Abstract
This study aimed to define the origin of Salmonella contamination on swine carcasses and the distribution of Salmonella serotypes in two commercial slaughterhouses during normal activity. Salmonellae were isolated from carcasses, from colons and mesenteric lymph nodes of individual pigs, and from the slaughterhouse environment. All strains were serotyped; Salmonella enterica serotype Typhimurium and Salmonella enterica serotype Derby isolates were additionally typed beyond the serotype level by pulsed-field gel electrophoresis (PFGE) and antibiotic resistance profiling (ARP); and a subset of 31 serotype Typhimurium strains were additionally phage typed. PFGE and ARP had the same discriminative possibility. Phage typing in combination with PFGE could give extra information for some strains. In one slaughterhouse, 21% of the carcasses were contaminated, reflecting a correlation with the delivery of infected pigs. Carcass contamination did not result only from infection of the corresponding pig; only 25% of the positive carcasses were contaminated with the same serotype or genotype found in the corresponding feces or mesenteric lymph nodes. In the other slaughterhouse, 70% of the carcasses were contaminated, and only in 4% was the same genotype or serotype detected as in the feces of the corresponding pigs. The other positive carcasses in both slaughterhouses were contaminated by genotypes present in the feces or lymph nodes of pigs slaughtered earlier that day or from dispersed sources in the environment. In slaughterhouses, complex contamination cycles may be present, resulting in the isolation of many different genotypes circulating in the environment due to the supply of positive animals and in the contamination of carcasses, probably through aerosols.
The genus Salmonella includes more than 2,400 different serotypes (19). The Kauffmann-White scheme that is used to serotype salmonellae is based on antigenic polymorphisms of the lipopolysaccharides (LPS) (O) and the flagella (H). A second level of characterization is based on phage typing. By use of 37 different phages, serotype Typhimurium can be divided in more than 210 phage types (1). Besides serotyping and phage typing, powerful bacterial molecular typing methods, such as plasmid profiling, pulsed-field gel electrophoresis (PFGE), IS200 typing, ribotyping, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism, are used for epidemiological investigation of salmonellae (3, 7, 8, 13, 16). These techniques are useful for defining clonal relationships between strains (17) and for assessing the distribution of Salmonella strains within food-processing environments (10, 15).
Within Salmonella and especially Salmonella enterica serotype Typhimurium, multiple-antibiotic-resistant strains are isolated with increased frequency. Serotype Typhimurium definitive type 104 (DT104) and serotype Typhimurium DT204b are virulent pathogens for humans and animals, with many strains showing multiple drug resistance characteristics (2, 14, 24). These multiple-antibiotic-resistant serotype Typhimurium strains cause particular concern because of their increasing prevalence in humans. Most of the strains typically carry resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (R type), but resistance to 9 or 10 different antibiotics also occurs (23). The selective pressure created by widespread use of antimicrobial agents in pigs during rearing may have contributed to the dissemination of these multidrug-resistant bacterial strains (7). It is well established that the distribution of antimicrobial resistance is often plasmid and/or transposon mediated. Integrons, a novel group of mobile DNA elements, have the potential to incorporate several antibiotic resistance genes by site-specific recombination (20).
In a previous study the prevalences of salmonellae in several slaughterhouses were determined (6). Salmonellae were isolated from 37% of the carcass samples collected during eight slaughterhouse visits. Overall 28% of the animals carried salmonellae in the feces and/or mesenteric lymph nodes. Swanenburg (21) found only 1.4% of the carcasses to be positive whereas 25.6% of the fecal samples were positive. At the slaughterhouse, two major contamination parameters are important: the status of the pigs supplied and slaughterhouse hygiene. Berends et al. (4) and Borch et al. (5) considered infected pigs to be the initial source of carcass contamination (calculated at 70%) and estimated that cross-contamination accounted for 30%.
In this study Salmonella strains, isolated from pigs and carcasses during a whole slaughtering day and from the slaughterhouse environment, were characterized. By using PFGE (serotype Typhimurium, serotype Ohio, and serotype Derby isolates) and phage typing (serotype Typhimurium isolates), the origin of Salmonella contamination in the carcasses and in the slaughterhouse environment was determined. Also, antibiotic resistance profiles (ARPs) for six antibiotics were determined in order to monitor the spread of multidrug-resistant strains in the feces of the pigs, on the carcasses, and in the slaughterhouse environment.
MATERIALS AND METHODS
Salmonella isolates.
All the strains used in this study were collected during a whole slaughtering day in two different commercial slaughterhouses (A2 and D1 [slaughterhouse D, time point 1]) (6). The slaughterhouse environments were sampled by taking overshoe samples at different time points (before and during slaughter in the morning and in the afternoon). A total of 17 environmental samples were taken in slaughterhouse A2, while 22 were taken in slaughterhouse D1. Swabs of the slaughterhouse equipment, knives, and splitting machine were collected (5 at A2 and 14 at D1). Animal samples (colon contents and mesenteric lymph nodes) were collected in A2 (240 samples) and D1 (220 samples) from each 25th and 50th pig, respectively; the pig carcass was swabbed according to the method of Korsak et al. (12). A feces sample (5 g of colon contents) and the mesenteric lymph nodes (10 g) were analyzed. This method of sampling resulted in corresponding carcass, fecal, and mesenteric lymph node samples originating from individual pigs. During a second visit to slaughterhouse D, 1 month later, samples were taken at random (D2).
All the samples were first incubated in buffered peptone water (1/10) (Oxoid Ltd., Basingstoke, United Kingdom) at 37°C overnight. For the isolation of Salmonella strains, a selective enrichment step in 10 ml of Rappaport-Vassiliadis enrichment broth (RV; Oxoid) and on the semisolid medium Diassalm (LabM, Bury, United Kingdom) at 42°C followed. After 24 h of incubation, a 10-μl loop of the RV culture or a 1-μl loop from a purple migration zone present on Diassalm was streaked onto xylose lysine desoxycholate agar (XLD; Oxoid) and incubated at 37°C for 24 h. Presumptive Salmonella colonies (black) on XLD were confirmed by PCR (6). All the Salmonella isolates were initially typed to the serotype level by repetitive extragenic palindromic sequence PCR (6).
ARP.
The ARPs of all the Salmonella isolates were determined. Six different antibiotics were tested: ampicillin (A) (A-9518), tetracycline (T) (T-3383), streptomycin (S) (S-6501), nalidixic acid (Nal) (N-4382), chloramphenicol (Ch) (C-0378), and sulfadiazine (Su) (S-8626) (Sigma-Aldrich, St. Louis, Mo.). The MIC was determined by making a twofold serial dilution of the antibiotic in H2O in a microtiter plate. An overnight Salmonella culture grown in Mueller-Hinton broth (Oxoid) at 37°C was diluted to 10−4 CFU/ml in Mueller-Hinton broth, and 100 μl of this dilution was exposed to different antibiotic concentrations at 37°C for 18 h (ampicillin, 6,666 to 0.06 μg/ml; tetracycline and streptomycin, 666 to 0.6 μg/ml; nalidixic acid and chloramphenicol, 66 to 0.06 μg/ml; sulfadiazine, 66,666 to 606 μg/ml). The MIC determined was the lowest concentration of antibiotics with which no Salmonella growth was visible. Resistant, susceptible, and intermediate Salmonella phenotypes were based on the population distribution of all MICs. Isolates were susceptible to ampicillin when the MIC was <8 μg/ml and resistant at a MIC of >32 μg/ml; susceptible, intermediate, or resistant to tetracycline at a MIC of <4, 8, or >16 μg/ml, respectively; resistant to streptomycin at a MIC of >16 μg/ml; susceptible or resistant to nalidixic acid at a MIC of <16 or >32 μg/ml, respectively; susceptible or resistant to chloramphenicol at a MIC of <8 or >32 μg/ml, respectively; and susceptible or resistant to sulfadiazine at a MIC of <256 or >512 μg/ml, respectively.
Phage typing.
A selection of 31 serotype Typhimurium isolates with different ARPs was sent to the Institute Pasteur—Brussels (Brussels, Belgium) for phage typing.
PFGE.
All serotype Typhimurium, serotype Derby, and serotype Ohio isolates were genotyped by PFGE after single digestion with XbaI and BlnI for serotypes Typhimurium and Ohio and with XbaI and SpeI for serotype Derby.
After growth with shaking in Luria broth (Invitrogen Ltd., Paisley, United Kingdom) at 37°C overnight, genomic DNAs of the Salmonella isolates were prepared in agarose plugs. The PFGE protocol described by Pasmans et al. (18) was followed. A slice of the plug was digested separately with XbaI and BlnI for serotypes Typhimurium and Ohio and with XbaI and SpeI for serotype Derby (Amersham Pharmacia Biotech, Uppsala, Sweden) at 37°C for 4 h in a water bath according to the manufacturer's instructions. The resulting genomic fragments were separated by PFGE using the contour-clamped homogeneous electric field method (CHEF-DRII; Bio-Rad Laboratories, Richmond, Calif.). The fragments were separated in a 1% (wt/vol) GTG agarose (FMC Bioproducts) gel in 0.5× TBE buffer (0.09 M Tris, 2 mM disodium EDTA [pH 8.5], 0.09 M boric acid) at a constant temperature of 14°C at 6 V/cm, with pulse times of 4 s for11 h and of 40 s for 13 h for the restriction enzymes XbaI and SpeI and with pulse times of 7 s for 11 h and of 65 s for 13 h for BlnI. Lambda DNA concatemers (size range, 50 to 1,000 kb) (New England Biolabs, Beverly, Mass.) were used as molecular size standards. To prevent DNA degradation during electrophoresis for typing of the serotype Ohio strains, 100 μM thiourea (Calbiochem, Darmstadt, Germany) was added to the running buffer.
The high-molecular-weight DNA fragments were visualized by ethidium bromide staining, and images were digitized under UV illumination by the GelDoc 2000 System (Bio-Rad Laboratories).
Numerical analysis of PFGE profiles.
PFGE patterns of serotype Typhimurium, serotype Ohio, and serotype Derby digested with the different restriction enzymes were analyzed separately. Together with visual analyses of the PFGE profiles, a numerical analysis after conversion, normalization, and analysis of similarity in band patterns was performed using the GelCompar II software package (Applied Maths, Ghent, Belgium). Similarities between profiles were calculated using the Dice coefficient, with a maximum position tolerance of 1%. PFGE patterns obtained with the two different restriction enzymes were clustered separately by the unweighted pair group method using arithmetic averages (UPGMA). The capital letter T, O, or D combined with a numerical suffix was used to designate the different PFGE groups for serotype Typhimurium, serotype Ohio, or serotype Derby, respectively.
RESULTS
ARP and phage typing.
For 263 Salmonella isolates collected in slaughterhouses A2 (101 isolates), D1 (140 isolates), and D2 (22 isolates), the resistance to six antibiotics was determined (Table 1). In total, 129 Salmonella isolates were susceptible to all the antibiotics tested. Antibiotic resistance was found for isolates belonging to serotypes Typhimurium, Brandenburg, Derby, Havana, and Rissen. Resistance to one antibiotic was found for tetracycline (10 strains resistant and 8 intermediate), sulfadiazine (5 strains resistant), and ampicillin (1 strain resistant). Resistance to two antibiotics was found for two antibiotic combinations, tetracycline with sulfadiazine (six strains) and chloramphenicol with sulfadiazine (one strain). Thirty strains were resistant to three antibiotics: chloramphenicol (29 strains) or streptomycin (1 strain) with tetracycline and sulfadiazine. Of the strains that were resistant to four antibiotics (25 of 36 strains), most were resistant to ampicillin, chloramphenicol, sulfadiazine, and tetracycline. The other profiles ware ASuST (10 strains) and ChSuST (1 strain). Two ARPs, AChSuST and AChSuTNal, were found for the strains resistant to five antibiotics. The ARP AChSuST was found in 36 strains, all isolated in the same slaughterhouse (D1). One serotype Derby strain with the ARP AChSuTNal was isolated from feces. The greatest variation was observed within serotype Typhimurium, with nine different ARPs, and serotype Derby, with eight.
TABLE 1.
Origins and characterization of Salmonella isolates from two different slaughterhouses
| Source | Serotype | ARPa | PFGE profileb
|
Phage typec | ||
|---|---|---|---|---|---|---|
| XbaI | BlnI | SpeI | ||||
| A2 | ||||||
| Carcasses | Typhimurium | Susceptible | T8 | T12 | ||
| T | T11 | T11/T11a | RDNC/P50 | |||
| TSu | T5 | T5 | 208 | |||
| ASuST | T4 | T4/T4a | 193 | |||
| AChSuT | T9 (3)/T8 | T9 (3)/T8 | RDNC/P169 (T9) | |||
| Brandenburg | Susceptible | |||||
| (T)I | ||||||
| Derby | Susceptible | D3 | D3 | |||
| T | D2a | D2a | ||||
| Su | D4 | D4 | ||||
| SuST | D2b | D2b | ||||
| Infantis, Livingstone, Virchow, Ohio | Susceptible | O1 | O1 | |||
| Colon contents | Typhimurium | Susceptible | T3/T8/T6 (4) | T3/T12/T6 (4) | ||
| T | T11 | T11 | RDNC/P50 | |||
| ASuST | T4 | T4 (3)/T4a (2) | 120 (T4, T4a); 193 (T4, T4a) | |||
| Brandenburg | Susceptible | |||||
| Su | ||||||
| (T)I | ||||||
| Derby | Susceptible | D1a/D3 | D1a/D3 | |||
| Livingstone, London, Anatum, Ohio, Infantis | Susceptible | O1 | O1 | |||
| Rissen | T | |||||
| Mesenteric lymph nodes | Typhimurium | Susceptible | T11/T3/T8/T6 | T11/T3/T12/T6 | ||
| (T)I | T1 | T1 | ||||
| ASuST | T4 | T4 | 193 | |||
| Brandenburg | Susceptible | |||||
| Su | ||||||
| (T)I | ||||||
| Derby | Susceptible | D1 | D1 | |||
| T | D2a | D2a | ||||
| Livingstone, London, Infantis, Poona, Virchow, Ohio, Goldcoast | Susceptible | O1 | O1 | |||
| Rissen | T | |||||
| Environment | Livingstone, Ohio, Brandenburg | Susceptible | O1 | O1 | ||
| Typhimurium | T | T11 | T11 | RDNC/P50 | ||
| D1 | Derby | Susceptible | D3 | D3 | ||
| Carcasses | Typhimurium | Susceptible | T2 | T2 | 12 | |
| ChSuT | T10 | T10 | 193 | |||
| ChSuST | ND | ND | ||||
| AChSuT | T8 | T8 | ||||
| AChSuST | T8 | T8 | 104L | |||
| Infantis, Anatum | Susceptible | |||||
| Colon contents | Typhimurium | Susceptible | T2 | T2 | 12 | |
| ChSuT | T10 | T10 | 193 | |||
| AChSuST | T10/T8 | T10/T8 | 104L (T8) | |||
| Derby | A | D1 | D1 | |||
| SuT | D2a | D2a | ||||
| AChSuTNal | D1b | D1b | ||||
| Anatum, Livingstone, Panama, 47:Z4Z13:− | Susceptible | |||||
| Mesenteric lymph nodes | Typhimurium | Susceptible | T2 | T2 | ||
| ChSu | T2a | T2a | ||||
| ChSuT | T10 | T10 | ||||
| AChSuT | T8 | T8 | ||||
| Livingstone, Anatum, Agona | Susceptible | |||||
| Derby | SuT | D2 | D2 | |||
| Havana | ChSuST | |||||
| Environment | Typhimurium | Susceptible | T2 | T2 | ||
| ChSuT | T10 | T10 | 193; 21 | |||
| AChSuST | T8 | T8 | 104L | |||
| AChSuT | T8 | T8 | 104L | |||
| Derby | Susceptible | D5 | D5 | |||
| SuT | D2 | D2 | ||||
| ChSuT | ||||||
| D2 | Anatum | Susceptible | ||||
| Carcasses | Typhimurium | AChSuST | T8 | T8 | 104L | |
| Derby | Susceptible | D5 | D5 | |||
| SKGII machine | Derby | Susceptible | D5 | D5 | ||
| Knives | Derby | Susceptible | D5 | D5 | ||
| Environment | Derby | Susceptible | D1 | D1 | ||
| Derby | ChSuT | D1c | D1c | |||
(T)I, intermediate for tetracycline. T4a, T11a, and T2a each show a one-band difference in the PFGE profile after restriction with BlnI (see Fig. 2).
Numbers in parentheses show the number of isolates with a given PFGE type.
ND, not determined. RDNC, phage type not defined (reacts but does not conform to a known type).
Thirty-one serotype Typhimurium isolates could be assigned to nine different phage types; the most dominant types were RDNC/P50, 193, 120, 12, and 104L.
Genotyping. (i) Salmonella serotype Derby.
Twenty-six serotype Derby isolates were digested separately with XbaI and SpeI; six main PFGE profiles were identified with both enzymes (same similarity factor) (Fig. 1; Table 1). Within the D1 and D2 genotype groups, small differences in the restriction pattern could be observed after restriction with both enzymes, resulting in a total of 10 different profiles. These small profile differences correspond with different ARPs: susceptible strains cluster with D1 and D1a, strains with the AChSuTNal ARP cluster with D1b, and D1c corresponds to strains with the ChSuT ARP. The D2 group could be divided into two subgroups after restriction with both enzymes: D2 corresponds to strains with the SuT ARP, D2a corresponds to strains with the T ARP, and D2b corresponds to strains with the SuST ARP.
FIG. 1.
Dendrogram based on combination of the PFGE patterns of Salmonella serotype Derby isolates after XbaI and SpeI DNA digestion. The main PFGE types are numbered; subtypes are designated by lowercase letters. Arrows indicate small differences within a genotype.
(ii) Salmonella serotype Typhimurium.
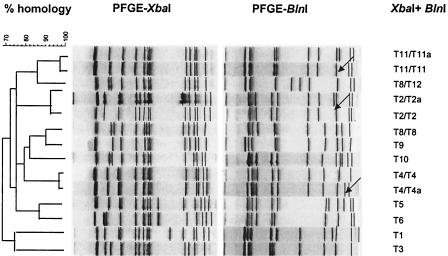
Ten different genotypes were identified among the 140 serotype Typhimurium isolates digested with XbaI (Fig. 2; Table 1). With BlnI digestion, 11 genotypes were identified. After restriction with BlnI, subgroups could be identified within the XbaI T8 genotype: T8/T8 and T8/T12 (Table 1). All the isolates of slaughterhouse A2 that were susceptible to all the antibiotics tested were clustered together in a subgroup of T8 with genotype T8 (XbaI) combined with T12 (BlnI). After BlnI digestion, two subgroups could be distinguished within genotypes T2 (T2/T2 and T2/T2a), T4 (T4/T4 and T4/T4a), and T11 (T11/T11 and T11/T11a).
FIG. 2.
Dendrogram based on combination of the PFGE patterns of a selection of Salmonella serotype Typhimurium isolates after XbaI and BlnI DNA digestion. The main PFGE-types are numbered; subtypes are designated by lowercase letters. Arrows indicates small differences within a genotype.
Genotype T11 corresponds with the tetracycline resistance profile and phage type RDNC/P50; genotype T8/T8 corresponds with the multiple-antibiotic-resistant phenotypes AChSuST and AChSuT and with phage type 104L; genotype T2 corresponds with the susceptible ARP and phage type 12. Phage typing could provide extra discrimination for some PFGE genotypes. Within genotype T10 two different phage types, designated 193 and 21, could be detected. On the other hand, within phage type 193, two different genotypes, T4 and T10, were distinguished.
In slaughterhouse A2, there was great variation among the different serotype Typhimurium strains (genotypes T1, T3, T4/T4, T4/T4a, T5, T6, T8/T8, T8/T12, T9, T11/T11, T11/T11a, and T12), whereas in slaughterhouse D, T2/T2, T2/T2a, T8/T8, and T10 were the most frequently isolated serotype Typhimurium genotypes. Only in genotype T8/T8 were isolates from both slaughterhouses clustered together.
(iii)Salmonella serotype Ohio.
PFGE of the serotype Ohio isolates after restriction with XbaI or BlnI provided no extra discrimination. The same genotype, O1, was recovered from all the different samples, and all isolates were susceptible to all antibiotics tested.
Epidemiological study.
In the two slaughterhouses, carcass contamination followed totally different courses relative to the Salmonella status of the incoming animals (Fig. 3). In slaughterhouse A2, an average of 26% of the carcasses were contaminated, with a slight increase toward the end of the day. The supply of pigs with Salmonella-positive feces ranged from 5 to 30%, with an average of 21% for that day. The percentage of pigs with Salmonella-positive feces and/or Salmonella-positive mesenteric lymph nodes ranged from 25 to 35%, with an average of 31.6%. In slaughterhouse D1, a nearly constant but high level of carcass contamination was noticed from the beginning to the end of the slaughter day: an average of 70% of carcass samples were found positive for salmonellae. The proportion of incoming animals with Salmonella-positive feces fluctuated between 0 and 20% during that day, with an average of 13%, and the proportion with Salmonella-positive feces and/or mesenteric lymph nodes fluctuated between 0 and 30%, with an average of 21.8%.
FIG. 3.
Correlation in time among Salmonella-positive carcasses (▵) and Salmonella positivity of feces (○) or of feces and/or mesenteric lymph nodes (•) of incoming animals in slaughterhouse A2 (left) and slaughterhouse D1 (right).
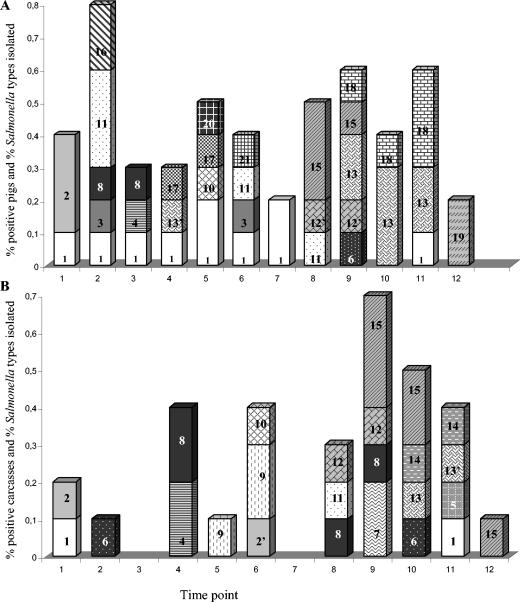
In slaughterhouse A2, the different serotypes and genotypes found on the carcasses corresponded partially with the serotypes and genotypes isolated from the feces and mesenteric lymph nodes of incoming pigs (Fig. 4). The diversity of Salmonella serotypes and genotypes was greater in the pig samples (the feces and mesenteric lymph nodes) than on the carcasses: serotype London, serotype Derby D1 and D1a, serotype Typhimurium T3, T1, and T6, serotype Goldcoast, and serotype Rissen were isolated only from the pig samples. On the other hand, serotype Typhimurium T9 and T5 and serotype Derby D4 were isolated from the carcasses only, not from pig or environmental samples taken on that day (Table 1; Fig. 4). Only 8 of the 32 positive carcasses sampled during that day were contaminated with the same Salmonella type as that found in the feces or mesenteric lymph nodes of the corresponding pig. Other carcasses were contaminated by Salmonella types isolated from previously slaughtered pigs. For example, serotype Ohio was introduced into the slaughter line at time points 8 and 9 (Fig. 4). The same serotype and genotype (O1) were detected on the carcasses at time points 9 and 10 but also at time point 12, after more than 700 pigs had passed the slaughter line. Also, serotype Infantis (susceptible to all antibiotics tested) was introduced at time points 2 and 3 in the slaughter line and was identified on the carcasses at time point 4 but also at later time points, 8 and 9, during slaughter activities. Two serotype Typhimurium genotypes (T4/T4 and T4/T4a) were delivered that day at different time points (time points 4, 9, 10, and 11) during slaughter. Both genotypes were detected on the carcasses at time points 10 and 11. The serotypes and genotypes found in the environment (Table 1) (serotypes Livingstone, Ohio, and Brandenburg, serotype Typhimurium T11/T11, and serotype Derby D3) were all isolated from the carcasses and the pig samples as well.
FIG. 4.
Distribution of Salmonella types at each time point when 250 pigs passed the slaughter line in slaughterhouse A2. (A) Positive samples from feces and/or mesenteric lymph nodes of pigs; (B) positive carcasses. Numbers in bars represent serotypes and genotypes as follows: 1, serotype Brandenburg; 2, serotype Typhimurium T11/T11; 2′, serotype Typhimurium T11/T11a; 3, serotype Derby D1; 4, serotype Derby D2a; 5, serotype Derby D2b; 6, serotype Derby D3; 7, serotype Derby D4; 8, serotype Infantis; 9, serotype Typhimurium T9; 10, serotype Virchow; 11, serotype Livingstone; 12, serotype Typhimurium T8/T8; 12′, serotype Typhimurium T8/T12; 13, serotype Typhimurium T4/T4; 13′, serotype Typhimurium T4/T4a; 14, serotype Typhimurium T5; 15, serotype Ohio O1; 16, serotype London; 17, serotype Typhimurium T3; 18, serotype Typhimurium T6; 19, serotype Rissen; 20, serotype Typhimurium T1; 21, serotype Goldcoast.
In slaughterhouse D1, with a carcass contamination level of about 70%, aerosols were generated during evisceration and the carcasses were frequently rinsed with water at different points along the slaughter line. Especially after the polishing step, the removal of the rectum and the viscera, the carcasses were rinsed with water, in contrast with the practice in slaughterhouse A, where the carcasses were not washed with water, except when feces contaminated the carcasses. Contamination cycles among pigs, the environment, and carcasses were defined. A total of seven Salmonella types were isolated from the carcasses (Table 1; Fig. 5). Serotype Anatum (susceptible) and four serotype Typhimurium types (T2/T2 [susceptible], T10 [ChSuT], T8/T8 [AChSuST] and T8/T8 [AChSuT]) were isolated from both pig and environmental samples (Table 1). Serotype Anatum was the most prevalent serotype in the pig samples (29.1% of infected pigs) but was found in only 1.3% of contaminated carcasses (Fig. 5). Serotype Typhimurium genotypes T2/T2, T8/T8, and T10, isolated from 33% of infected pigs, were responsible for 96% of the carcass contamination (Fig. 5). Genotype T2/T2 was introduced into the slaughter line at time point 6, and from that moment the same genotype was detected on the carcasses till the end of the day (Fig. 5). Serotype Typhimurium T8 (AChSuT) was found on the carcasses during the whole day and was never supplied by positive pigs during that day (Fig. 5). The serotype Typhimurium genotype T10 was introduced in the beginning, and from that moment the same genotype was identified in carcass samples taken at different time points (time points 1, 2, 3, 5, 6, and 9). Probably these strains persisted in the environment and continuously contaminated the carcasses during slaughter activity.
FIG. 5.
Distribution of Salmonella types at each time point when 350 pigs passed the slaughter line in slaughterhouse D1. (A) Positive samples from feces and/or mesenteric lymph nodes of pigs; (B) positive carcasses. Numbers in bars represent serotypes and genotypes as follows: 1, serotype Typhimurium T8/T8 (AChSuST); 2, serotype Typhimurium T10 (ChSuT); 3, serotype Typhimurium T8/T8 (AChSuT); 4, serotype Typhimurium TND (ChSuST); 5, serotype Typhimurium T2/T2 (susceptible); 5′, serotype Typhimurium T2a/T2a (ChSu); 6, serotype Infantis; 7, serotype Anatum; 8, serotype Havana; 9, serotype Derby D2a (SuT); 10, serotype Agona; 11, serotype Derby D1b (AChSuTNal); 12, serotype 47:Z4213:-; 13, serotype Panama; 14, serotype Derby D1 (A); 15, serotype Livingstone.
On the second sampling day, 1 month later, serotype Derby was the most common serotype found on the carcasses killed during the day and on the knives and SKGII machine (measuring meat quality). One carcass hanging in the refrigerator and killed the week before carried serotype Typhimurium with the same ARP and genotype (T8/T8) as those of the strains isolated a month before.
Figure 6 presents the numbers of isolates of different genotypes or serotypes present in the incoming pigs, on the carcasses, and in the environmental samples for the two slaughterhouses. In slaughterhouse A2, 17 different genotypes or serotypes were delivered during the day via positive pigs. Ten of these genotypes or serotypes were recovered from the carcasses, and five were recovered from the slaughterhouse environment. The ratio between strains originating from Salmonella-positive pigs and strains from carcass contamination for slaughterhouse A2 was greater than 1. In slaughterhouse D1, serotype Anatum and three different serotype Typhimurium genotypes (T2/T2, T8/T8 [AChSuST and AChSuT], and T10) were all isolated from positive pigs, carcasses, and the environment. Although these serotypes and genotypes were delivered during that day, the ratio between infected pigs and carcasses was less than 0.2. Genotype T8/T8 was the only genotype within serotype Typhimurium that was recovered in both slaughterhouses. Although the supply of serotype Typhimurium genotype T8/T8 by positive pigs was the same for both slaughterhouses, 60% of the carcasses were contaminated with this genotype in slaughterhouse D1, whereas in slaughterhouse A2 the percentage was 10 times lower. Comparison of the two slaughterhouses shows a correlation between the high percentage of contaminated carcasses in slaughterhouse D1 and a constant and high prevalence of certain Salmonella strains on the carcasses.
FIG. 6.
Number of isolates for every genotype or serotype recovered from pig samples (feces and mesenteric lymph nodes) (open bars), carcasses (shaded bars), and the environment (solid bars) for slaughterhouses A2 (A) and D1 (B).
The results in this study show no perfect match between Salmonella strains present in the mesenteric lymph nodes and the feces. At slaughterhouse A2, 21% of the pigs had Salmonella-positive feces whereas a total of 31.6% had positive feces or mesenteric lymph nodes. For 12 pigs, Salmonella was isolated from the mesenteric lymph nodes but not from the corresponding feces. For three pigs, the serotype or genotype isolated from the mesenteric lymph nodes was different from the serotype or genotype isolated from the feces. Serotype Virchow, serotype Goldcoast, serotype Poona, and serotype Derby D2a/D2a were additional serotypes or genotypes not isolated from the feces during that sampling day. In slaughterhouse D1, 21.8% of the pigs had either positive feces or positive mesenteric lymph nodes; 10 of the 14 positive pigs showed Salmonella-positive mesenteric lymph nodes while their feces were negative.
DISCUSSION
Bacterial typing methods, such as PFGE, antibiotic resistance profiling, and phage typing, were used to elucidate the origin and the contamination cycles of salmonellae in pig slaughterhouses. Within serotype Typhimurium, 10 different PFGE restriction profiles could be distinguished after restriction with XbaI and 11 could be distinguished after restriction with BlnI. Within serotype Derby, 10 different PFGE genotypes were identified with either XbaI or SpeI; these clustered into six main groups. The diversity in these two serotypes was also reflected by antibiotic resistance profiling, with nine different ARPs found for serotype Typhimurium and eight for serotype Derby. Antibiotic resistance profiling provided no further discrimination for most of the genotypes; genotypes T8/T8 and T10 were exceptions. Phage typing revealed an extra degree of difference for some genotypes: genotypes T10 and T4 within serotype Typhimurium. This high diversity in serotype Typhimurium was also found in isolates from human sporadic and outbreak cases in Norway after PFGE (11).
In the literature the carrier pig itself is mentioned as the predominant source of carcass contamination. Live animals that carry salmonellae are 3 to 4 times more likely to end up as positive carcasses (4, 5, 9). This study shows that more-complex contamination cycles between infected pigs and the slaughterhouse environment are determinative for the origin and prevalence of contamination on carcasses. In slaughterhouse A2 an average of 26% of the carcasses were contaminated, whereas in the other slaughterhouse an average of 70% of the carcasses were positive. For 25% of the positive carcasses in slaughterhouse A2, the same Salmonella type was isolated on the carcass as in the feces or the mesenteric lymph node samples from the same pigs. Probably the evisceration technique is the reason for this contamination. Forty-seven percent of the contaminated carcasses were contaminated by strains of other, previously slaughtered pigs. This corresponds with the results obtained by Wonderling et al. (25), who found that 54% of the carcasses were contaminated with Salmonella types not found in the feces of the same animal. The remaining contaminated carcasses (28%) were contaminated by strains originating from the slaughterhouse environment. The transmission of Salmonella contamination from the environment could occur via the slaughterhouse personnel or the equipment. At slaughterhouse A2 the carcass contamination and the environmental contamination reflected the supply of positive pigs during that day, and the ratio between the number of isolates delivered that day and the number recovered on the carcasses was greater than 1. In the other slaughterhouse, D1, where the prevalence of carcass contamination was very high, the origin of the carcass contamination was only partially related to the corresponding pig (4%). Here three different genotypes were mainly recovered from the carcasses (serotype Typhimurium T2, T8/T8 [corresponding to phage type 104L], and T10). Seventy percent of the positive carcasses were contaminated by strains isolated from previously slaughtered pigs, but the number of isolates recovered from the carcasses was at least 5 times higher than the number of isolates in feces or mesenteric lymph nodes of incoming pigs. Serotypes and genotypes not present in incoming pigs were isolated from 26% of the carcasses.
The main contamination source was probably a continuous contamination cycle between slaughtered pigs, the environment, and the carcasses. From the beginning to the end of slaughter activity in slaughterhouse D1, the same multiresistant genotype, T8/T8, corresponding to phage type 104L, could be detected on the carcasses and in the environment. We could not find the same strain on the equipment, indicating no direct contamination source in the slaughter line. In that slaughterhouse, however, an aerosol was generated by washing the carcasses with water when the carcasses were eviscerated; this aerosol could probably function as the most important vehicle for transmission of the strains to the carcasses. In the slaughtering process, the evisceration step is a critical point. Paying close attention to that step can reduce fecal contamination of the carcasses and makes rinsing with water unnecessary. Swanenburg et al. (22) and Giovannacci et al. (10) also found differences in contamination sources between slaughterhouses and slaughter days. Swanenburg et al. (22) identified the carcass splitter as an important contamination source and proposed that cross-contamination between carcasses may occur upon manipulation by slaughter personnel. Wonderling et al. (25) suggested that approximately 50% of the contaminated carcasses were likely contaminated through carcass-to-carcass or feces-to-carcass contact; this cross-contamination indicates that the presence of a pathogen in the feces of only a few pigs can contaminate different carcasses during processing.
Only in 30.6% of positive pigs was a correlation found between the contamination in the feces and that in the mesenteric lymph nodes of the same pig. In 4.8% of infected pigs, different Salmonella types were isolated in the two kinds of samples, and multiple Salmonella types were isolated from each of six samples. In the mesenteric lymph nodes, rarer serotypes such as serotype Agona, serotype Poona, serotype Goldcoast, and serotype Havana were isolated. This is probably due either to the higher sensitivity attained by the Salmonella isolation method in the mesenteric lymph nodes (because of lower background flora) than in the feces or to the fact that not every carrier pig is shedding salmonellae at the moment of sampling. The combination of both samples reflects the supply of positive animals more sensitively. To avoid having positive carcasses at the end of the slaughter line, it is important that the supply of carrier pigs be reduced. With the help of molecular or phenotypic typing methods, it is shown clearly that every positive animal supplied to the slaughterhouse represents a potential risk for contaminating its own carcass as well as carcasses at a later time point in the production chain, by direct cross-contamination or by contamination cycles through the environment.
Acknowledgments
This work was financially supported by the Flemish government (IWT).
We thank P. Vanmol and E. Engels for excellent technical assistance and A. Vanhee, A. Van de Bossche, L. Duboccage, J. Depuydt, and P. Delputte, whose help was crucial for sampling at the slaughterhouses. We also thank the personnel of slaughterhouses A2 and D1 for their participation in this study. We thank C. Godard and C. Wildemauwe for phage typing results and spontaneous help.
REFERENCES
- 1.Anderson, E. S., L. R. Ward, M. J. Saxe, and J. D. de Sa. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. (London) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baggesen, D. L., and F. M. Aarestrup. 1998. Characterisation of recently emerged multiple antibiotic-resistant Salmonella enterica serovar Typhimurium DT104 and other multiresistant phage types from Danish pig herds. Vet. Rec. 25:95-97. [DOI] [PubMed] [Google Scholar]
- 3.Baggesen, D. L., D. Sandvag, and F. M. Aarestrup. 2000. Characterization of Salmonella enterica serovar Typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J. Clin. Microbiol. 38:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berends, B. R., F. Van Knapen, J. M. Snijders, and D. A. Mossel. 1997. Identification and quantification of risk factors regarding Salmonella spp. on pork carcasses. Int. J. Food Microbiol. 36:199-206. [DOI] [PubMed] [Google Scholar]
- 5.Borch, E., T. Nesbakken, and H. Christensen. 1996. Hazard identification in swine slaughter with respect to foodborne bacteria. Int. J. Food Microbiol. 30:9-25. [DOI] [PubMed] [Google Scholar]
- 6.Botteldoorn, N., M. Heyndrickx, N. Rijpens, K. Grijspeerdt, and L. Herman. 2003. Salmonella on pig carcasses: positive pigs and cross contamination in the slaughterhouse. J. Appl. Microbiol. 95:891-903. [DOI] [PubMed] [Google Scholar]
- 7.Daly, M., J. Buckley, E. Power, C. O'Hare, M. Cormican, B. Cryan, P. G. Wall, and S. Fanning. 2000. Molecular characterization of Irish Salmonella enterica serotype Typhimurium: detection of class I integrons and assessment of genetic relationships by DNA amplification fingerprinting. Appl. Environ. Microbiol. 66:614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebner, P. D., and A. G. Mathew. 2001. Three molecular methods to identify Salmonella enterica serotype Typhimurium DT104: PCR fingerprinting, multiplex PCR and rapid PFGE. FEMS Microbiol. Lett. 205:25-29. [DOI] [PubMed] [Google Scholar]
- 9.Fedorka-Cray, P. J., S. C. Whipp, R. E. Isaacson, N. Nord, and K. Langer. 1994. Transmission of Salmonella typhimurium to swine. Vet. Microbiol. 41:333-344. [DOI] [PubMed] [Google Scholar]
- 10.Giovannacci, I., S. Queguiner, C. Ragimbeau, G. Salvat, J. L. Vendeuvre, V. Carlier, and G. Ermel. 2001. Tracing of Salmonella spp. in two pork slaughter and cutting plants using serotyping and macrorestriction genotyping. J. Appl. Microbiol. 90:131-147. [DOI] [PubMed] [Google Scholar]
- 11.Heir, E., B.-A. Lindstedt, I. Nygård, T. Vardund, V. Hasseltvedt, and G. Kapperud. 2002. Molecular epidemiology of Salmonella typhimurium isolates from human sporadic and outbreak cases. Epidemiol. Infect. 128:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korsak, N., G. Daube, Y. Ghafir, A. Chahed, S. Jolly, and H. Vindevogel. 1998. An efficient sampling technique used to detect four foodborne pathogens on pork and beef carcasses in nine Belgian abattoirs. J. Food Prot. 61:535-541. [DOI] [PubMed] [Google Scholar]
- 13.Liebana, E., L. Garcia-Migura, C. Clouting, C. A. Cassar, F. A. Clifton-Hadley, E. A. Lindsay, E. J. Threlfall, S. A. Chappell, and R. H. Davies. 2002. Investigation of the genetic diversity among isolates of Salmonella enterica serovar Dublin from animals and humans from England, Wales and Ireland. J. Appl. Microbiol. 93:732-744. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay, E. A., A. J. Lawson, R. A. Walker, L. R. Ward, H. R. Smith, F. W. Scott, S. J. O'Brien, I. S. T. Fisher, P. D. Cook, D. Wilson, D. J. Brown, H. Hardardottir, W. J. B. Wannet, H. Tschape, and E. J. Threlfall. 2002. Molecular characterisation of multiresistant strain of Salmonella enterica serotype Typhimurium DT204b responsible for an international outbreak of salmonellosis: importance of electronic exchange of gel data for outbreak investigations, p. 43-49. In P. Colin and G. Clément (ed.), Proceedings of the International Symposium on Salmonella and Salmonellosis. ISPAIA-ZOOPOLE développement, Ploufragan, France.
- 15.Millemann, Y., M.-C. Lesage, E. Chaslus-Dancla, and J.-P. Lafont. 1995. Value of plasmid profiling, ribotyping, and detection of IS200 for tracing avian isolates of Salmonella typhimurium and S. enteritidis. J. Clin. Microbiol. 33:173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsen, J. E., M. N. Skov, Ø. Angen, E. J. Threlfall, and M. Bisgaard. 1997. Genomic relationships between selected phage types of Salmonella enterica subsp. enterica serotype Typhimurium defined by ribotyping, IS200 typing and PFGE. Microbiology 143:1471-1479. [DOI] [PubMed] [Google Scholar]
- 17.On, S. L. W., and D. L. Baggesen. 1997. Determination of clonal relationships of Salmonella typhimurium by numerical analysis of macrorestriction profiles. J. Appl. Microbiol. 83:699-706. [DOI] [PubMed] [Google Scholar]
- 18.Pasmans, F., F. Van Immerseel, M. Heyndrickx, A. Martel, C. Godard, C. Wildemauwe, R. Ducatelle, and F. Haesebrouck. 2003. Host adaptation of pigeon isolates of Salmonella enterica subsp. enterica serovar Typhimurium variant Copenhagen phage type 99 is associated with enhanced macrophage cytotoxicity. Infect. Immun. 71:6068-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popoff, M. Y., J. Bockemuhl, F. W. Brenner, and L. L. Gheesling. 2000. Supplement 2000 (no. 44) to the Kauffmann-White scheme. Res. Microbiol. 152:907-909. [DOI] [PubMed] [Google Scholar]
- 20.Recchia, G. D., and R. M. Hall. 1997. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 5:389-394. [DOI] [PubMed] [Google Scholar]
- 21.Swanenburg, M. 2000. Salmonella in the pork production chain: sources of Salmonella on pork. Ph.D. thesis. Utrecht University, Utrecht, The Netherlands.
- 22.Swanenburg, M., H. A. P. Urlings, J. M. A. Snijders, D. A. Keuzenkamp, and F. van Knapen. 2001. Salmonella in slaughter pigs: prevalence, serotypes and critical control points during slaughter in two slaughterhouses. Int. J. Food Microbiol. 70:243-254. [DOI] [PubMed] [Google Scholar]
- 23.Threlfall, E. J., J. A. Skinnner, A. Graham, and L. R. Ward. 2002. Antimicrobial drug resistance in nontyphoidal salmonellas from humans in England and Wales, 2000: dramatic increase in multiple resistances in Salmonella enterica serotypes Typhimurium and Virchow, p. 445-449. In P. Colin and G. Clément (ed.), Proceedings of the International Symposium on Salmonella and Salmonellosis. ISPAIA-ZOOPOLE développement, Ploufragan, France.
- 24.Wall, P. G., D. Morgan, K. Lamden, M. Ryan, M. Griffin, E. J. Threlfall, L. R. Ward, and B. Rowe. 1994. A case-control study of infection with an epidemic strain of multiresistant Salmonella typhimurium DT104 in England and Wales. Commun. Dis. Rep. 4:R130-R135. [PubMed] [Google Scholar]
- 25.Wonderling, L., R. Pearce, M. Wallace, J. E. Call, I. Feder, M. Tamplin, and J. B. Luchansky. 2003. Use of pulsed-field gel electrophoresis to characterize the heterogeneity and clonality of Salmonella isolates obtained from the carcasses and feces of swine at slaughter. Appl. Environ. Microbiol. 69:4177-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]