Abstract
Porphyrin-phospholipid can chelate Mn ions in the hydrophobic portion of bilayers, giving rise to magnetic resonance (MR) contrast. However, limited water content within the bilayer diminishes longitudinal MR contrast. We used molecular dynamics (MD) simulations to show that an amine modified porphyrin-phospholipid (“N-HPPH-lipid”) produces bilayers with increased hydration. Following synthesis, N-HPPH-lipid (but not the original porphyrin-phospholipid) could be readily hydrated into bilayers that formed liposome-like structures. X-ray scattering and electron paramagnetic resonance measurements were consistent with a bilayer perturbed with water. Despite the presence of intrabilayer water, N-HPPH-lipid liposomes were resistant to ester hydrolysis and, if used in small amounts, could be formulated to successfully entrap cargo. When chelated with Mn, N-HPPH-lipid generated stronger water relaxation (compared to the original porphyrin-phospholipid) in intact, but not detergent-disrupted, liposomes. The suitability of Mn-N-HPPH liposomes for in vivo MR imaging is demonstrated in mice.
Keywords: Porphyrin-phospholipid, MD simulations, Water permeation, Bilayer, Contrast agents
Porphyrin-phospholipid (PoP) conjugates have been developed for a range of multimodal imaging and therapeutic applications.[1–6] 2-[1-hexyloxyethyl]-2-devinyl pyropheophorbide-a (HPPH) is a porphyrin derivative being assessed in clinical trials for photodynamic therapy.[7] Liposomes formed from HPPH-lipid PoP have been reported for chemophototherapy.[8,9] Manganese (Mn) is a paramagnetic contrast agent used for magnetic resonance imaging (MRI).[10–13] Mn has been chelated within PoP bilayers of liposome-like porphysome nanovesicles for MRI.[14,15] There is a caveat with this approach however: Mn chelated in PoP is located inside the hydrophobic bilayer, where only a small number of water molecules are accessible, thereby reducing longitudinal MR contrast.
Bilayer water permeability depends on numerous factors including the type of lipid head groups and fatty acyl chains, membrane thickness and surface density.[16–21] Neutron and X-ray scattering, magic-angle spinning nuclear magnetic resonance, and femtosecond infrared spectroscopy have been used to study bilayer structures and intra-bilayer distribution of water, which decreases rapidly as a function of distance from the aqueous interface toward the central hydrophobic core of the bilayer.[22–26] Hydrogen bonding is critical in determining the properties of water[27] and the penetration of water and other molecules into bilayers[28,29]. In bilayers, amine-rich lipids such as phophatidylethanolamines are capable of forming complex hydrogen bonding networks involving other lipids and water.[30]
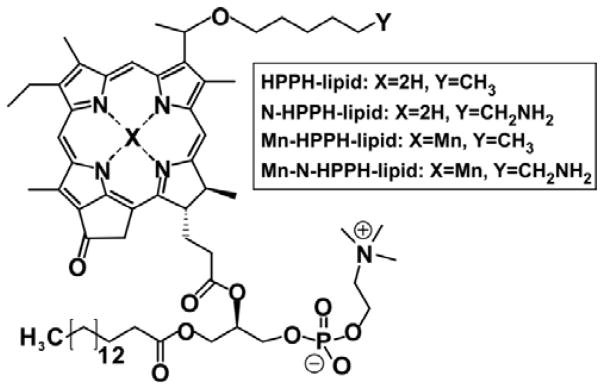
In this work, we examined an amine-modified version of HPPH-lipid, referred to as N-HPPH-lipid. Based on the propensity for amines to undergo hydrogen bonding with water, we hypothesized that intrabilayer water content could be enhanced, thereby improving relaxivity for MR contrast generation in Mn-PoP bilayers. Molecular dynamic (MD) simulations were used to initially test this hypothesis. Simulations were based on our previously developed PoP force fields.[8] The chemical structures of HPPH-lipid and amine-modified N-HPPH-lipid, as well as their Mn-chelated analogs are shown in Figure 1. Synthetic details and analytical information are provided in the Supporting Methods and Figures S1–S3 (Supporting Information) Both the N-HPPH-lipid and Mn-N-HPPH-lipid were capable of forming nanoparticles. The absorbance spectra of the liposomes in aqueous solution and the lipids themselves in organic solvent are shown in Figure S4 (Supporting Information).
Figure 1.
Structure of the porphyrin-phospholipids (PoPs) used in this study.
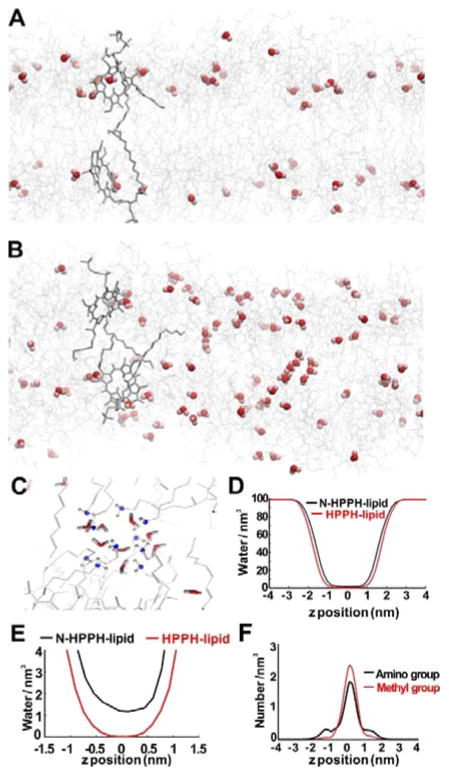
MD simulations were used to predict the bilayer behavior of N-HPPH-lipid bilayers. Relative to HPPH-lipid, the addition of the amino group in N-HPPH-lipid resulted in a substantial increase in water into the hydrophobic bilayer. As shown in Figures 2A and 2B, water molecules penetrated into the middle of the N-HPPH-lipid bilayer while no water molecules were observed in the middle of the HPPH-lipid bilayer. Inter-leaflet hydrogen bonds formed by amino groups accounted for more than 50% of the total inter-leaflet hydrogen bonds for the N-HPPH bilayer (Figure 2C). The numbers of water molecules per nm3 in both cases are shown in Figure 2D, where the z-axis represents the bilayer normal direction from the center of the bilayer. In between the bilayers, at every z-coordinate, water density in the N-HPPH-lipid system was higher than that in the HPPH-lipid one. Within 1 nm from the center of the bilayer, there were no water molecules in the HPPH-lipid bilayer, whereas there was more than one water per nm3 in N-HPPH-lipid bilayers (Figure 2E). Figure 2F shows the density of either amino groups in N-HPPH-lipid bilayers or methyl groups in the HPPH-lipid bilayers. The methyl groups of the HPPH-lipids were distributed mostly at the vertical center of the two leaflets while the amino groups of the N-HPPH-lipids were more disordered with some of them pointing out to the solvent to form hydrogen bonds with water molecules. The more disordered state of the amino groups also caused the area per lipid of the N-HPPH-lipid system to be larger than that of the HPPH one (0.95±0.01 versus 0.89±0.01 nm2). At the same time, the thickness of the simulated N-HPPH-lipid bilayer was slightly thinner than that of the HPPH-lipid one (4.45 nm vs 4.49 nm; Figure S5, Supporting Information). The thickness between the most highly electron distributed layers was 2.80 nm for N-HPPH-lipid and 2.99 nm for HPPH-lipid. The center of the bilayer had the highest free energy barrier against penetration of water molecules. The amino group lowers the barrier and allowed more water molecules to stay in this area. Hydrogen bonding between water molecules and the amino groups was the cause of water permeation.
Figure 2.
PoP MD simulations. A snapshot of a bilayer composed of A) HPPH-lipid and B) N-HPPH-lipid. Water molecules are shown as red spheres and lipids using grey wires. C) Close-up of the N-HPPH-liposome showing a cluster of water bonding with the amine group (blue-grey) in the lipid tail region. D), E) Water density profiles of N-HPPH-lipid and HPPH-lipid bilayers along the bilayer z position. F) Amino or methyl group bilayer density for PoP bilayers formed from N-HPPH-lipid or HPPH-lipid, respectively.
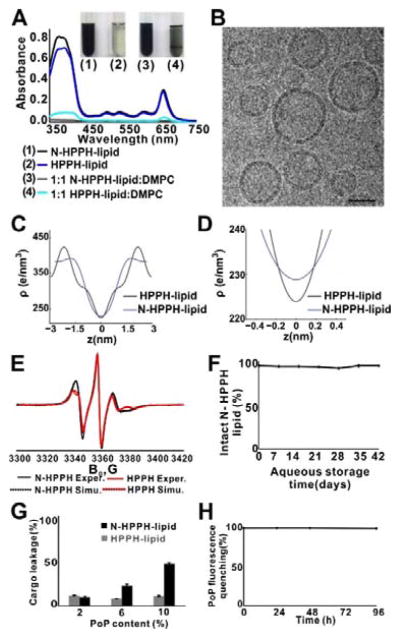
Having established a theoretical basis for bilayer hydration in silico, we examined these compounds experimentally. Compared to HPPH-lipid, dry films of N-HPPH-lipid were rapidly solubilized following water addition. For both a 100% PoP and a 1:1 molar ratio PoP:DMPC formulation, only N-HPPH-lipid could be fully dissolved after brief water addition and shaking. The N-HPPH-lipid sample became a dark solution free of aggregation after half a minute of vortex shaking, but HPPH-lipid barely dissolved. For the HPPH-lipid, a 1:1 PoP:DMPC formulation showed only slightly improved rapid dissolution. After brief hydration with water, intense Soret and Q band absorption peaks were observed with N-HPPH-lipid, but not HPPH-lipid (Figure 3A). N-HPPH-lipid was nearly completely solubilized, as compared to less than 10% of HPPH-lipid (Figure S6, Supporting Information). The average diameter of N-HPPH-lipid structures was about 80 nm based on light scattering after extrusion (Figure S7, Supporting Information). Vesicular, liposome-like structures were observed by cryo-electron microscopy (Figure 3B) and confirmed that bilayers are formed by N-HPPH-lipid.
Figure 3.
N-HPPH-lipid bilayers with enhanced hydration. A) Optical absorption of hydrated PoP lipid films shortly after water addition. B) Representative cryo-electron micrographs showing structures formed by N-HPPH-lipid at a 1:1 DMPC:PoP molar ratio. 40 nm scale bar is shown. C), D) Bilayer electron density determined by X-ray diffraction in bilayers formed with a 1:1 DMPC:PoP molar ratio. E) Electron spin resonance spectra showing differences in lipid ordering between N-HPPH-lipid and HPPH-lipid bilayers (containing DMPC and PoP and probed with 0.5 molar % 10 PC spin label). F) Hydrolysis of N-HPPH-lipid liposomes during aqueous storage. G) Sulforhodamine B leakage over 24 hr in saline, in liposomes containing indicated molar percentage of PoP, together with 35 mol. % Chol and the remaining composition of DMPC. H) Structural intactness of cargo-loaded, N-HPPH-liposomes (PoP:DMPC:Chol; 10:35:55 molar ratio) based on fluorescence self-quenching. Data show mean ± s.d. for n=3.
Out-of-plane X-ray diffraction PoP bilayer measurements were taken at a high hydration level at 97% relative humidity (RH). As demonstrated in Figure S8a (Supporting Information), in pure PoP samples, only broad, diffuse scattering was observed, indicating the lipids did not form bilayers under these conditions. However, the membrane profile of a 1:1 molar ratio HPPH-lipid:DMPC showed a series of evenly spaced Bragg-peaks indicating a well-ordered, lamellar structure with spacing of dz = 6.4 nm. The presence of DMPC, which is in its solid ordered (gel) phase under the experimental conditions, likely favored formation of stacked bilayers. However, N-HPPH-lipid:DMPC did not show a stable, ordered lamellar sample during hydration (Supporting Figure S8b). While Bragg-peaks were initially present, the sample quickly swelled (increasing dz) until the Bragg-peak disappeared and only diffuse features remained. PoP:DMPC samples were therefore scanned at a reduced hydration at 75% RH to stabilize a lamellar structure for the structural analysis (Figure S8c, Supporting Information). A number of Bragg peaks were observed with lamellar spacings of dz = 5.98 nm and 5.36 nm for HPPH-lipid and N-HPPH-lipid, respectively.
The observed X-ray diffraction bilayer thickness of N-HPPH-lipid is consistent with cryo-TEM measurements of the liposomal bilayer of the same composition (5.36 and 5.34 nm, respectively). Consistent with the X-ray diffraction data, previous cryo-TEM measurements of bilayers containing conventional PoPs (e.g. HPPH-lipid) gave rise to thicker bilayers.[31,32]
Braggs peaks were used to produce electron-density-profiles for an analysis of the real-space membrane structure. The electron density profiles are shown in Figure 3C. The peak in the profile at z~2.0 nm indicates the position of the electron richer head groups; z = 0 nm represents the center of the bilayer. From the peak, the bilayer head-to-head distance,dHH, was determined. The width of the water layer was calculated using the relation dW = dz − dHH. dW for 1:1 HPPH-lipid:DMPC bilayers was 1.6 nm; and dW was 1.92 nm for 1:1 N-HPPH-lipid:DMPC bilayers. The N-HPPH-lipid bilayer attracted an additional 10 waters/lipid compared to HPPH-lipid. In addition, there was a difference in electron density at the bilayer center caused by additional water molecules. At z = 0 nm, there is an additional 5 e−/nm3 in electron density, corresponding to about 0.5 water molecules per lipid molecule at 75% RH (Figure 3D).
Electron spin resonance (ESR) can analyze lipid bilayer structures.[33–36] In accordance with the MD results, ESR showed more lipid layer disturbance in the N-HPPH-liposome hydrophobic layer suggesting that more water molecules can enter it. Experiments with spin-labeled phospholipids, 10 and 16-PC spin labels (1-acyl-2-[n-(4,4-dimethyloxazolidine-N-oxyl)stearoyl]-sn-glycero-3-phosphocholine with n=10 and 16) showed that at DMPC/PoP ratio equal or above 3:1, the mixed bilayer behaved similarly to a pure DMPC bilayer. Spectra were generally similar to pure DMPC although differ in mobility and ordering parameters. The main chain transition could be detected by quick spectral changes within a narrow temperature range and was slightly shifted down in temperature (e.g. ~< 2°C at 7:1 ratio) and broadened. Samples with a 1:1 ratio, though, showed spectra indicative of much slower rate of molecular motion for the nitroxide reporter group and substantial exchange broadening indicative of poor mixing of spin-labeled phosphatidylcholines with the rest of lipid phase causing formation of their aggregates. N-HPPH-lipid and HPPH-lipid showed noticeable difference in the ESR lineshapes for the whole range of DMPC/porphyrin-lipid ratios studied. To simulate the ESR spectra and to extract ordering information from them we used NLSL program.[37] The order parameter S0 corresponds to the rotation of the molecular long axis in the liquid crystal restricted within an orienting potential that can be simply approximated as: U(θ) = λ·cos2θ, where λ is the strength of the potential. The ordering of the lipid chain relative to the bilayer normal can then be expressed as canonically weighted average value of the 2nd order Legendre polynomial[38] as:
The best fits of ESR spectra obtained using a simple ordering potential with only one coefficient for 10PC in DMPC with 25 mol. % of either N-HPPH-lipid or HPPH-lipid yield the order parameter S0 = 0.22 and 0.3 respectively. However, to better simulate the shape of the hyperfine component with IN=−1 one may need to introduce an additional coefficient assuming that the most favored direction of the diffusion axis forms a cone relative to the membrane normal (Figure 3E).[39] Also in this case the best fit for N-HPPH-lipid corresponds to lower chain order as well as to a larger value of the cone angle (0.19 vs 0.29 and 44° vs 39°) compared to HPPH-lipid, indicating substantial bilayer disturbance by the amino compound.
Despite the presence of intrabilayer water in N-HPPH-lipid liposomes, as determined by multiple lines of experimental evidence, no hydrolysis of the ester linkage between porphyrin and lipid was detected for over a month of storage in aqueous solution (Figure 3F and Figure S9, Supporting Information).
Liposome composition is a key factor to determine its stability, clearance time and cargo release rate.[40–42] Due to a more hydrophilic bilayer, N-HPPH-lipid resulted in easier cargo leakage from liposomes. Water soluble sulforhodamine B was used as a model cargo. For HPPH-lipid liposomes, only minor leakage was observed after 24 hr incubation with 2%, 6% and 10% PoP in a conventional liposome formulation consisting of PEG-lipid, DMPC and cholesterol (Chol). However, as the N-HPPH-lipid composition increased, the liposomes became markedly leaky (Figure 3G). Despite leakage, the liposome structure remained stable in a self-assembled state, based on the fluorescence self-quenching of the PoP (Figure 3H). Although N-HPPH-lipid resulted in more cargo leakage, when containing just 2% N-HPPH-lipid, liposomes stably entrapped cargo. In 50% serum, cargo leakage was just 12 % over 24 hours (Figure S10, Supporting Information). Thus, N-HPPH-liposome has the potential to form stable or leaky liposomes depending on the PoP content in the liposome. Self-quenching of N-HPPH-lipid and Mn-HPPH-lipid was assessed in different buffers as shown in Figure S11 (Supporting Information). In serum-free buffers (PBS and cell media), no unquenching was observed, indicating the bilayer remains intact. In fetal bovine serum, a limited degree (<20%) of unquenching occurred, indicating that the PoPs might partially exchange with serum components with this formulation. Because detergent-solubilized Mn-PoPs are substantially less fluorescent compared to free base PoPs, the propensity for the Mn to leave the macrocycle was assessed by fluorescence during incubation in various buffers, followed by detergent disruption to avoid self-quenching fluorescence effects. As shown in Figure S12 (Supporting Information), no detectable de-chelation of the Mn was observed during up to 24 hours incubation in low pH or serum, demonstrating the Mn is stable in the macrocycle in those conditions.
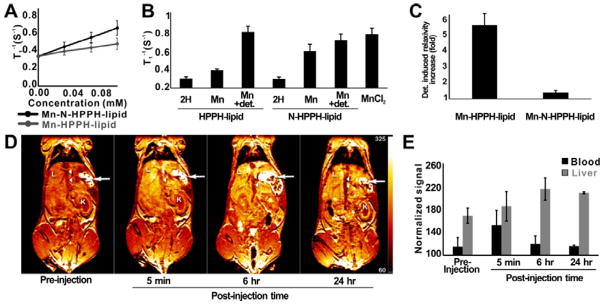
PoP nanostructures have been explored for MRI contrast when Mn2+ is inserted into the porphyrin. However, bilayer hydrophobicity restricts the interaction of water molecules with the paramagnetic metal.[14] Improved access of water molecules to the Mn ion is predicted to increase the effectiveness of Mn-PoP as an MRI contrast agents. With Mn chelation of the PoP, the amine modification induced a 150% higher T1 relaxivity (mM • s)−1, going from 0.98 for Mn-HPPH-lipid liposomes to 2.46 for Mn-N-HPPH-lipid liposomes (Figure 4A and Figure S13, Supporting Information). However, with the addition of TX-100 detergent, both samples demonstrated nearly equal T1 relaxation rates (Figure 4B) indicating it is the bilayer structure and water therein that causes the T1 relaxivity differences between the two types of Mn-PoPs. Mn-HPPH-lipid showed over a 5 fold change in relaxivity with detergent disruption, whereas N-HPPH-lipid changed less than 1.5 fold (Figure 4C).
Figure 4.
MR contrast with Mn-N-HPPH liposomes. A) T1 relaxivity for indicated PoP liposomes. B) T1 rates for PoP liposomes at 0.1 mM PoP. TX-100 detergent (“+ det.”) lysed the liposomes. C) Detergent-induced relaxivity enhancement of PoP liposomes. D) Representative T1-weighted MRI of a BALB/c mouse IV injected with Mn-N-HPPH liposomes. Immediate and sustained signal enhancement was seen in the liver (L) for up to 24 hr, while enhancement in the inferior vena cava (arrow heads) peaked immediately after injection but returned to baseline. Stomach (arrows) and kidneys (K) are labelled for anatomical reference. Scale is arbitrary units following signal normalization by phantoms. E) MR signal in blood and liver of mouse. In vitro experiments used 1:1 PoP:DMPC and in vivo experiments used [5:45:50] [DSPE-PEG2K:N-HPPH-lipid:DMPC]. Data show mean ± SD, n=3.
The suitability of Mn-N-HPPH-liposomes as an MRI contrast agent was investigated in mice following intravenous injection. An increase of MR signal in blood and liver was visible post-injection (Figure 4D). The signal in blood increased immediately post injection and subsequently decreased to near baseline levels by 6 hr. Accumulation and retention in the liver lasted up to 24 hr, presumably due to uptake of the liposomes by the reticuloendothelial system (Figure 4E).
No acute toxicity was observed during MR imaging of the amine-modified PoP liposomes. Additional characterization of N-HPPH-lipid in vitro and in vivo was assessed. In vitro, in Caco-2 cells, no cytotoxicity trends were observed with 48 hour incubation of Caco-2 cells at increasing PoP or Mn-PoPs at concentrations us to 0.2 mM (Figure S14, Supporting Information). Using the same formulation used for MR imaging, PEGylated N-HPPH-lipid liposomes exhibited a 8.2 hour half-life, based on a non-compartment model (Figure S15, Supporting Information). To further probe the toxicity of N-HPPH-liposome, the body weight of mice was monitored following intravenous administration of N-HPPH-liposomes with an equivalent formulation and dose as the MR imaging study. Over the course of the 10 day monitoring period, no behavioral changes or body weight loss was observed in the N-HPPH-lipid liposome treatment group, compared to a PBS control group (Figure S16, Supporting Information). Although more in depth toxicity studies are required, it appears that N-HPPH-lipid does not induce acute toxicity at functional doses.
In summary, MD simulations were used to evaluate an amino-modified porphyrin-phospholipid; N-HPPH-lipid. Simulations predicted enhanced water distribution within the bilayer. This was supported by multiple lines of experimental evidence. N-HPPH-lipid liposomes gave rise to superior MR contrast when chelated with Mn and could be used safely for MRI in mice.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01EB017270, DP5OD017898) and the National Science Foundation (1555220). ACERT is supported by the NIH/NIGMS (P41GM103521). The Roswell Park imaging shared resource is supported by the NIH/NCI (P30CA016056).
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Dedicated to Professor Gang Zheng on the occasion of his 50th birthday.
Contributor Information
Shuai Shao, Department of Biomedical Engineering, University at Buffalo, State University of New York, Buffalo, New York 14260, USA.
Trang Nhu Do, Department of Chemistry and Waterloo Institute for Nanotechnology, University of Waterloo, 200 University Avenue West, Waterloo, Ontario N2L3G1, Canada.
Aida Razi, Department of Biochemistry and Biomedical Sciences and M. G. DeGroote Institute for Infectious Diseases Research, McMaster University, Hamilton, Ontario L8S4L8, Canada.
Upendra Chitgupi, Department of Biomedical Engineering, University at Buffalo, State University of New York, Buffalo, New York 14260, USA.
Jumin Geng, Department of Biomedical Engineering, University at Buffalo, State University of New York, Buffalo, New York 14260, USA.
Richard J. Alsop, Department of Physics and Astronomy, McMaster University, Hamilton, Ontario L8S4M1, Canada
Dr. Boris G. Dzikovski, Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA
Prof. Maikel C Rheinstädter, Department of Physics and Astronomy, McMaster University, Hamilton, Ontario L8S4M1, Canada
Prof. Joaquin Ortega, Department of Biochemistry and Biomedical Sciences and M. G. DeGroote Institute for Infectious Diseases Research, McMaster University, Hamilton, Ontario L8S4L8, Canada
Prof. Mikko Karttunen, Department of Chemistry and Waterloo Institute for Nanotechnology, University of Waterloo, 200 University Avenue West, Waterloo, Ontario N2L3G1, Canada. Department of Mathematics and Computer Science & Institute for Complex Molecular Systems, Eindhoven University of Technology, Eindhoven, Netherlands
Dr. Joseph A. Spernyak, Department of Cell Stress Biology, Roswell Park Cancer Institute Buffalo, NY 14263, USA
Prof. Jonathan F. Lovell, Department of Biomedical Engineering, University at Buffalo, State University of New York, Buffalo, New York 14260, USA.
References
- 1.Lovell JF, Jin CS, Huynh E, Jin H, Kim C, Rubinstein JL, Chan WCW, Cao W, Wang LV, Zheng G. Nat Mater. 2011;10:324. doi: 10.1038/nmat2986. [DOI] [PubMed] [Google Scholar]
- 2.Lovell JF, Jin CS, Huynh E, MacDonald TD, Cao W, Zheng G. Angew Chem Int Ed. 2012;51:2429. doi: 10.1002/anie.201108280. [DOI] [PubMed] [Google Scholar]
- 3.Rieffel J, Chen F, Kim J, Chen G, Shao W, Shao S, Chitgupi U, Hernandez R, Graves SA, Nickles RJ, Prasad PN, Kim C, Cai W, Lovell JF. Adv Mater. 2015;27:1785. doi: 10.1002/adma.201404739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao S, Geng J, Ah Yi H, Gogia S, Neelamegham S, Jacobs A, Lovell JF. Nat Chem. 2015;7:438. doi: 10.1038/nchem.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo D, Li N, Carter KA, Lin C, Geng J, Shao S, Huang WC, Qin Y, Atilla-Gokcumen GE, Lovell JF. Small. 2016;12:3039. doi: 10.1002/smll.201503966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huynh E, Zheng G. Nano Today. 2014;9:212. [Google Scholar]
- 7.Ethirajan M, Chen Y, Joshi P, Pandey RK. Chem Soc Rev. 2011;40:340. doi: 10.1039/b915149b. [DOI] [PubMed] [Google Scholar]
- 8.Carter KA, Shao S, Hoopes MI, Luo D, Ahsan B, Grigoryants VM, Song W, Huang H, Zhang G, Pandey RK, Geng J, Pfeifer BA, Scholes CP, Ortega J, Karttunen M, Lovell JF. Nat Commun. 2014;5:3546. doi: 10.1038/ncomms4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo D, Carter KA, Miranda D, Lovell JF. Adv Sci. 2016 doi: 10.1002/advs.201600106. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takehara Y, Sakahara H, Masunaga H, Isogai S, Kodaira N, Sugiyama M, Takeda H, Saga T, Nakajima S, Sakata I. Magn Reson Med. 2002;47:549. doi: 10.1002/mrm.10109. [DOI] [PubMed] [Google Scholar]
- 11.Pan D, Caruthers SD, Hu G, Senpan A, Scott MJ, Gaffney PJ, Wickline SA, Lanza GM. J Am Chem Soc. 2008;130:9186. doi: 10.1021/ja801482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan D, Caruthers SD, Senpan A, Schmieder AH, Wickline SA, Lanza GM. WIREs Nanomed Nanobiotechnol. 2011;3:162. doi: 10.1002/wnan.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H-LM, Haedicke IE, Cheng W, Tchouala Nofiele J, Zhang X. J Magn Reson Imaging. 2014;40:1474. doi: 10.1002/jmri.24483. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald TD, Liu TW, Zheng G. Angew Chem. 2014;126:7076. doi: 10.1002/anie.201400133. [DOI] [PubMed] [Google Scholar]
- 15.Keca JM, Chen J, Overchuk M, Muhanna N, MacLaughlin CM, Jin CS, Foltz WD, Irish JC, Zheng G. Angew Chem. 2016;128:6295. doi: 10.1002/anie.201600234. [DOI] [PubMed] [Google Scholar]
- 16.Paula S, Volkov AG, Van Hoek AN, Haines TH, Deamer DW. Biophys J. 1996;70:339. doi: 10.1016/S0006-3495(96)79575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathai JC, Tristram-Nagle S, Nagle JF, Zeidel ML. J Gen Physiol. 2008;131:69. doi: 10.1085/jgp.200709848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang TX, Anderson BD. J Membr Biol. 1995;148:157. doi: 10.1007/BF00207271. [DOI] [PubMed] [Google Scholar]
- 19.Lande MB, Donovan JM, Zeidel ML. J Gen Physiol. 1995;106:67. doi: 10.1085/jgp.106.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansen M, Blume A. Biophys J. 1995;68:997. doi: 10.1016/S0006-3495(95)80275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurtovenko AA, Anwar J, Vattulainen I. Chem Rev. 2010;110:6077. doi: 10.1021/cr1000783. [DOI] [PubMed] [Google Scholar]
- 22.Kučerka N, Nagle JF, Sachs JN, Feller SE, Pencer J, Jackson A, Katsaras J. Biophys J. 2008;95:2356. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tristram-Nagle S, Kim DJ, Akhunzada N, Kucerka N, Mathai JC, Katsaras J, Zeidel M, Nagle JF. Chem Phys Lipids. 2010;163:630. doi: 10.1016/j.chemphyslip.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitter J, Lechner RE, Dencher NA. J Phys Chem B. 1999;103:8036. [Google Scholar]
- 25.Zhou Z, Sayer BG, Hughes DW, Stark RE, Epand RM. Biophys J. 1999;76:387. doi: 10.1016/S0006-3495(99)77205-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volkov VV, Palmer DJ, Righini R. J Phys Chem B. 2007;111:1377. doi: 10.1021/jp065886t. [DOI] [PubMed] [Google Scholar]
- 27.Titantah JT, Karttunen M. Soft Matter. 2015;11:7977. doi: 10.1039/c5sm00930h. [DOI] [PubMed] [Google Scholar]
- 28.Boonnoy P, Jarerattanachat V, Karttunen M, Wong-ekkabut J. J Phys Chem Lett. 2015;6:4884. doi: 10.1021/acs.jpclett.5b02405. [DOI] [PubMed] [Google Scholar]
- 29.Yang K, Ma YQ. Nat Nanotechnol. 2010;5:579. doi: 10.1038/nnano.2010.141. [DOI] [PubMed] [Google Scholar]
- 30.Suits F, Pitman MC, Feller SE. J Chem Phys. 2005;122:244714. doi: 10.1063/1.1899152. [DOI] [PubMed] [Google Scholar]
- 31.Luo D, Carter KA, Razi A, Geng J, Shao S, Lin C, Ortega J, Lovell JF. J Controlled Release. 2015;220(Part A):484. doi: 10.1016/j.jconrel.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo D, Carter KA, Razi A, Geng J, Shao S, Giraldo D, Sunar U, Ortega J, Lovell JF. Biomaterials. 2016;75:193. doi: 10.1016/j.biomaterials.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa EJX, Shida CS, Biaggi MH, Ito AS, Lamy-Freund MT. FEBS Lett. 1997;416:103. doi: 10.1016/s0014-5793(97)01178-2. [DOI] [PubMed] [Google Scholar]
- 34.Man D, Olchawa R, Kubica K. J Liposome Res. 2010;20:211. doi: 10.3109/08982100903286485. [DOI] [PubMed] [Google Scholar]
- 35.Heberle FA, Wu J, Goh SL, Petruzielo RS, Feigenson GW. Biophys J. 2010;99:3309. doi: 10.1016/j.bpj.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stimson L, Dong L, Karttunen M, Wisniewska A, Dutka M, Róg T. J Phys Chem B. 2007;111:12447. doi: 10.1021/jp0746796. [DOI] [PubMed] [Google Scholar]
- 37.Budil DE, Lee S, Saxena S, Freed JH. J Magn Reson A. 1996;120:155. [Google Scholar]
- 38.Schneider DJ, Freed JH. Spin Labeling : Theory and Application, Biological Magnetic Resonance. Plenum; New York: 1989. [Google Scholar]
- 39.Dzikovski BG, Borbat PP, Freed JH. Biophys J. 2004;87:3504. doi: 10.1529/biophysj.104.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veiga MP, Arrondo JLR, Goñi FM, Alonso A, Marsh D. Biochemistry (Mosc) 2001;40:2614. doi: 10.1021/bi0019803. [DOI] [PubMed] [Google Scholar]
- 41.Mombelli E, Morris R, Taylor W, Fraternali F. Biophys J. 2003;84:1507. doi: 10.1016/S0006-3495(03)74963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tardi PG, Gallagher RC, Johnstone S, Harasym N, Webb M, Bally MB, Mayer LD. Biochim Biophys Acta BBA - Biomembr. 2007;1768:678. doi: 10.1016/j.bbamem.2006.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.