Abstract
Iron-reducing enrichments were obtained from leachate ponds at the U.S. Borax Company in Boron, Calif. Based on partial small-subunit (SSU) rRNA gene sequences (approximately 500 nucleotides), six isolates shared 98.9% nucleotide identity. As a representative, the isolate QYMF was selected for further analysis. QYMF could be grown with Fe(III)-citrate, Fe(III)-EDTA, Co(III)-EDTA, or Cr(VI) as electron acceptors, and yeast extract and lactate could serve as electron donors. Growth during iron reduction occurred over the pH range of 7.5 to 11.0 (optimum, pH 9.5), a sodium chloride range of 0 to 80 g/liter (optimum, 20 g/liter), and a temperature range of 4 to 45°C (optimum, approximately 35°C), and iron precipitates were formed. QYMF was a strict anaerobe that could be grown in the presence of borax, and the cells were straight rods that produced endospores. Sodium chloride and yeast extract stimulated growth. Phylogenetic analysis of the SSU rRNA gene indicated that the bacterium was a low-G+C gram-positive microorganism and had 96 and 92% nucleotide identity with Alkaliphilus transvaalensis and Alkaliphilus crotonatoxidans, respectively. The major phospholipid fatty acids were 14:1, 16:1ω7c, and 16:0, which were different from those of other alkaliphiles but similar to those of reported iron-reducing bacteria. The results demonstrated that the isolate might represent a novel metal-reducing alkaliphilic species. The name Alkaliphilus metalliredigens sp. nov. is proposed. The isolation and activity of metal-reducing bacteria from borax-contaminated leachate ponds suggest that bioremediation of metal-contaminated alkaline environments may be feasible and have implications for alkaline anaerobic respiration.
Bacterial metal reduction has widened the realm of life-supporting biological reactions (29) and has been implicated as an important biochemical process on early Earth (20, 36, 39). The ability to reduce metals can be exploited for the bioreduction or immobilization of many toxic metals, including cobalt, chromium, uranium, and technetium (6). However, metal reduction in alkaline environments has not been well documented.
Concentrated deposits of boron exist in some arid regions (e.g., Turkey and the United States) and are economically exploited for products including fiberglass, ceramics, glass, laundry bleaches, fire retardants, insecticides, and semiconductors (25, 42). Borax is a common detergent ingredient and is sometimes used as a mild disinfectant. The toxicity is low, but boron is not completely harmless. Boron can potentially affect reproductive organs of males and impair fetal development in pregnant females, as well as be phytotoxic (27, 30). In addition, sodium perborate has been shown to be an in vitro mutagen (34).
Metal-reducing bacteria can be isolated from a variety of habitats, and much work has focused on the metal-reducing bacteria Shewanella oneidensis (28) and Geobacter spp. (22). Sites contaminated with toxic metals can have drastically different environmental conditions, and the biological reduction of most metals has commonly been studied at circumneutral pH values (36). Little is known about metal reduction under different extreme conditions, and only recently has bacterial Fe(III) reduction been demonstrated under thermophilic (20, 39), psychrophilic (45), or acidic (18) conditions. However, metal reduction under alkaliphilic growth conditions has not been demonstrated. We report the isolation and partial characterization of a novel alkaliphilic microorganism that can reduce metals at pH values up to 11.0 in the presence of elevated salt levels. Phylogenetic analysis of the small-subunit (SSU) rRNA gene indicated that the isolate was a low-G+C, gram-positive bacterium but, in addition to phenotypic differences, had only 96% sequence identity with the closest known relative. The name Alkaliphilus metalliredigens sp. nov. is proposed for the microorganism.
MATERIALS AND METHODS
Site characteristics and water chemistry.
Leachate ponds at U.S. Borax Company (Boron, Calif.) were chosen for the study because of alkaline pH. The pond water had prominent blooms of both algae and cyanobacteria at the time of sampling (March 2001). Samples were collected into sterile 50-ml tubes from water in the pond and partially dried soft sediments along the pond embankments. The water samples were filtered onto 0.4-μm-pore-size Gelman filter disks, and supernatants were saved for elemental analyses. Supernatants were analyzed by inductively coupled plasma mass spectroscopy (Activation Labs Limited).
Bacterial enrichments and isolation.
Sediment slurry samples were enriched for microbial growth in the presence of acetate, lactate, and yeast extract and of iron-citrate as the electron acceptor at pH values corresponding to individual samples (pH 8.0 to 9.5). Ferric citrate solution (pH 7.0) was prepared with ferric citrate hydrate, and the pH was modified with NaOH. Anaerobic culture techniques were used throughout the study with anoxic medium in tubes (18 by 150 mm) or serum bottles with butyl rubber stoppers and aluminum crimp seals under an N2-CO2 (80:20) atmosphere. All media and solutions were boiled under O2-free nitrogen gas or an N2-CO2 gas mix and dispensed into tubes or bottles under streams of the corresponding gas.
Sediment slurries were diluted 10-fold into anaerobic growth medium with lactate, acetate, and iron-citrate. Cultures were incubated at 20°C, and iron precipitation was observed within 5 days. The medium for enrichments contained the following ingredients: 4.3 mM K2HPO4, 9.4 mM (NH4)2SO4, 50 mM Tris, 6.1 μM Na2SeO4, 4 mM NaHCO3, 7 mM sodium lactate, 7 mM sodium acetate, 15 mM iron-citrate, 0.025 g of yeast extract (Difco) per liter, 10 ml of mineral stock solution (100×), and a headspace of N2-CO2 (80:20). The mineral stock contained the following ingredients per liter: 1.5 g of nitrilotriacetic acid, 3.0 g of MgSO4, 0.5 g of MnSO4, 1.0 g of NaCl, 0.1 g of FeSO4, 0.1 g of CaCl2, 0.1 g of CoCl2, 0.13 g of ZnCl, 0.01 g of CuSO4, 0.01 g of AlK(SO4), 0.01 g of H3BO3, 0.03 g of Na2MoO4, 0.02 g of NiCl2.6H2O, and 0.03 g of Na2WO4.
The growth medium contained the following ingredients: K2HPO4, 5.7 mM; (NH4)2SO4, 12.5 mM; NaCl, 327 mM; Na2CO3, 19.8 mM; Na2B4O7, 10 mM; yeast extract, 0.025 g/liter, 100× mineral solution, 10 ml. The pH was adjusted to approximately 9.5. The headspace was N2-CO2 (80:20). Aerobic growth was tested in the above-mentioned medium as well as a medium used for the isolation and growth of different soil microorganisms, MR2A. MR2A contains peptone, tryptone, yeast extract, glucose, pyruvate, starch, and trace minerals (7). A significant decrease in growth rate was not observed up to 25 mM borax, and the samples from the leachate ponds were normally 10 to 15 mM borax. Therefore, the bacterium was cultured in medium with 10 mM borax.
Bacterial isolates were obtained from solid (2% agar) enrichment medium plates in an anaerobic glove bag (Coy Laboratories) with an N2-H2 (97%:3%) atmosphere. The enrichment culture was transferred twice before being used as an inoculum for agar plates. Colonies appeared dark brown after growth on Fe(III)-citrate solid medium and became hard. Isolated colonies were transferred to liquid medium with sterile Pasteur pipettes as agar bores after being restreaked at least four times.
Morphology.
Bacterial cell morphology was examined under scanning electron microscopy (SEM). Cell cultures (2 ml) were harvested by centrifugation (16,000 × g; 5 min), washed in fresh anoxic medium, and centrifuged again (16,000 × g; 5 min). Preparation of each sample for SEM, including glutaraldehyde fixation and osmium tetroxide staining, was performed as previously described (1). At least two separately stained and fixed samples were viewed with a JEOL 5500 scanning electron microscope (JEOL Inc., Peabody, Mass.).
Growth studies.
Bacterial growth with Fe(III), Co(III), and Cr(VI) was quantified by direct cell counting with epifluorescence microscopy of acridine orange-stained samples and statistical analysis as described previously (9, 15). Inocula were 10% or less of starting culture volumes. Light microscopy was used to visualize wet mounts and acridine orange-stained filters with a Nikon Phase Contrast Optiphot microscope (Nikon). Growth with the humic acid analog 2,6-anthraquinone disulfonate (AQDS) was monitored with a spectrophotometer, and an increase in cell number was monitored via direct counts. All growth studies were done in duplicate, and the error bars in figures indicate standard deviations.
Possible electron donors and acceptors were tested in the Na2CO3-buffered medium at pH 9.5 and 20°C and were added from sterile anoxic stock solutions. Lactate (10 mM) and/or yeast extract (0.25 g/liter) was routinely used as the electron donor for testing electron acceptors. Soluble Fe(III) was provided as Fe(III)-citrate or Fe(III)-EDTA (10 mM), Cr(VI) was supplied as K2CrO7 (100 μM), and Co(III) was added as Co(III)-EDTA (3 mM). The other tested electron acceptors were used at the concentrations in parentheses: akaganeite (1 mM), manganese oxides (1 mM), trimethylamine oxide (10 mM), dimethyl sulfoxide (10 mM), thiosulfate (10 mM), fumarate (10 mM), and nitrate (1 and 15 mM). Chemicals were purchased from Sigma Chemical Co., except for akaganeite and manganese oxide, which were prepared as previously described (21, 31). The effects of pH, temperature, and sodium chloride levels on the growth of isolate QYMF were determined in the Na2CO3-buffered medium by varying the respective parameter. The pH was adjusted by decreasing the Na2CO3 levels or by the addition of KOH as previously described (37).
Chemical and mineralogical analyses.
Fe(II) production was monitored with a spectrophotometer by the ferrozine method and acid extraction (21). The decrease in Co(III)-EDTA was monitored using a spectrophotometer at 535 nm (3), and the reduction of AQDS was monitored at 450 nm (24). The decrease in Cr(VI) was determined with a spectrophotometer from H2SO4-extractable Cr(VI) with a symdiphenylcarbazide reagent (0.25% acetone) at 540 nm (23). All analyses were done in duplicate.
Precipitates in each sample were collected on a 0.22-μm-pore-size membrane (Millipore; polyvinylidene fluoride) and washed three times with deionized water. The filtered precipitates were dried under an N2 atmosphere to prevent oxidation. The filtered and dried precipitates were carbon coated with a Ted Pella carbon sputter coater and immediately examined. The morphology, mineralogy, and chemistry of precipitated phases of iron were determined with a JEOL JSM-35CF (JEOL Ltd., Tokyo, Japan) scanning electron microscope with energy-dispersive X-ray analysis (EDX). The mineralogical composition of the precipitated or transformed phases was determined using X-ray diffraction performed on a Scintag (Scintag, Inc., Sunnyvale, Calif.) XDS 2000 diffractometer (40 kV, 35 mV) equipped with Co-Kα radiation (λ = 0.17889 nm).
DNA extraction, amplification, and sequence determination.
DNA was isolated with a Promega Wizard kit and the addition of lysostaphin (10 mg/liter; Sigma Chemical Co.) and freeze-thawing according to the manufacturer's instructions. The SSU rRNA genes were amplified from isolates with the FD1 and 1540R primers (Table 1), and PCR products were purified with Montage PCR 96 filter plates (Millipore, Inc.) or treated with ExoSAP-IT (U.S. Biochemical Corporation) according to the manufacturer's instructions. PCR parameters for the amplification of the SSU rRNA gene sequences were described previously (40, 48). DNA sequences were determined with a BigDye Terminator kit (Applied Biosystems) with a 3700 DNA analyzer (Perkin-Elmer) according to the manufacturer's instructions with the SSU rRNA gene-specific primers 350r, 519f, 529r, 788f, 925r, and 1099f (Table 1, Escherichia coli designations) and compared with sequences from GenBank.
TABLE 1.
Nucleotide sequences for PCR primers
| Designation | Sequence (5′ to 3′) |
|---|---|
| FD1 | AGA GTT TGA TCC TGG CTC AG |
| 519f | CAG CAG CCG CGG TAA |
| 788f | ATT AGA TAC CCT GGT A |
| 1099f | GCA ACG AGC GCA ACC C |
| 350r | CTG CTG C(gc)(ct) CCC GTA G |
| 529r | CGC GGC TGC TGG CAC |
| 925r | CCG TCA ATT C(ac)T TT(ag) AGT TT |
| 1540R | AAG GAG GTG ATC CAG CC |
Raw data were assembled into contiguous sequences and edited with Sequencher software (Gene Codes Corp.). The nearly complete SSU rRNA gene sequences (approximately 1,500 nucleotides) were aligned with ClustalW (38). Phylogenetic analyses of SSU rRNA gene sequences were conducted using MEGA version 2.1 (17). Neighbor-joining and maximum-parsimony phylogenies were constructed from dissimilatory distances and pairwise comparisons and did not differ significantly.
Lipid analysis.
Cells (5.0 × 1010) cultured in Na2CO3-buffered medium with yeast extract as an electron donor and 10 mM Fe(III)-EDTA as an electron acceptor were harvested (4,000 × g, 30 min), frozen at −70°C, and lyophilized. Lyophilized bacterial cell pellets (approximately 20 mg) were extracted by a modified Bligh and Dyer single-phase organic solvent system that consisted of 142.5 ml of chloroform, methanol, and aqueous 50 mM phosphate buffer (1:2:0.8, vol/vol/vol) as previously described (2, 41). The lipid phase was collected and fractionated on a silicic acid column into neutral lipids, glycolipids, and polar lipids. The polar phospholipid fatty acids were treated using a mild alkaline methanolysis to produce fatty acid methyl esters. Methyl esters were analyzed with an Agilant 6890 series gas chromatograph interfaced to an Agilant 5973 mass selective detector with a 20-m nonpolar column (46). Mass spectra were determined by electron impact at 70 eV. Monounsaturated fatty acid methyl ester double-bond position was determined by gas chromatography-mass spectrometry analysis of the dimethyl disulfide adducts (46).
Nucleotide sequence accession number.
The GenBank accession number assigned to the SSU 16S rRNA gene sequence for isolate QYMF is AY137848.
RESULTS
Water chemistry.
The hydrogeochemical analysis of water samples from the leachate ponds indicated that Na, Mg, K, and Ca were the major elements, and sodium concentrations ranged from 0.04 to 0.53 M. Boron was between 0.19 and 0.28 M, which corresponded to approximately 2,000 to 3,000 ppm. Iron concentrations were approximately 0.1 to 0.2 mM, chromium ranges were 3.8 to 4.5 μM, and cobalt was present up to concentrations of 0.2 μM. Uranium concentrations ranged from 0.6 to 1.3 μM, arsenic concentrations were approximately 1.7 mM, and selenium was not detected.
Microbial enrichment and isolation.
Positive enrichments displayed signs of iron reduction within 5 days. Colonies could be picked and restreaked upon initial appearance, but colonies became hard to the touch of a needle or loop after significant reduction of Fe(III) in the medium. After restreaking, the colonies were transferred to liquid medium via an agar bore from a sterile Pasteur pipette. Seven bacterial isolates were obtained, visualized by phase-contrast microscopy, and compared by partial SSU rRNA gene sequence determination. The seven isolates displayed at least 98.9% sequence identity and had a mean sequence dissimilarity of 0.004% ± 0.002% based on partial (V2-V6 region) SSU rRNA gene sequences (approximately 500 nucleotides). Differences in phenotypic characteristics can sometimes be observed in microorganisms with >98% sequence identity in the SSU rRNA gene, but the isolates had similar metal-reducing capacities. Isolate QYMF is described in detail.
Cell morphology.
QYMF cells were straight, with some cells being slightly curved, and had a mean length of 3 to 6 μm and an approximate width of 0.5 μm (Fig. 1). The cells were Gram stain positive. Wet mounts of active cultures visualized with a phase-contrast microscope indicated that the cells were motile, but flagella were not observed by SEM. Exponentially growing cells were often observed as single cells, and few chains were formed as the culture entered stationary phase. As the cells entered stationary-phase growth, terminal endospores were observed (Fig. 1b).
FIG. 1.
Scanning electron micrographs of isolate QYMF grown with Fe3+-EDTA at ×17,670 (a) and ×13,020 (b) magnifications. The arrow denotes an endospore. Bars, 1.0 μm.
Growth characteristics.
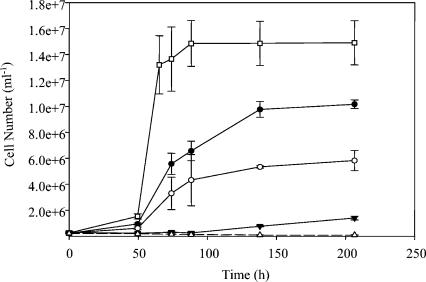
Isolate QYMF was a strict anaerobe and did not display aerobic or fermentative growth with glucose, starch, or sucrose. The isolate did not display significant growth in MR2A medium (7) in the presence or absence of oxygen. Growth was not observed in aerobic growth medium. Yeast extract could be utilized as an electron donor in the presence of Co(III), and when yeast extract levels were increased the Co(III) decreased and the cell numbers increased, respectively (Fig. 2). Similar results were observed with Fe(III) (data not shown). However, little growth was observed when the yeast extract concentration was 0.01 g/liter or less without an additional electron donor, and Co(III) was not reduced at a significant amount (Fig. 2 and 3).
FIG. 2.
Cell growth in the presence of Co(III) and 0.25 (□), 0.1 (•), 0.05 (○), or 0.01 (▾) g of yeast extract per liter determined via direct cell counts. The control with yeast extract only (▵) did not show significant growth. The medium included 2 g of borate per liter and 20 g of sodium chloride per liter, and the pH value was 9.6. The error bars indicate standard deviations of duplicates.
FIG. 3.
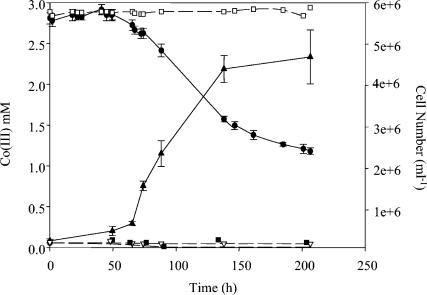
Cell growth in the presence of Co(III), yeast extract (0.01 g/liter), and lactate (10 mM) determined via a decrease in Co(III) (•) and an increase in cell number (▴). The controls included the following: Co(III) only (□), yeast extract only (▿), and yeast extract plus lactate (▪). The medium included 2 g of borate per liter and 20 g of sodium chloride per liter, and the pH value was 9.6. The error bars indicate standard deviations of duplicates.
Growth was not observed when cells were provided with yeast extract alone, nor was growth observed in the presence of yeast extract and lactate (Fig. 3). Small amounts of yeast extract (i.e., 0.01 g/liter) stimulated growth in the presence of lactate and Co(III), and significant growth was not observed unless the lactate was present (Fig. 2). However, when the yeast extract levels were 0.01 g/liter, little growth was observed compared to that with higher levels of yeast extract. Similar results were observed with Fe(III) and Cr(VI) (data not shown). Tryptone, but not Casamino Acids, could be substituted for yeast extract. The final pH of the medium was not significantly changed, and significant growth was observed only with the metals Fe(III), Co(III), and Cr(VI).
The isolate could only be grown in the presence of complexed Fe(III) (citrate or EDTA), Cr(VI), and Co(III)-EDTA, as well as the humic acid analog AQDS. Lactate could serve as the electron donor with the metals listed above as electron acceptors, but fumarate, glycine, histidine, alanine, or arginine did not support growth. Nor did the oxidation and reduction of glycine-alanine or arginine-histidine appear to support growth. Growth was not observed with the following electron acceptors: akaganeite, manganese oxides, trimethylamine oxide, dimethyl sulfoxide, thiosulfate, fumarate, and nitrate. Cells could reduce AQDS in the presence of yeast extract, but the addition of AQDS did not promote the reduction of akaganeite.
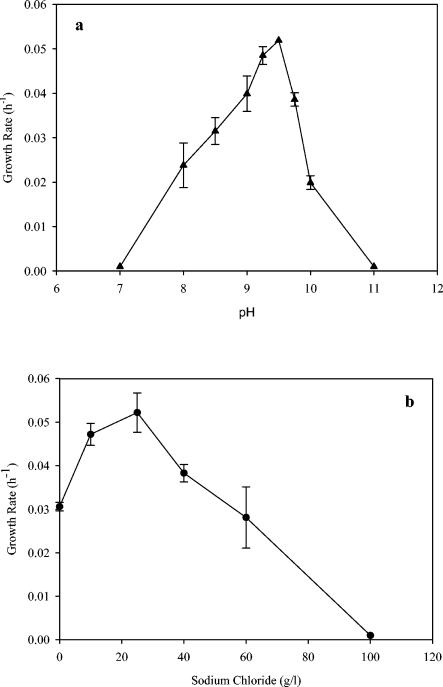
The temperature range for growth was between 4 and 45°C, and the optimum temperature was approximately 35°C. The generation time was approximately 2.3 h at 35°C and approximately 4 h at 22°C when cells were grown in the presence of AQDS (5 mM) at pH 9.5. Little growth was observed above 45°C. When the isolate was grown in the presence of Fe(III)-citrate and yeast extract at 22°C, the pH optimum was approximately 9.5, and growth at pH 7.0 or 11.0 was negligible (Fig. 4a). The salt NaCl was not essential for growth, but the optimal concentration was approximately 20 g/liter (Fig. 4b). The growth rate was decreased threefold when cells were grown in the presence of 80 g/liter, and growth was negligible at 100 g/liter.
FIG. 4.
Relationship between sodium chloride concentration (b) or pH (a) and growth rate when yeast extract and Fe(III)-citrate were provided as electron donor and acceptor, respectively. The borate concentration was 2 g/liter. The pH value was 9.6 for the determination of the pH optimum, and the salt concentration was 20 g/liter for the optimal pH determination. The error bars indicate standard deviations of duplicates. It should be noted that, when sodium chloride was not added, the medium still contained approximately 8 mM sodium from sodium carbonate.
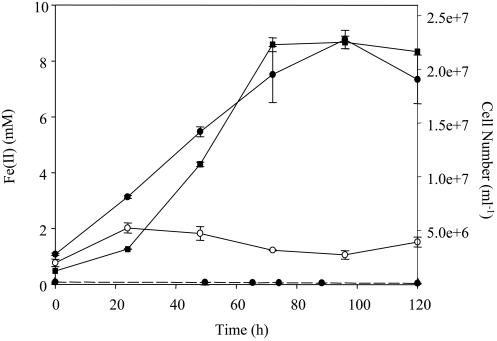
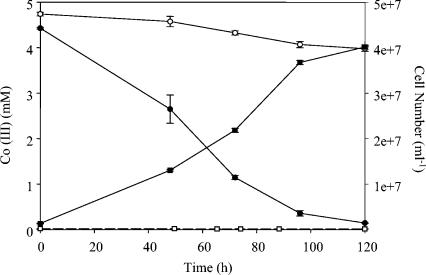
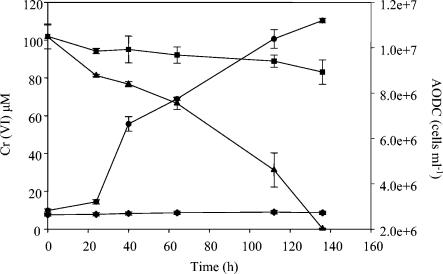
QYMF could reduce Fe(III), Co(III), and Cr(VI) with a concomitant increase in cell number. When Fe(III)-citrate served as the electron acceptor and yeast extract was the electron donor, the growth rate was approximately 0.05 h−1 and a brown precipitate was formed (Fig. 5). Complexed Fe(III)-EDTA could also support growth in a similar fashion, but growth resulted in the formation of a white precipitate when the headspace was H2-CO2 (80:20). The growth rate in the presence of 4.5 mM Co(III) was more than twofold lower than with 15 mM Fe(III) (Fig. 6), but increased growth rates were observed with lower Co(III)-EDTA concentrations. An increase in cell number was also observed during the reduction of 100 μM Cr(VI) and yeast extract (Fig. 7).
FIG. 5.
Production of Fe(II) (•) and increase in cell number (▪) when cells were grown with Fe(III), yeast extract, and 20 g of sodium chloride per liter at pH 9.6. The controls included Fe(III) only (○) and yeast extract only (• with dashed line). The error bars indicate standard deviations of duplicates.
FIG. 6.
Decrease in Co(III)-EDTA (•) and increase in cell number (▪) when cells were grown with Co(III), yeast extract, and 20 g of sodium chloride per liter at pH 9.6. The controls included Co(III) only (○) and yeast extract only (□). The error bars indicate standard deviations of duplicates.
FIG. 7.
Decrease in Cr(VI) (▴) and increase in cell number (•) when cells were grown with Cr(VI), yeast extract, and 20 g of sodium chloride per liter at pH 9.6. The controls included Cr(VI) only (▪) and yeast extract only (♦). The error bars indicate standard deviations of duplicates.
Mineralogy.
Cells grown at pH 9.0 with Fe(III)-EDTA as an electron acceptor and yeast extract and lactate as electron donors in medium with a carbonate buffer formed a white precipitate. Analysis with SEM and EDX showed that the precipitated phase contained iron and phosphorus with keg or barrel morphology and a pronounced platy habit (data not shown). X-ray diffraction analysis of the precipitate displayed sharp and intense reflection patterns, and the crystalline precipitates had X-ray diffraction patterns indicative of Fe3(PO4)2 · 8H2O (vivianite).
SSU rRNA gene-based relationships.
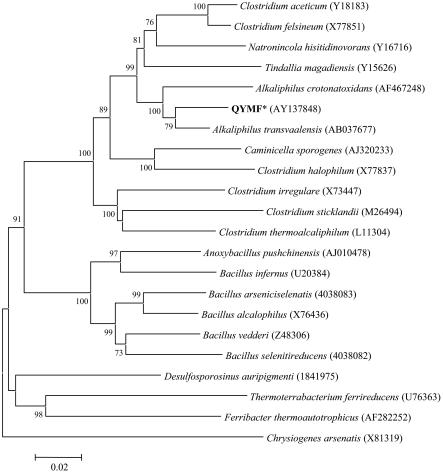
The SSU rRNA gene was amplified from genomic DNA, and the sequence was determined with multiple primers. The isolate was a low-G+C gram-positive bacterium (Fig. 8). The SSU rRNA gene of the isolate formed a cluster with two other sequences from the recently named genus Alkaliphilus. The sequence of isolate QYMF had 96 and 92% nucleotide identity with Alkaliphilus transvaalensis and Alkaliphilus crotonatoxidans, respectively. Both A. transvaalensis and A. crotonatoxidans are reported to be strict anaerobes that form endospores (4, 37). The next closest related sequences were from Clostridium aceticum and Clostridium felsineum (91% identity). The strain QYMF also clustered with Tindallia magadiensis and Natronincola histidinovorans, two alkaliphilic bacteria isolated from a soda lake in Kenya (14, 47).
FIG. 8.
Phylogenetic relationships between isolate QYMF and other alkaliphilic or metal-reducing bacteria based on nearly complete SSU rRNA gene sequences. Sequences were aligned with ClustalW, and pairwise deletions were used for neighbor joining. Bootstrap (n = 500) values below 50 are not shown, and Chrysiogenes arsenatis was used as the outgroup. GenBank accession numbers are in parentheses.
Polar lipid fatty acid analysis.
Lipid extraction and gas chromatography analysis detected the following cellular fatty acids at the given molar percentages: 0.6% i14:0, 14% 14:1, 0.3% 14:1ω5c, 5.4% 14:0, 0.8% br14:1, 0.8% i15:0, 1% a15:0, 3% i15:1, 2.2% 15:1, 1.5% 15:0, 0.6% br15:1, 2.3% 16:1, 27% 16:1ω7c, 1.1% 16:1, 28% 16:0, 0.5% 17:1, 0.8% 17:0, 1.1% 18:1ω9c, 0.4% 18:1ω7c, and 4.1% 18:0. The terminal branched fatty acid, 16:1ω7c; 16:0; and the monounsaturated acid, 14:1, were the most predominant fatty acids, constituting almost 70% of the fatty acid composition. The unbranched saturated acids, 14:0 and 18:0, constituted 9.5% of the total.
DISCUSSION
By definition, strict alkaliphiles should have an optimum pH above 9.0 and display little or no growth at a near-neutral pH of 6.5 (10, 12). Thus, QYMF can be described as an alkaliphile that grew between the pH values of 7.5 and 11. The optimum pH value for growth was approximately 9.5, and this value is similar to the pH values observed with the original samples. The closest known relative, A. transvaalensis, could grow in the pH range of 8.5 to 12.5 and had a slightly higher pH optimum of 10.0 (37). The other known Alkaliphilus species, A. crotonatoxidans, has a pH range of 5.5 to 9.0 but has a pH optimum of 7.5 (4). T. magadiensis, an alkaliphilic bacterium isolated from Lake Magadi, Kenya, had a pH growth range of 7.5 to 10.5 and had an optimum of pH 8.5 (47). Another alkaliphile isolated from Lake Magadi, N. histidinovorans, had a pH profile more similar to that of strain QYMF and grew at pH values from 8.0 to 10.5 with an optimum of 9.4. However, for both T. magadiensis and N. histidinovorans, only the fermentation of a limited number of amino acids supported growth with the production of ammonia.
Isolate QYMF had a decreased growth rate when sodium chloride was not added to the medium, and the growth rate was significantly diminished when the salt concentration was >6%. These results suggested that the microorganism was moderately halophilic, and that the sodium chloride requirement and sensitivity determined in pure culture were similar to the measured sodium levels from the leachate ponds (between 2.0 and 30 g/liter). The alkaliphile A. transvaalensis was reported to have an optimal growth rate with 0.5% sea salts and could tolerate 3.3% sea salts; however, values with sodium chloride were not reported for the microorganism (37). The most closely related Clostridium spp., C. aceticum and C. felsineum, are not halophilic; N. histidinovorans has a higher salinity optimum of 9% (14), and T. magadiensis had a similar range of tolerance between 3 and 6% (47).
The terminal branched fatty acid, 16:1ω7c, and 16:0 were two of the three most predominant lipids, comprising 55% of the fatty acid composition for strain QYMF when cells were grown with Fe(III). The unbranched saturated acids, 14:0 and 18:0, constituted 9.5% of the total and are fatty acids commonly identified in bacteria. The major lipids for the most closely related microorganism, A. transvaalensis, were i15:0, 14:0, and 16:0, and this is similar to those observed for thermophilic clostridia (37) as well as A. crotonatoxidans (4). The fatty acids i15:0 and i17:0 were not detected in strain QYMF at significant levels, and these results indicated that strain QYMF had a different membrane composition than did the previously observed Alkaliphilus spp. The strain QYMF also had a dramatically different lipid profile compared to alkaliphilic Bacillus spp., namely, the differences in 14:0, 16:0, and 15:0 derivatives; 16:1ω7c content; and overall total unsaturated lipids (43).
Previous studies have indicated that Shewanella species grown in the presence of different substrates have different fatty acid compositions (see discussion in reference 46). The two reported Alkaliphilus species were grown with different electron acceptors, and the different growth substrates could have affected the fatty acid composition of the respective cells. Metal reduction in the other two Alkaliphilus spp. (A. transvaalensis and A. crotonatoxidans) has not been reported. Isolate QYMF displayed a lipid profile different from those of other alkaliphilic microorganisms and had a higher percentage of total unsaturated fatty acids, similar to gram-negative microorganisms. Interestingly, the distantly related metal-reducing bacteria Shewanella algae and Geobacter metallireducens both had levels of 16:1ω7c similar to that of strain QYMF when grown with Fe(III) (46). The results suggested that 16:1ω7c could be a common membrane component observed in metal-reducing bacteria, but further work is needed. The lipid 16:1ω7c has not been reported in other alkaliphilic species; however, the microorganisms were not or could not be grown with Fe(III).
The majority of metal-reducing bacteria are classified as δ- and γ-Proteobacteria and, to a lesser extent, gram-positive bacteria. The closest gram-positive metal reducers were T. magadiensis (91%), Bacillus infernus (83%), and “Ferribacter thermoautotrophicus” (81%) based on SSU rRNA gene sequences. Iron reduction was reported in T. magadiensis, but the pH of the medium was not stated, the optimum pH for T. magadiensis is only 8.5, and T. magadiensis has only 91% SSU rRNA gene sequence identity with isolate QYMF (14). Current data suggest that these other gram-positive bacteria do not reduce metals above pH 9.0 under tested growth conditions, and these results suggested that isolate QYMF was a novel and unique metal-reducing bacterium based on SSU rRNA gene sequence and phenotypes.
Growth of QYMF cells increased with increasing concentrations of yeast extract, and a similar result was observed with A. transvaalensis (37). A. crotonatoxidans is a strict anaerobe that can utilize yeast extract, peptone, tryptone, and some sugars, but growth was not stimulated in the presence of fumarate, sulfur, and thiosulfate (4). A. transvaalensis is also a strict anaerobe that can utilize proteinaceous substrates, and growth was improved with thiosulfate, sulfur, or fumarate (37). Isolate QYMF could utilize yeast extract in the presence of metals, but thiosulfate, fumarate, dimethyl sulfoxide, or sulfate was not used. At the time of sample collection, prominent blooms of algae and cyanobacteria were noted at the leachate ponds, and these might serve as a source of proteinaceous material for strain QYMF and other microbiota. The genus Bacillus contains alkaliphilic organisms that can reduce oxyanions (arsenate and/or selenate) (35), but little information exists in the literature regarding anaerobic alkaliphilic respiration with sulfur compounds or metals. The genus Alkaliphilus may represent a novel group of anaerobic alkaliphilic bacteria that can utilize proteinaceous material and reduce metals and sulfur compounds.
The growth medium contained phosphate and most likely explained the production of vivianite, as confirmed with EDX. When the buffer was changed from carbonate to Tris or the headspace was changed from nitrogen to hydrogen, the precipitates changed size and morphology, suggesting that variation in the chemical milieu altered the types of observed precipitates. These results indicated that alkaline mineral precipitation may vary according to the dynamics of the chemical environments, but further work is needed to discern the relationships between mineralogy and biochemistry.
Shewanella putrefaciens has been shown to reduce Fe(III)-citrate but not Fe(III)-EDTA (9). The authors suggested the possible importance of ligands to promote bioavailability of Fe(III) from ferrihydrite or other iron oxides in natural habitats (8). Based on the solubility and free energy of ferrihydrite, terrestrial seawater at pH 8.1 can potentially have approximately 20 nM dissolved iron (19). Isolate QYMF could reduce Fe(III)-citrate and Fe(III)-EDTA at significant rates above pH values of 9.0, but insoluble iron was not reduced at elevated pH values. These results suggested that the reduction of insoluble iron was limited by the unavailability of soluble iron at pH values above 9.0; however, further work is needed to characterize the metal-reducing capacity of isolate QYMF and the effects of culture conditions including pH.
Soda lakes, with elevated pH and salinity, are typically located in arid environments and constitute a habitat similar to the borax leachate ponds (12). Both cultivation-based and molecular studies have been done with samples from soda lakes (5, 11, 12), and these environments can contain extreme metabolic diversity within an autonomous microbial community. Zavarzin (44) has argued that continental soda lakes were a possible origin of bacterial diversity. Although present-day soda lakes and other alkaline environments are geologically recent, alkaline habitats have most likely existed since archaean times (11).
In the context of astrobiology, the chemical energy derived from metal redox cycles has been recognized as a plausible source for extraterrestrial microbial life (29, 32, 33). Recently, Kempe and Kazmierczak (13) hypothesized that the Jovian moon Europa had low initial Ca(II) levels that would have promoted biogenesis and that an alkaline saline ocean currently exists (26). The identification of microorganisms from environments possibly analogous to extraterrestrial habitats (e.g., in redox cycles, pH, salinity, and temperature) is ever important (16) and can provide data to improve models for exobiological evolution and detection.
Isolate QYMF is a unique bacterium classified in the recently named Alkaliphilus genus and was able to reduce metals in alkaline pH. Future work is needed to assess the possible niches of anaerobic respiring microorganisms in alkaline environments, to more fully understand the role of metal-reducing microorganisms in alkaline environments, and to assess the potential applicability for metal reduction during bioimmobilization of toxic heavy metals. Isolate QYMF was recently selected for whole-genome sequence determination at the Joint Genome Institute, and the genome sequence data will provide insight into the novel and unique lifestyle of this microorganism.
Description of sp. nov. Alkaliphilus metalliredigens.
Alkaliphilus metalliredigens (metallum, L., metal; redigere, L., to reduce to a specific state or condition). Cells are straight, with some cells being slightly curved, and have a mean length of 3 to 6 μm and an approximate width of 0.5 μm. The cells stain gram positive and form terminal endospores. The strain QYMF is an obligate anaerobe and has a pH optimum of 9.5, a temperature optimum of 35°C, and sodium chloride optimum of approximately 2%. The organism can grow in the presence of 10 mM sodium borate. Yeast extract can be used as an electron donor, and small amounts stimulate growth in the presence of lactate or acetate. The electron donors yeast extract and lactate or acetate can be utilized with a reduction of Fe(III)-citrate or Fe(III)-EDTA. Co(III)-EDTA, Cr(VI), or AQDS can also be reduced. Strain QYMF does not utilize fumarate, nitrate, dimethyl sulfoxide, trimethylamine oxide, thiosulfate, sulfate, glycine, arginine, histidine, or alanine as electron acceptors under the tested conditions. The type strain was isolated from a borax-contaminated alkaline leachate pond in Boron, Calif. The type strain is A. metalliredigens strain QYMF.
Acknowledgments
We thank T. J. Phelps and R. Lauf at ORNL and Robert Bates at the U.S. Borax Company for samples from leachate ponds and S. C. Brooks for helpful discussions.
Y. Roh was supported by Seed Money Project 01-3210-006L. This work was supported by the United States Department of Energy via the Natural and Accelerated Bioremediation Research Program, Biological Investigation-Ocean Margin Program, and the Microbial Genomes Program, Office of Biological and Environmental Research, Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for DOE under contract no. DE-AC05-96OR22464.
REFERENCES
- 1.Blair, B. G., and K. L. Anderson. 1998. Comparison of staining techniques for scanning electron microscopic detection of ultrastructural protuberances on cellulolytic bacteria. Biotech. Histochem. 73:107-113. [DOI] [PubMed] [Google Scholar]
- 2.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 3.Caccavo, F., Jr., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60:3752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, X., X. Liu, and X. Dong. 2003. Alkaliphilus crotonatoxidans sp. nov., a strictly anaerobic, crotonate-dismutating bacterium isolated from a methanogenic environment. Int. J. Syst. E vol. Microbiol. 53:971-975. [DOI] [PubMed] [Google Scholar]
- 5.Duckworth, A. W., W. D. Grant, B. E. Jones, and R. van Steenbergen. 1996. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol. Ecol. 19:181-191. [Google Scholar]
- 6.Fredrickson, J. K., and Y. A. Gorby. 1996. Environmental processes mediated by iron-reducing bacteria. Curr. Opin. Biotechnol. 7:287-294. [DOI] [PubMed] [Google Scholar]
- 7.Fries, M. R., J. Zhou, J. Chee-Sanford, and J. M. Tiedje. 1994. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl. Environ. Microbiol. 60:2802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, J. R., and T. J. Dichristina. 2002. Effects of Fe(III) chemical speciation on dissimilatory Fe(III) reduction by Shewanella putrefaciens. Environ. Sci. Technol. 36:373-380. [DOI] [PubMed] [Google Scholar]
- 9.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of nucleopore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horikoshi, K. 1999. Alkaliphiles: some application of their products for biotechnology. Microbiol. Mol. Biol. Rev. 63:735-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, B. E., W. D. Grant, A. W. Duckworth, and G. G. Owenson. 1998. Microbial diversity of soda lakes. Extremophiles 2:191-200. [DOI] [PubMed] [Google Scholar]
- 12.Jones, B. E., and W. D. Grant. 2000. Microbial diversity and ecology of alkaline environments, p. 171-190. In J. Seckbach (ed.), Journey to diverse microbial worlds. Kluwer Academic Publishers, Dordrecht, the Netherlands.
- 13.Kempe, S., and J. Kazmierczak. 2002. Biogenesis and early life on Earth and Europa: favored by an alkaline ocean. Astrobiology 2:123-130. [DOI] [PubMed] [Google Scholar]
- 14.Kevbrin, V. V., T. N. Zhilina, F. A. Rainey, and G. A. Zavarzin. 1998. Tindallia magadii gen. nov., sp. nov., an alkaliphilic anaerobe ammonifier from soda lake deposits. Curr. Microbiol. 37:94-100. [DOI] [PubMed] [Google Scholar]
- 15.Kirchman, D. L. 1993. Statistical analysis of direct counts of microbial abundance, p. 117-119. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, New York, N.Y.
- 16.Knauth, L. P., and D. M. Burt. 2001. Follow the water, beware the brine: astrobiological implications of aqueous seeps on Mars. Astrobiology 1:350-381. [Google Scholar]
- 17.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 18.Kusel, K., T. Dorsch, G. Acker, and E. Stackebrandt. 1999. Microbial reduction of Fe(III) in acidic sediments: isolation of Acidiphilum cryptum JF-5 capable of coupling the reduction of Fe(III) to the oxidation of glucose. Appl. Environ. Microbiol. 65:3633-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmuir, D. 1997. Aqueous environmental geochemistry, p. 431-485. Prentice-Hall, Old Tappan, N.J.
- 20.Liu, S. V., J. Zhou, C. Zhang, D. R. Cole, M. Gajdarziska-Josifovska, and T. J. Phelps. 1997. Thermophilic Fe(III)-reducing bacteria from the deep subsurface: the evolutionary implications. Science 277:1106-1109. [Google Scholar]
- 21.Lovley, D. R., and E. J. P. Phillips. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovley, D. R. 1993. Dissimilatory metal reduction. Annu. Rev. Microbiol. 47:263-290. [DOI] [PubMed] [Google Scholar]
- 23.Lovley, D. R., and E. J. P. Phillips. 1994. Reduction of chromate by Desulfovibrio vulgaris and its c3 cytochrome. Appl. Environ. Microbiol. 60:726-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovley, D. R., J. D. Coates, E. L. Blunt-Harris, E. J. P. Phillips, and J. C. Woodward. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445-448. [Google Scholar]
- 25.Matterson, K. J. 1980. Borate ore discovery, mining and beneficiation, section A3, vol. 5. In R. Thompson and A. J. E. Welch (ed.), Supplement to Mellor's comprehensive treatise on inorganic and theoretical chemistry. Longman, New York, N.Y.
- 26.McCord, T. B., G. B. Hansen, F. P. Fanale, R. W. Carlson, D. L. Matson, T. V. Johnson, W. D. Smythe, J. K. Crowley, P. D. Martin, A. Ocampo, C. A. Hibbitts, and J. C. Granahan. 1998. Salts on Europa's surface detected by Galileo's near infrared mapping spectrometer. Science 280:1242-1245. [DOI] [PubMed] [Google Scholar]
- 27.Mills, W. B., J. Y. Loh, M. C. Bate, and K. M. Johnson. 1999. Evaluation of potential risks from ash disposal site leachate. J. Environ. Eng. 125:306-313. [Google Scholar]
- 28.Nealson, K. H., and C. R. Myers. 1992. Microbial reduction of manganese and iron: new approaches to carbon cycling. Appl. Environ. Microbiol. 58:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nealson, K. H., and B. L. Cox. 2002. Microbial metal-ion reduction and Mars: extraterrestrial expectations? Curr. Opin. Microbiol. 5:296-300. [DOI] [PubMed] [Google Scholar]
- 30.Ohlendorf, H. M., D. J. Hoffman, M. K. Saiki, and T. W. Aldrich. 1986. Embryonic mortality and abnormalities of aquatic birds: apparent impacts of selenium from irrigation drainage water. Sci. Total Environ. 52:49-63. [Google Scholar]
- 31.Roh, Y., R. J. Lauf, A. D. McMillan, C. Zhang, C. J. Rawn, J. Bai, and T. J. Phelps. 2001. Microbial synthesis and the characterization of some metal-doped magnetite. Solid State Commun. 110:529-534. [Google Scholar]
- 32.Schulze-Makuch, D. 2002. At the crossroads between microbiology and planetology. ASM News 68:364-365. [Google Scholar]
- 33.Schulze-Makuch, D., and L. N. Irwin. 2002. Energy cycling and hypothetical organisms in Europa's ocean. Astrobiology 2:105-121. [DOI] [PubMed] [Google Scholar]
- 34.Seiler, J. P. 1989. The mutagenic activity of sodium perborate. Mutat. Res. 224:219-227. [DOI] [PubMed] [Google Scholar]
- 35.Stolz, J. F., and R. S. Oremland. 1999. Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23:615-627. [DOI] [PubMed] [Google Scholar]
- 36.Straub, K. L., M. Benz, and B. Schink. 2001. Iron metabolism in anoxic environments at near neutral pH. FEMS Microbiol. Rev. 34:181-186. [DOI] [PubMed] [Google Scholar]
- 37.Takai, D. P. Moser, T. C. Onstott, N. Spolelstra, S. M. Pfiffner, A. Dohnalkova, and J. K. Fredrickson. 2001. Alkaliphilus transvaalensis gen. nov., sp. nov., an extremely alkaliphilic bacterium isolated from a deep South African gold mine. Int. J. Syst. E vol. Microbiol. 51:1245-1256. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weigel, J., and J. Hanel. 2002. Chemolithoautotrophic thermophilic iron (III) reducer, p. 235-251. In L. Ljungdahl, M. W. W. Adams, M. Johnson, and T. L. Baxton (ed.), Biochemistry and physiology of anaerobic bacteria. Springer-Verlag, New York, N.Y.
- 40.Weisburg, W. W., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White, D. C., W. M. Davis, J. S. Nickels, J. D. King, and R. J. Bobbie. 1979. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 40:51-62. [DOI] [PubMed] [Google Scholar]
- 42.Woods, W. G. 1994. An introduction to boron: history, sources, uses, and chemistry. Environ. Health Perspect. 102:5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yumoto, I., K. Yamazaki, M. Hishinuma, Y. Nodasaka, N. Inoue, and K. Kawasaki. 2000. Identification of facultatively alkaliphilic Bacillus sp. strain YN-2000 and its fatty acid composition and cell-surface aspects depending on culture pH. Extremophiles 4:285-290. [DOI] [PubMed] [Google Scholar]
- 44.Zavarzin, G. A. 1993. Epicontinental soda lakes as probable relict biotopes of terrestrial biota formation. Microbiology 62:473-479. [Google Scholar]
- 45.Zhang, C., R. D. Stapleton, J. Zhou, A. V. Palumbo, and T. J. Phelps. 1999. Iron reduction by psychrotrophic enrichment cultures. FEMS Microbiol. Ecol. 30:367-371. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, C. L., Y. Li, E. Yi, J. Fong, A. D. Peacock, E. Blunt, D. Lovley, and D. C. White. 2002. Carbon isotope signatures of fatty acids in Geobacter metallireducens and Shewanella algae. Chem. Geol. 195:17-28. [Google Scholar]
- 47.Zhilina, T. N., E. N. Detkova, F. A. Rainey, G. A. Osipov, A. M. Lysenko, N. A. Kostrikina, and G. A. Zavarzin. 1998. Natronoincola histidinovorans gen. nov., sp. nov., a new alkaliphilic acetogenic anaerobe. Curr. Microbiol. 37:177-185. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, J.-Z., M. R. Fries, J. C. Chee-Sanford, and J. M. Tiedje. 1995. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene and description of Azoarcus tolulyticus sp. nov. Int. J. Syst. Bacteriol. 45:500-506. [DOI] [PubMed] [Google Scholar]