Abstract
The predominance of Geobacter species in environments in which Fe(III) reduction is important has suggested that Fe(III) reduction rates might be estimated in Geobacter-dominated environments by assessing in situ activity with molecular techniques. To determine whether mRNA levels of key respiratory genes might be correlated with respiration rates in Geobacter sulfurreducens, studies were conducted with fumarate as the electron acceptor and acetate as the limiting electron donor in anaerobic continuous cultures. Levels of mRNA for a fumarate reductase gene, frdA, quantified by real-time reverse transcription-PCR were directly correlated with fumarate reduction rates. In similar studies with Fe(III) as the electron acceptor, mRNA levels for omcB, a gene for an outer membrane c-type cytochrome involved in Fe(III) reduction, were positively correlated with Fe(III) reduction rates. Levels of mRNA for frdA and omcB were also positively correlated with fumarate and Fe(III) reduction rates, respectively, when growth was limited by the availability of fumarate or Fe(III), but mRNA levels were higher than in acetate-limited cultures. Levels of mRNA for omcC, which encodes a c-type cytochrome highly similar to OmcB but not necessary for Fe(III) reduction, followed patterns different than those of omcB. This agrees with the previous finding that OmcC is not involved in Fe(III) reduction and suggests that changes in mRNA levels of omcB are related to its role in Fe(III) reduction. These results demonstrate that mRNA levels for respiratory genes might be used to estimate in situ Fe(III) reduction rates in Geobacter-dominated environments but suggest that information on environmental conditions and/or the metabolic state of Geobacter species is also required for accurate rate estimates.
Dissimilatory Fe(III) reduction is one of the most geochemically significant processes in anoxic sedimentary environments, but estimating in situ rates of Fe(III) reduction is difficult. Dissimilatory Fe(III) reduction plays an important role in the anaerobic degradation of organic matter and influences sediment chemistry through the dissolution of Fe(III) oxides and the resultant release of soluble Fe(II), phosphate, and trace metals, as well as the production of Fe(II) minerals (20, 21). The ability of some dissimilatory Fe(III) reducers to degrade organic contaminants (6, 22, 29) and the abundance of Fe(III) oxides in many subsurface environments (17) can lead to significant removal of organic contaminants from groundwater via Fe(III) reduction. Fe(III)-reducing microorganisms can reduce toxic metals such as uranium (28), chromium (18), cobalt (3), and technetium (13). Stimulating the growth and activity of Fe(III)-reducing microorganisms in uranium-contaminated subsurface sediments can be an effective strategy for removing uranium from contaminated groundwater (1, 5).
Estimates of the in situ rates of Fe(III) reduction in subsurface environments could provide helpful insights into sediment biogeochemistry and would aid in modeling bioremediation by Fe(III)-reducing microorganisms. However, suitable methods for accurately measuring in situ rates of Fe(III) reduction are not yet available. This contrasts with most other anaerobic respiratory processes, for which good assessment techniques are available. For example, rates of sulfate reduction are typically estimated by monitoring the reduction of tracer quantities of 35SO42− (8), but reduction of Fe(III) cannot be measured by tracer techniques because there is rapid isotope exchange between Fe(III) and Fe(II) forms (39). Rates of Fe(III) reduction can be estimated from the accumulation of Fe(II) in sediments over time (24, 46). However, most of the Fe(II) produced is in solid phases and most sediments are sufficiently heterogeneous that sediments must be mixed prior to sampling in order to obtain reliable estimates of Fe(II) accumulation. This mixing can affect rates of microbial metabolism, especially in subsurface environments (4). Furthermore, long incubation times in sealed containers are typically required to detect significant increases in Fe(II) (24), which raises serious questions about the relationship of the measured rates to in situ activity.
An alternative strategy for measuring Fe(III) reduction might be to monitor mRNA levels of key genes involved in Fe(III) reduction. In some instances, environmental mRNA levels have been correlated with rates of microbial activity. For example, it has been shown that nahA transcript levels correlated positively with [14C]naphthalene mineralization rates, soil naphthalene concentration, and nahA gene frequency in soils (7). In some, but not all, instances direct correlations between levels of mercuric reductase (merA)-specific transcripts and Hg(II) volatilization rates have been observed (11, 33). However, it is less clear whether a direct correlation between rates of anaerobic respiration and levels of mRNA for key respiratory genes can be expected. For example, studies with Escherichia coli found that expression of genes for the reduction of fumarate, dimethyl sulfoxide, and nitrate changed little with increased growth rates or actually decreased (9, 47). Furthermore, unlike some respiratory process that are highly conserved, the mechanisms for Fe(III) reduction may be significantly different in phylogenetically distinct organisms (19, 35, 36).
Molecular analyses of the structure of the microbial community in a variety of subsurface environments in which Fe(III) reduction is important have demonstrated that Geobacter species are typically the predominant Fe(III)-reducing microorganisms (1, 10, 40, 41, 43, 44). This enrichment of a single group of microorganisms contrasts with the much greater diversity of microorganisms found in many other environments. Thus, if it is feasible to determine the rates of Fe(III) reduction in Geobacter species in situ, it might be possible to estimate rates of Fe(III) reduction in these Geobacter-dominated subsurface environments.
Geobacter sulfurreducens serves as a pure-culture model for the Geobacter species that predominate in subsurface environments (15, 16, 32). Since current methods in common use for mRNA quantification are relatively insensitive (2, 31), we developed real-time reverse transcription (RT)-PCR assays to quantify the expression levels of respiratory genes in G. sulfurreducens. Here we report that there is a direct correlation between rates of Fe(III) and fumarate reduction in G. sulfurreducens and levels of mRNA for key genes involved in Fe(III) and fumarate respiration.
MATERIALS AND METHODS
Culturing conditions and growth media.
G. sulfurreducens strain PCA (3) was grown in continuous culture at various growth rates under strict anaerobic conditions (N2/CO2 [80:20, vol/vol]) in a cysteine-free freshwater medium as previously described (A. Esteve-Núñez et al., submitted for publication). The acetate-limited cultures contained 5.5 mM acetate and either 30 mM fumarate or 56 mM Fe(III) citrate. For growth under electron acceptor-limiting conditions the acetate concentration was 10 mM while fumarate or Fe(III) citrate was supplied at 10 or 20 mM, respectively. Cells were grown in 200-ml water-jacketed glass vessels, maintained at 30°C, and stirred at a constant speed of 600 rpm with a magnetic bar (Esteve-Núñez et al., submitted). The flow rate was maintained constant with a peristaltic pump. Steady-state cell growth was expected after the medium input reached 5 volumes of the culture vessel, and this was confirmed by measuring cell density and fumarate, succinate, and Fe(II) concentrations.
Total RNA extraction and RT-PCR conditions.
Cells were harvested from each chemostat vessel by centrifugation at 5,000 × g for 15 min at 4°C. Total RNA was isolated immediately with a QIAGEN RNA Midi kit (QIAGEN Inc., Valencia, Calif.), and the extracted RNA was treated with RNase-free DNase (Ambion Inc., Austin, Tex.). DNA contamination was checked by agarose gel electrophoresis following RT-PCR by performing control experiments in which no reverse transcriptase was added to extracted RNA prior to the PCR step. RNA concentration was determined by measuring A260 with a Biophotometer (Eppendorf, Hamburg, Germany) and additionally verified by fluorometry with the RiboGreen RNA quantitation kit (Molecular Probes, Eugene, Oreg.). Purified RNA was stored at −80°C.
One microgram of total RNA served as the template for cDNA synthesis. For reverse transcription reactions SuperScript III RNase H− reverse transcriptase (200 U; Invitrogen Life Technologies, Rockville, Md.) was used in accordance with the manufacturer's instructions. Either random hexamers (2.5 μM final concentration; Invitrogen Life Technologies) or gene-specific antisense primers (1 μM; Sigma Genosys, The Woodlands, Tex.) (Table 1) were used for cDNA synthesis. The cDNA synthesis conditions were as follows: incubation at 55°C for 60 min (25°C for 5 min prior to incubation at 55°C for random primers) and enzyme inactivation at 70°C for 15 min. To remove RNA complementary to the cDNA, E. coli RNase H (2 U) was added and the reaction mixture was incubated at 37°C for 20 min, followed by rapid cooling to 4°C. The cDNA samples were stored at −20°C before PCR analysis. Two cDNA syntheses per RNA sample were performed.
TABLE 1.
Primers used for real-time PCR assays for quantitative detection of respiratory genes
| Target gene | Primera | Sequence | Length of amplicon (bp) | Reference |
|---|---|---|---|---|
| omcB | 8912 | 5′-CCCACTTCGACAACTATTCG-3′ | 212 | This study |
| 8908-2 | 5′-GGTCAGCAGGCCACCGG-3′ | 12 | ||
| omcC | 8917 | 5′-GGTCTTCACCCAGATCTCG-3′ | 232 | This study |
| 8915 | 5′-GGGTGTTGTGGTAGAAGGG-3′ | This study | ||
| frdA | frdA-2 | 5′-GAACTCGGTTACAACGTTG-3′ | 144 | This study |
| frdA-Q5 | 5′-GATGGTGTCATAGAAGAGACG-3′ | This study |
Sense and antisense primers for each gene are listed in sequence.
Primer design and optimization.
Primers used to amplify G. sulfurreducens sequences were designed in accordance with the desired criteria by the TaqMan system (GeneAmp 5700 SDS user manual; Applied Biosystems, Foster City, Calif.) and the G. sulfurreducens genome sequence (32). Primers were further evaluated and optimized for the application with the TaqMan system with the Primer Express software (version 1.5; Applied Biosystems). For each assay, primer concentrations and annealing temperatures were experimentally optimized to obtain specific amplification. The resulting conditions were experimentally checked with genomic DNA isolated from G. sulfurreducens cultures. All primers were synthesized by Sigma Genosys Oligofactory.
In order to determine whether the designed primers were suitable, gene-specific qualitative PCR (without addition of SYBR green) was performed prior to quantitative PCR. PCR products were amplified from cDNA generated by reverse transcription with the appropriate primers (Table 1) under the following conditions: 96°C for 40s; 27 cycles of 96°C for 40s, 55°C (frdA) or 58°C (omcB and omcC) for 1 min, and 72°C for 1 min; and a final extension at 72°C for 10 min. The PCR products were checked with ethidium bromide-stained agarose gel, and the specificity of PCR products was verified by sequence analysis.
Real-time PCR quantification.
Real-time RT-PCR is the most sensitive and flexible method for the detection of low-abundance mRNA (49). The assays were performed with a GeneAmp 5700 Sequence Detection System (Applied Biosystems). Quantification of samples was carried out by determining the threshold cycle value and by comparing results to a standard curve to determine the starting copy number. The threshold cycle was proportional to the logarithm of the target molecule number. The number of target molecules was automatically calculated by the GeneAmp 5700 SDS software (version 1.3). The principles of real-time PCR and more details have been described by Bustin, Raeymaekers, and Suzuki et al. (2, 38, 45). The precision and reproducibility of quantification were carefully optimized. In order to eliminate intra-assay and interassay variations, the same PCR run was performed in four replicates and the separate PCR runs were performed in triplicate, respectively. In addition, correct lengths of PCR products were checked by agarose gel electrophoresis to ensure that the fluorescent signal obtained in the real-time PCR originated from specific PCR products and not from artifacts.
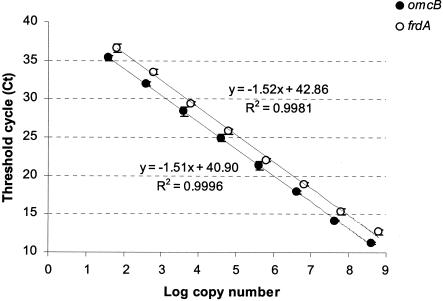
Dilution series of purified RT-PCR products were used as calibration standards for the real-time PCR quantification. The cDNAs, which were generated as described above, were amplified with the assay primers (Table 1), and the resulting amplicons were purified with a QIAquick PCR purification kit in accordance with the manufacturer's (QIAGEN) instructions. The purified RT-PCR products were then quantified by measurement of A260 with a Biophotometer (Eppendorf), and the concentration was verified by fluorometry with the PicoGreen double-stranded DNA quantitation kit (Molecular Probes). The concentration of RT-PCR products was converted to numbers of target molecule per microliter, and the omcB or frdA standards were prepared for serial dilution and stored at −20°C. The detection limits of all PCR assays were determined from four independent measurements with dilution series of purified RT-PCR products (standards) in a real-time PCR (109 to 101 target molecules per reaction). All assays had a minimum sensitivity of 101 to 102 target molecules per reaction (Fig. 1).
FIG. 1.
Standard curves for target amplicons omcB and frdA. The symbols represent means ± standard deviations of four replicates of PCR amplifications of the standard samples. The lines are trend lines from the linear regression. Amplicon lengths are 212 (omcB) and 144 (frdA) bp. The lower detection limit was about 40 to 70 copies of target mRNA. The copy numbers of unknown samples were calculated after real-time amplification from the linear regression of the standard curve.
PCR was performed with 96-well optical reaction plates (Applied Biosystems) sealed with optical caps on the GeneAmp 5700 Sequence Detection System. A 5-μl sample of known (standards) or unknown (cDNA) amounts of template DNA was added to 25 μl of 2 × TaqMan PCR Master mix (Applied Biosystems) and a 400 nM final concentration of each amplification primer in a final volume of 50 μl.
The temperature profile was composed of an initial incubation step of 2 min at 50°C (activation of the polymerase), followed by a denaturation step of 10-min at 95°C; 40 cycles of denaturation for 45 s at 95°C, annealing for 1 min at 55 or 58°C, and elongation for 1 min at 72°C; and a final elongation step of 6 min at 72°C. Four negative controls (reaction mixtures with no template) were included in each real-time PCR run. The size of the PCR products was verified by agarose gel electrophoresis.
Analytical techniques.
Cell density of G. sulfurreducens during growth on fumarate was monitored at 600 nm with a Spectronic Genesys 2 spectrophotometer (Spectronic Instruments, VWR, Boston, Mass.). Reduction of Fe(III) citrate was assessed by monitoring Fe(II) concentrations with ferrozine assays as previously described (26). Total iron concentrations were determined with hydroxylamine as previously described (27). Fumarate, succinate, and acetate concentrations were measured by high-pressure liquid chromatography on a Hewlett Packard series 1100 (Agilent Technologies, Inc., Albany, N.Y.) with a Bio-Rad Aminex HPX-87H column (300 by 7.8 mm) and a mobile phase of 8 mM H2SO4. Protein concentrations were determined by the bicinchoninic acid method with bovine serum albumin as the standard (42).
The specific fumarate reduction rate or Fe(III) reduction rate at each growth rate was calculated from the protein concentration and the amount of succinate production or Fe(II) production, respectively.
RESULTS
Expression of frdA and omcB under acetate-limited conditions.
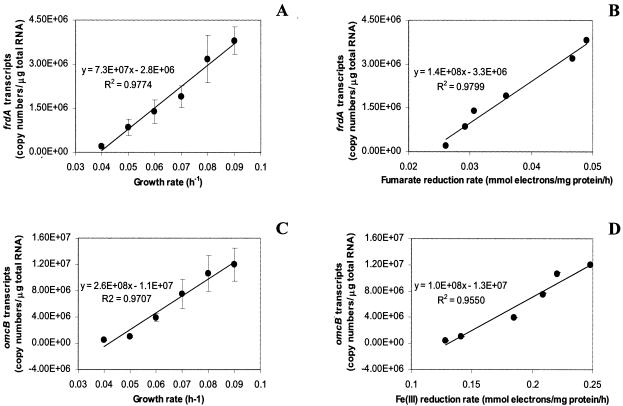
When G. sulfurreducens was grown with acetate as the electron donor and fumarate as the electron acceptor in continuous cultures in which acetate availability limited growth, levels of mRNA for the fumarate reductase gene, frdA, were directly correlated (R2 = 0.99) with rates of fumarate reduction and growth rates (Fig. 2A and B). The number of copies of frdA transcripts increased about 100-fold relative to the total RNA over the range of growth rates evaluated (Fig. 2A).
FIG. 2.
Expression of respiratory genes in G. sulfurreducens grown at various growth rates with fumarate (30 mM) (A, B) or ferric citrate (56 mM) (C, D) as the terminal electron acceptor and acetate (5.5 mM) as the limiting electron donor. Data are means ± standard deviations of triplicates.
In a similar manner, there was a strong positive correlation (R2 = 0.94) between levels of mRNA for omcB, a gene for an outer membrane c-type cytochrome required for Fe(III) reduction (12, 30) and the rate of Fe(III) reduction (Fig. 2D). Levels of mRNA for omcB increased more than 60-fold over the range of growth rates evaluated (Fig. 2C).
Expression of frdA and omcB under electron acceptor-limited conditions.
In order to determine whether levels of mRNA during growth when the electron acceptor was limiting were similar to those when the electron donor was limiting, G. sulfurreducens cells were grown with excess electron donor (acetate) and either limited fumarate or Fe(III) citrate. The levels of mRNA for frdA and omcB were positively correlated with the rates of fumarate and Fe(III) reduction, respectively (Fig. 3). The number of copies of frdA mRNA was ca. threefold higher relative to the total RNA under the fumarate-limited conditions (Fig. 3A) versus the acetate-limited conditions (Fig. 2A). The relative levels of omcB mRNA in Fe(III)-limited cultures were ca. 20-fold higher (Fig. 3C) than those in acetate-limited cultures (Fig. 2C).
FIG. 3.
Expression of respiratory genes in G. sulfurreducens grown at various growth rates with fumarate (10 mM) (A, B) or ferric citrate (20 mM) (C, D) as the limiting terminal electron acceptor and acetate (10 mM) as the electron donor. Data are means ± standard deviations of triplicates.
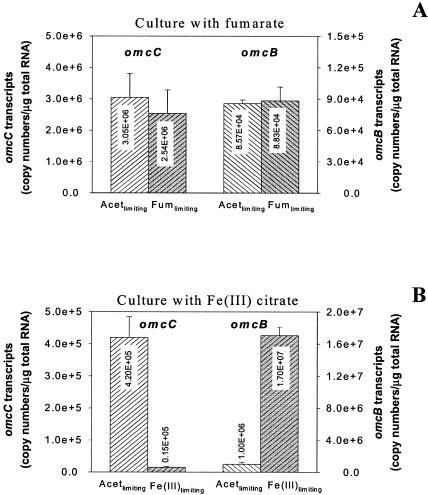
Differential expression of omcB and omcC.
In order to further evaluate whether expression of omcB could be specifically related to Fe(III) reduction, the expression of omcC was also evaluated. Although omcC and omcB are 79% identical in nucleotide sequences (73% identity in peptide sequences) and may have arisen from a gene duplication event, OmcB is required for Fe(III) reduction but OmcC is not (12). At the same dilution rate, omcB transcript levels were lower than omcC transcript levels during growth on fumarate but were higher during growth on Fe(III) (Fig. 4). The difference in omcB and omcC transcript levels during growth on Fe(III) was particularly pronounced under Fe(III)-limited conditions, in which levels of omcB mRNA increased in comparison to those in acetate-limited cultures whereas omcC transcript levels in Fe(III)-limited cultures were substantially lower than those under acetate-limited conditions (Fig. 4).
FIG. 4.
Differential expression of respiratory genes in G. sulfurreducens grown at the same growth rate (μ = 0.08 h−1) under acetate (Acet)-limiting versus terminal electron acceptor-limiting conditions: A, omcC and omcB gene expression in culture with fumarate (Fum); B, omcC and omcB gene expression in culture with Fe(III). Data are means ± standard deviations of triplicates.
DISCUSSION
The results demonstrate that there is a positive correlation between levels of mRNA for key respiratory genes and rates of reduction of the relevant electron acceptor in G. sulfurreducens. As detailed below, this provides the possibility of estimating rates of Fe(III) reduction, and possibly other types of respiration, in Geobacter-dominated subsurface environments. However, the finding that the levels of mRNA associated with a given rate of Fe(III) is substantially influenced by whether metabolism is limited by electron donor or electron acceptor availability suggests that it may be necessary to monitor the pattern of expression of several genes in order to evaluate whether metabolism is electron donor or electron acceptor limited before it will be possible to accurately estimate in situ respiration rates.
Fumarate respiration.
Initial studies of fumarate respiration were conducted because this is the best-understood form of respiration in G. sulfurreducens. A cluster of three genes, frdCAB, with high homology to the three-gene operon for the fumarate reductase in Wolinella succinogenes is present in the G. sulfurreducens genome, and disruption of frdA eliminates the capacity for fumarate reduction in G. sulfurreducens (J. E. Butler and D. R. Lovley, unpublished data).
The strong correlation between mRNA levels for frdA and rates of fumarate reduction further verify that frdA is involved in fumarate respiration. The regulatory mechanisms that account for the apparent increase in the transcription of frdA, and presumably other genes involved in fumarate reduction, are unknown. Whether there was an increase in the levels of the fumarate reductase associated with higher levels of frdA mRNA was not documented. However, producing more of a terminal reductase when conditions favor higher rates of reduction of the electron acceptor could be an adaptive response. In a similar manner, it has been noted that levels of mRNA for the dsrAB genes, the key, highly conserved genes coding for the dissimilatory sulfite reductase in sulfate reduction (37, 48), increase in dissimilatory sulfate-reducing microorganisms as rates of sulfate reduction increase (34; L. Hayes and D. R. Lovley, Abstr. 102nd ASM Gen. Meet. Am. Soc. Microbiol., abstr. N-95, p. 321-322, 2002).
In contrast, in E. coli, frdA expression changed little as the cell growth rate increased from 0.12 to 0.60 h−1 under anaerobic conditions and there was also no increase, or a slight decrease, in the expression of genes associated with the reduction of dimethyl sulfoxide or nitrate as growth rates were increased (47). The growth rates evaluated in E. coli were substantially higher than those at which G. sulfurreducens can be grown. This suggests that phylogenetically distinct microorganisms may use different mechanisms for controlling the production of respiratory proteins or that at high growth rates different forms of regulation are more effective.
Fe(III) reduction.
Mechanisms for electron transport to Fe(III) in G. sulfurreducens are not as well understood as those for fumarate reduction; however, it has been demonstrated that the gene encoding OmcB, an outer membrane c-type cytochrome, is required for Fe(III) reduction (12). Deletion of ppcA, a gene for a smaller, periplasmic c-type cytochrome, also had an effect on Fe(III) reduction (14), but Fe(III) reduction was only inhibited approximately 40% compared to nearly complete inhibition of Fe(III) reduction in the OmcB-deficient mutant (12). Furthermore, the OmcB protein is predicted to be localized in the outer membrane, where it might directly interact with Fe(III) (12), whereas PpcA is periplasmic and thus likely to be an intermediary in electron flow to Fe(III) and other extracellular electron acceptors (14). Thus, of the genes that encode proteins currently known to be important in Fe(III) reduction in G. sulfurreducens, the omcB gene is the one most specifically associated with Fe(III) reduction.
The direct correlation between levels of omcB mRNA and rates of Fe(III) reduction further suggests that OmcB is a key component in electron transfer to Fe(III), as does the finding that levels of omcB were substantially higher during growth on Fe(III) than on fumarate. It is notable that levels of mRNA for the closely related gene omcC were higher during growth on fumarate than during growth on Fe(III) and were lower than levels of omcB RNA during growth on Fe(III), especially when Fe(III) availability was the factor limiting growth. These results suggest that, in contrast to omcB, transcription of omcC is regulated by factors other than those involved in a physiological response to the use of Fe(III) as an electron acceptor. This is consistent with the finding that knocking out omcC does not affect Fe(III) reduction, indicating that, despite their high degree of similarity, OmcB plays a key role in electron transfer to Fe(III) whereas OmcC does not (12).
Implications for estimating in situ respiration rates.
The direct correlation between levels of mRNA for two key respiratory genes and rates of respiration suggests that it may eventually be possible to monitor in situ respiration in environments in which Geobacter species predominate. It is encouraging that a similar response was observed from two fundamentally different types of respiration. Whereas fumarate reduction takes place at the inner membrane, current evidence suggests that Fe(III) reduction takes place at or near the outer membrane.
The higher levels of omcB and frdA mRNA in cultures limited by electron acceptor [Fe(III) or fumarate] availability in comparison with cultures growing at the same rate but limited by electron donor (acetate) availability suggest that expression of the relevant terminal reductases is regulated in order to produce more of the appropriate terminal respiration genes as the availability of the electron acceptor decreases. This result has important implications for evaluating rates of respiration in Geobacter-dominated communities because the relative levels of mRNA for respiratory genes are affected not only by the rate of electron acceptor reduction but also by the availability of the electron acceptor. Thus, it may not be possible to specify Fe(III) reduction rates based on levels of mRNA for a single respiratory gene. Initial studies were conducted with acetate availability as the limiting factor because in many subsurface environments respiration is likely to be limited by the availability of the primary electron donor for anaerobic respiration, acetate. However, in some instances, Fe(III) could be the limiting factor. For example, as Fe(III) is depleted from sediments as a result of Fe(III) reduction, Fe(III) eventually becomes limiting and the predominant terminal electron-accepting process switches from Fe(III) reduction to sulfate reduction (23, 25). Furthermore, when acetate is added to the subsurface in millimolar quantities in order to stimulate dissimilatory metal reduction in uranium-contaminated aquifers, the availability of Fe(III) rather than acetate might limit the activity of Fe(III)-reducing microorganisms (1). Determining unequivocally whether a Geobacter population is either electron donor or Fe(III) limited from standard geochemical measurements would be difficult. Therefore, analysis of mRNA levels for a suite of genes, some of which could provide information on the factors limiting growth and activity, might be necessary in order to estimate in situ rates of respiration from levels of mRNAs for key respiratory genes.
Acknowledgments
This research was funded by the Office of Biological and Environmental Research, U.S. Department of Energy, under the Natural and Accelerated Bioremediation (DE-FG02-97ER62475) and Genomics: GTL (DE-FC02-02ER63446) programs, and grant N000140310315 from the Office of Naval Research. A.E.-N. was the recipient of a postdoctoral fellowship from the Secretaría de Estado de Educación y Universidades (Spain), cofunded by the European Social Fund.
We are grateful for the excellent technical support of Manju Sharma.
REFERENCES
- 1.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 3.Caccavo, F., Jr., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60:3752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapelle, F. H., and D. R. Lovley. 1990. Rates of microbial metabolism in deep coastal plain aquifers. Appl. Environ. Microbiol. 56:1865-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finneran, K. T., M. E. Housewright, and D. R. Lovley. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 4:510-516. [DOI] [PubMed] [Google Scholar]
- 6.Finneran, K. T., and D. R. Lovley. 2001. Anaerobic degradation of methyl tert-butyl ether (MTBE) and tert-butyl alcohol (TBA). Environ. Sci. Technol. 35:1785-1790. [DOI] [PubMed] [Google Scholar]
- 7.Fleming, J. T., J. Sanseverino, and G. S. Sayler. 1993. Quantitative relationship between naphthalene catabolic gene frequency and expression in predicting PAH degradation in soils at town gas manufacturing sites. Environ. Sci. Technol. 27:1068-1074. [Google Scholar]
- 8.Fossing, H., and B. B. Jorgensen. 1989. Measurements of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry 8:205-222. [Google Scholar]
- 9.Gunsalus, R. P. 1992. Control of electron flow in Escherichia coli: coordinated transcription of respiratory pathway genes. J. Bacteriol. 174:7069-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffrey, W. H., S. Nazaret, and T. Barkay. 1996. Detection of the merA gene and its expression in the environment. Microb. Ecol. 32:293-303. [DOI] [PubMed] [Google Scholar]
- 12.Leang, C., M. V. Coppi, and D. R. Lovley. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd, J. R., J. Chesnes, S. Glasauer, D. J. Bunker, F. R. Livens, and D. R. Lovley. 2002. Reduction of actinides and fission products by Fe(III)-reducing bacteria. Geomicrobiol. J. 19:103-120. [Google Scholar]
- 14.Lloyd, J. R., C. Leang, A. L. Hodges Myerson, M. V. Coppi, S. Cuifo, B. Methe, S. J. Sandler, and D. R. Lovley. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovley, D. R. 2002. Analysis of the genetic potential and gene expression of microbial communities involved in the in situ bioremediation of uranium and harvesting electrical energy from organic matter. OMICS 6:331-339. [DOI] [PubMed] [Google Scholar]
- 16.Lovley, D. R. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 1:35-44. [DOI] [PubMed] [Google Scholar]
- 17.Lovley, D. R. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55:259-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovley, D. R. 1993. Dissimilatory metal reduction. Annu. Rev. Microbiol. 47:263-290. [DOI] [PubMed] [Google Scholar]
- 19.Lovley, D. R. 2002. Dissimilatory metal reduction: from early life to bioremediation. ASM News 68:231-237. [Google Scholar]
- 20.Lovley, D. R. 2000. Fe(III) and Mn(IV) reduction, p. 3-30. In D. R. Lovley (ed.), Environmental microbe-metal interactions. ASM Press, Washington, D.C.
- 21.Lovley, D. R. 1995. Microbial reduction of iron, manganese, and other metals. Adv. Agron. 54:175-231. [Google Scholar]
- 22.Lovley, D. R., M. J. Baedecker, D. J. Lonergan, I. M. Cozzarelli, E. J. P. Phillips, and D. I. Siegel. 1989. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature 339:297-299. [Google Scholar]
- 23.Lovley, D. R., and S. Goodwin. 1988. Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochim. Cosmochim. Acta 52:2993-3003. [Google Scholar]
- 24.Lovley, D. R., and E. J. Philips. 1986. Availability of ferric iron for microbial reduction in bottom sediments of the freshwater tidal Potomac River. Appl. Environ. Microbiol. 52:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovley, D. R., and E. J. Philips. 1987. Competitive mechanisms for inhibition of sulfate reduction and methane production in the zone of ferric iron reduction in sediments. Appl. Environ. Microbiol. 53:2636-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovley, D. R., and E. J. Philips. 1986. Organic matter mineralization with the reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovley, D. R., and E. J. P. Phillips. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 29.Lovley, D. R., J. C. Woodward, and F. H. Chapelle. 1994. Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe(III) ligands. Nature 370:128-131. [DOI] [PubMed] [Google Scholar]
- 30.Magnuson, T. S., N. Isoyama, A. L. Hodges-Myerson, G. Davidson, M. J. Maroney, G. G. Geesey, and D. R. Lovley. 2001. Isolation, characterization and gene sequence analysis of a membrane-associated 89 kDa Fe(III) reducing cytochrome c from Geobacter sulfurreducens. Biochem. J. 359:147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12:7035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Methé, B. A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. Van Aken, D. R. Lovley, and C. M. Fraser. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 33.Nazaret, S., W. H. Jeffrey, E. Saouter, R. Von Haven, and T. Barkay. 1994. merA gene expression in aquatic environments measured by mRNA production and Hg(II) volatilization. Appl. Environ. Microbiol. 60:4059-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neretin, L. N., A. Schippers, A. Pernthaler, K. Hamann, R. Amann, and B. B. Jorgensen. 2003. Quantification of dissimilatory (bi)sulphite reductase gene expression in Desulfobacterium autotrophicum using real-time RT-PCR. Environ. Microbiol. 5:660-671. [DOI] [PubMed] [Google Scholar]
- 35.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for accessing insoluble Fe(III) oxide during dissimilatory Fe(III) reduction by Geothrix fermentans. Appl. Environ. Microbiol. 68:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for Fe(III) oxide reduction in sedimentary environments. Geomicrobiol. J. 19:141-159. [Google Scholar]
- 37.Odom, J. M., and H. D. Peck, Jr. 1984. Hydrogenase, electron transfer proteins, and energy coupling in the sulfate-reducing bacteria Desulfovibrio. Annu. Rev. Microbiol. 38:551-592. [DOI] [PubMed] [Google Scholar]
- 38.Raeymaekers, L. 1999. General principles of quantitative PCR, p. 31-41. In B. K. U. Reischl (ed.), Quantitative PCR protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 39.Roden, E. E., and R. G. Wetzel. 2002. Kinetics of microbial Fe(III) oxide reduction in freshwater wetland sediments. Limnol. Oceanogr. 47:198-211. [Google Scholar]
- 40.Röling, W. F. M., B. M. van Breukelen, M. Braster, B. Lin, and H. W. van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 43.Snoeyenbos-West, O. L., K. P. Nevin, R. T. Anderson, and D. R. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]
- 44.Stein, L. Y., M. T. La Duc, and T. J. Grundl. 2001. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediment. Environ. Microbiol. 3:10-18. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, M. T., Lance T. Taylor, and Edward F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thamdrup, B. 2000. Bacterial manganese and iron reduction in aquatic sediments. Adv. Microb. Ecol. 16:41-84. [Google Scholar]
- 47.Tseng, C. P., A. K. Hansen, P. Cotter, and R. P. Gunsalus. 1994. Effect of cell growth rate on expression of the anaerobic respiratory pathway operons frdABCD, dmsABC, and narGHJI of Escherichia coli. J. Bacteriol. 176:6599-6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, T., and M. J. Brown. 1999. mRNA quantification by real time TaqMan polymerase chain reaction: validation and comparison with RNase protection. Anal. Biochem. 269:198-201. [DOI] [PubMed] [Google Scholar]