the E. coli population. Thus, there were phylogenetic differences between the Paris populations sampled in 1981 (9) and in 2001 (this work), with an increase in the strains of the B2 phylogenetic group from 10 to 37%. These changes in phylogenetic group distribution over a 20-year sampling period are not due to climatic changes but might be the result of social evolution involving changes in dietary habits and a better level of hygiene in France, as pointed out by others who studied different intestinal pathogens, such as Helicobacter pylori (2). Diet has been reported to be a key factor determining the relative abundance of E. coli phylogenetic groups in mammals (12). The level of hygiene might also favor the intraindividual isolate diversity that we observed in the tropical zone. For instance, there was a higher turnover rate for individual E. coli isolates in Pakistani infants than for Swedish infants (8.5 versus 1.5 strains during the first 6 months of life) (1). Furthermore, it seems that a change even occurs with time, as this turnover decreased in Sweden between the 1980s and 2003 (17).
Multinomial logistic regression analysis of the data from the matched French Guyana, PF, and BIW populations for which age and sex data were available showed that the geographic area (temperate versus tropical) was the explanatory variable and that the sex and age factors had no effect (data not shown).
Implications of population structure for the emergence of E. coli pathogenic clones.
Numerous studies have shown that isolates responsible for extraintestinal diseases belong mainly to the B2 group and, to a lesser extent, to the D group. Both groups have a higher prevalence of extraintestinal virulence determinants than the strains in the A and B1 groups (20). Principal-component analysis, using the data obtained from one isolate per individual (Table 1), showed a correlation between the frequencies of the B2 and D isolates (data not shown), suggesting that their commensal niches may be similar. In contrast, recent studies of the phylogenetic distribution of E. coli strains showing the pathogenic diversity of this species indicate that pathogenic strains associated with severe acute diarrhea are distributed outside the B2 and D groups (11). As tropical populations seem to preferentially harbor strains of group A and to a lesser extent B1, these strains might have the genetic background necessary for the emergence of pathogenic intestinal strains. This might be one of the factors explaining the higher incidence of diarrhea in tropical countries. In addition, the great diversity of E. coli microbiota may help hosts to fight intestinal pathogens more effectively by keeping the gut immune system active (16).
Acknowledgments
We are grateful to Olivier Clermont for the discussions that we had during the redaction of the paper.
This work was partly supported by a grant from Danone. P.E.-P. was awarded a grant from La Fondation pour la Recherche Médicale. The study of the Wayampi population and of BIW and PF populations was supported by a grant from INSERM, and the study of the BIW and PF populations was supported by a grant from the French Environment Ministry.
REFERENCES
- 1.Adlerberth, I., F. Jalil, B. Carlsson, L. Mellander, L. A. Hanson, P. Larsson, K. Khalil, and A. E. Wold. 1998. High turnover rate of Escherichia coli strains in the intestinal flora of infants in Pakistan. Epidemiol. Infect. 121:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banatvala, N., K. Mayo, F. Megraud, R. Jennings, J. J. Deeks, and R. A. Feldman. 1993. The cohort effect and Helicobacter pylori. J. Infect. Dis. 168:219-221. [DOI] [PubMed] [Google Scholar]
- 3.Caugant, D. A., B. R. Levin, G. Lidin-Janson, T. S. Whittam, C. Svanborg Eden, and R. K. Selander. 1983. Genetic diversity and relationships among strains of Escherichia coli in the intestine and those causing urinary tract infections. Prog. Allergy 33:203-227. [DOI] [PubMed] [Google Scholar]
- 4.Caugant, D. A., B. R. Levin, and R. K. Selander. 1984. Distribution of multilocus genotypes of Escherichia coli within and between host families. J. Hyg. 92:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caugant, D. A., B. R. Levin, and R. K. Selander. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98:467-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desjardins, P., B. Picard, B. Kaltenbock, J. Elion, and E. Denamur. 1995. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J. Mol. Evol. 41:440-448. [DOI] [PubMed] [Google Scholar]
- 8.Donnenberg, M. S. 2002. Escherichia coli: virulence mechanisms of a versatile pathogen, Elsevier Science Academic Press, San Diego, Calif.
- 9.Duriez, P., O. Clermont, S. Bonacorsi, E. Bingen, A. Chaventre, J. Elion, B. Picard, and E. Denamur. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671-1676. [DOI] [PubMed] [Google Scholar]
- 10.Dykhuizen, D. E., and L. Green. 1991. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 173:7257-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escobar-Páramo, P., O. Clermont, A. B. Blanc-Potard, H. Bui, C. Le Bouguenec, and E. Denamur. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085-1094. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, D. M., and A. Cowling. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575-3586. [DOI] [PubMed] [Google Scholar]
- 13.Goullet, P., and B. Picard. 1986. Comparative esterase electrophoretic polymorphism of Escherichia coli isolates obtained from animal and human sources. J. Gen. Microbiol. 132:1843-1851. [DOI] [PubMed] [Google Scholar]
- 14.Hartl, D. L., and D. E. Dykhuizen. 1984. The population genetics of Escherichia coli. Annu. Rev. Genet. 18:31-68. [DOI] [PubMed] [Google Scholar]
- 15.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellander, L., B. Carlsson, F. Jalil, T. Soderstrom, and L. A. Hanson. 1985. Secretory IgA antibody response against Escherichia coli antigens in infants in relation to exposure. J. Pediatr. 107:430-433. [DOI] [PubMed] [Google Scholar]
- 17.Nowrouzian, F., B. Hesselmar, R. Saalman, I. L. Strannegard, N. Aberg, A. E. Wold, and I. Adlerberth. 2003. Escherichia coli in infants' intestinal microflora: colonization rate, strain turnover, and virulence gene carriage. Pediatr. Res. 54:8-14. [DOI] [PubMed] [Google Scholar]
- 18.Obata-Yasuoka, M., W. Ba-Thein, T. Tsukamoto, H. Yoshikawa, and H. Hayashi. 2002. Vaginal Escherichia coli share common virulence factor profiles, serotypes and phylogeny with other extraintestinal E. coli. Microbiology 148:2745-2752. [DOI] [PubMed] [Google Scholar]
- 19.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 20.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watt, S., P. Lanotte, L. Mereghetti, M. Moulin-Schouleur, B. Picard, and R. Quentin. 2003. Escherichia coli strains from pregnant women and neonates: intraspecies genetic distribution and prevalence of virulence factors. J. Clin. Microbiol. 41:1929-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, L., B. Foxman, and C. Marrs. 2002. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 40:3951-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]